Abstract
Background
The muscle fiber number and fiber composition of muscle is largely determined during prenatal development. In order to discover genes that are involved in determining adult muscle phenotypes, we studied the gene expression profile of developing fetal bovine longissimus muscle from animals with two different genetic backgrounds using a bovine cDNA microarray. Fetal longissimus muscle was sampled at 4 stages of myogenesis and muscle maturation: primary myogenesis (d 60), secondary myogenesis (d 135), as well as beginning (d 195) and final stages (birth) of functional differentiation of muscle fibers. All fetuses and newborns (total n = 24) were from Hereford dams and crossed with either Wagyu (high intramuscular fat) or Piedmontese (GDF8 mutant) sires, genotypes that vary markedly in muscle and compositional characteristics later in postnatal life.
Results
We obtained expression profiles of three individuals for each time point and genotype to allow comparisons across time and between sire breeds. Quantitative reverse transcription-PCR analysis of RNA from developing longissimus muscle was able to validate the differential expression patterns observed for a selection of differentially expressed genes, with one exception. We detected large-scale changes in temporal gene expression between the four developmental stages in genes coding for extracellular matrix and for muscle fiber structural and metabolic proteins. FSTL1 and IGFBP5 were two genes implicated in growth and differentiation that showed developmentally regulated expression levels in fetal muscle. An abundantly expressed gene with no functional annotation was found to be developmentally regulated in the same manner as muscle structural proteins. We also observed differences in gene expression profiles between the two different sire breeds. Wagyu-sired calves showed higher expression of fatty acid binding protein 5 (FABP5) RNA at birth. The developing longissimus muscle of fetuses carrying the Piedmontese mutation shows an emphasis on glycolytic muscle biochemistry and a large-scale up-regulation of the translational machinery at birth. We also document evidence for timing differences in differentiation events between the two breeds.
Conclusion
Taken together, these findings provide a detailed description of molecular events accompanying skeletal muscle differentiation in the bovine, as well as gene expression differences that may underpin the phenotype differences between the two breeds. In addition, this study has highlighted a non-coding RNA, which is abundantly expressed and developmentally regulated in bovine fetal muscle.
Background
Genetic background and development prior to birth are known to influence the composition of bovine muscle tissue [1,2]. Prenatal muscle development is therefore a promising source of gene discovery for the molecular events that determine adult muscle phenotype.
Prenatal muscle development in mammals begins with the determination of myogenic cells (myoblasts) in the dermamyotome and their migration to their sites of differentiation [3]. Myoblasts then fuse to form multinucleated muscle cells, myotubes. In cattle, primary myotubes appear prior to 47 days of gestation [4]. Primary skeletal muscle fibers subsequently differentiate from this generation of myotubes [5]. Secondary muscle fibers are derived from a separate population of myotubes arising around day 90 [6].
The number of muscle fibers is roughly fixed at two thirds of bovine fetal development and muscle fibers differentiate before birth, making cattle a relatively mature species at birth compared to other mammals. During the final third of gestation in cattle, fetal isoforms of the muscle contractile proteins disappear and are replaced by adult isoforms. Also over the final third of gestation, metabolic differentiation takes place, with an increase in glycolytic and oxidative pathway enzymes [3]. Histologically, a steady decrease in the extracellular matrix contribution to muscle mass can be observed over fetal development in the bovine [7].
Large mammals such as pigs and cattle offer an advantageous model for the study of myogenesis in vivo, as individual muscles can be identified and reliably dissected from early stages. In addition, livestock breeds represent useful genetic models for muscle hypertrophy and other muscle traits, as these have been subject to intensive genetic selection due to their impact on meat quality. For example, array studies of pig fetal development have characterized the coordinate genome-wide expression changes during a developmental time course [8] and also exploited the contrasting muscle phenotypes of the Pietrain and Duroc pig breeds to compare prenatal events that may underlie the adult muscle phenotype [9].
Similarly, Sudre et al. [10] studied expression profiles of bovine fetal muscles using a human skeletal muscle macroarray. A number of human macroarray elements detected significant differential expression signals between fetal developmental stages. Their study highlighted the crucial developmental changes that occur during the final trimester of gestation, as most of the differentially expressed genes in this study were identified in the comparison between 210 and 260 d of gestation. A human microarray was employed in a study of fetal muscle from fetuses carrying myostatin loss-of-function mutations [11]. This work described breed-related differences in the expression of genes relevant to several skeletal muscle compartments (extracellular matrix, contractile cells and adipocytes).
In the present study, we used a bovine cDNA microarray derived from adult muscle and adipose tissue libraries to characterize genome-wide transcriptional changes that accompany muscle differentiation in cattle and to determine the impact of two contrasting genotypes on prenatal muscle development. We carried out a transcriptional profiling study of longissimus muscle from fetuses of both Wagyu × Hereford and Piedmontese × Hereford crosses, sampled at d 60, 135 and 195, as well as newborn calves. The double-muscled mutations of GDF8 carried by beef breeds such as Belgian Blue or Piedmontese cattle [1] have a large impact on muscle yield, and animals of the Japanese Black (or Wagyu) breed of cattle are genetically predisposed to accumulate intramuscular fat [12]. The postnatal phenotypes of offspring from Hereford dams sired by these breeds have been well-characterised and are highly divergent for muscle growth and intramuscular fat content [13]. The sampling time points chosen in this study represent primary myogenesis (d 60), secondary myogenesis (d 135), as well as beginning (d 195) and final stages (birth) of functional differentiation of muscle fibers.
Results
The entire set of expression data was deposited on Gene Expression Omnibus database [14] and can be accessed using accession numbers GSM132170 and GSM132194. In this microarray profiling experiment of developing bovine longissimus muscle, we detected 174 individual genes (617 array elements) that met the criteria for differential gene expression [see Additional file 1] either across developmental time or between the two sire breeds.
Temporal changes in gene expression
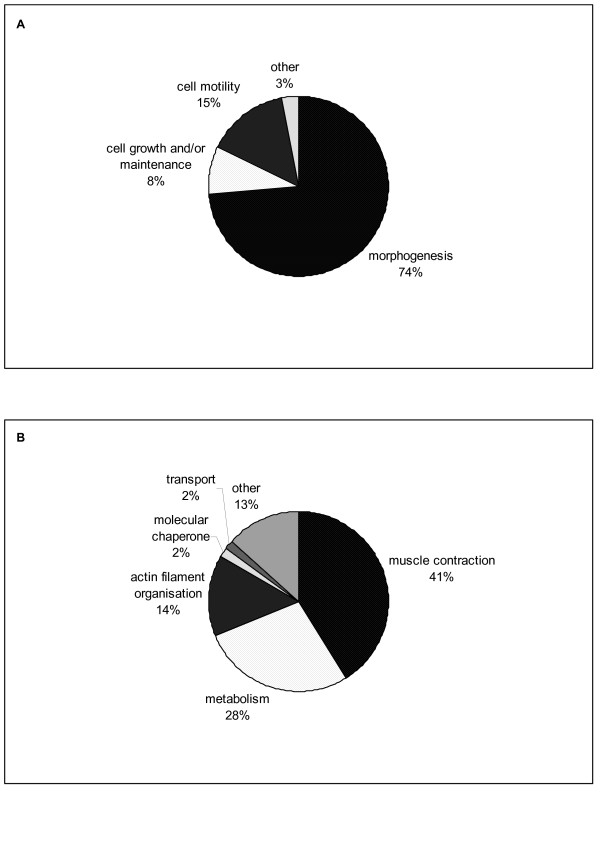
Tables 1 and 2 list expression values for differentially expressed (DE) genes for which reliable annotations could be obtained using the Basic Local Alignment Search Tool (BLASTN [15]). These genes were differentially expressed across developmental time, regardless of breed. Twenty genes showed decreasing expression over time and 35 genes showed increasing gene expression over time. All genes in Tables 1 and 2 showed expression changes of more than 2-fold over the time course examined. Gene Ontology annotation of the DE genes using EASE [16] showed that genes involved with the biological processes, morphogenesis and cellular growth were overrepresented in the genes that show decreasing expression over time (Figure 1A). When annotations with respect to cellular component were examined, 69% of the genes with decreasing expression could be classified as belonging to "extracellular matrix" (data not shown). Collagen genes alone represent 68% of the DE genes in Table 1. On the other hand, in the list of genes with increasing expression over time, genes with biological process annotations belonging to muscle cell contraction and metabolism are overrepresented (Figure 1B). The main observation from the examination of the developmental time course is therefore that transcripts associated with extracellular matrix synthesis are increasingly less represented as a proportion of total tissue RNA as muscle development proceeds, while at the same time transcripts of muscle fiber structural, contractile and metabolism genes are gaining in prevalence when maturation of muscle fibres takes place in preparation for postnatal life. In most instances, the most marked increase or decrease in gene expression can be observed between d 195 of fetal development and birth (Tables 1 and 2).
Table 1.
Genes showing decreasing expression in longissimus muscle of bovine fetuses over developmental time1
| Gene2 | GenBank Accession3 | Number of array elements4 | Gene expression ratio5 135 d/60 d | Gene expression ratio5 195 d/60 d | Gene expression ratio5 birth/60 d |
| cadherin 11, type 2, OB-cadherin (osteoblast) (CDH11) | DW521798 | 1 | 0.26** | 0.23** | 0.10** |
| collagen, type I, alpha 1 (COL1A1) | CF613531 | 14 | 0.87 | 0.74 | 0.16** |
| collagen, type I, alpha 2 (COL1A2) | CF613950 | 9 | 0.75 | 0.64** | 0.15** |
| collagen, type III, alpha 1 (COL3A1) | CF613798 | 29 | 0.72 | 0.78 | 0.15** |
| collagen, type XII, alpha 1 (COL12A1) | CF614200 | 1 | 0.48** | 0.23** | 0.12** |
| collagen, type XV, alpha 1(COL15A1) | DW521810 | 1 | 0.58** | 0.41** | 0.18** |
| fibrillin 1 (FBN1) | CF765332 | 4 | 0.57** | 0.60** | 0.23** |
| fibronectin 1 (FN1) | CF614211 | 7 | 0.39** | 0.36** | 0.08** |
| follistatin-like 1 (FSTL1) | CF613847 | 1 | 0.61** | 0.35** | 0.13** |
| guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 (GNAI1) | DW521797 | 1 | 0.58** | 0.50** | 0.20** |
| glypican 3 (GPC3) | DW521793 | 1 | 0.91 | 0.84 | 0.11** |
| Insulin-like growth factor binding protein 5 (IGFBP5) | CF614176 | 1 | 0.71 | 0.72 | 0.21** |
| lumican (LUM) | CF613654 | 2 | 0.79 | 0.64** | 0.16** |
| osteoglycin (OGN) | CF614268 | 1 | 0.59** | 0.67* | 0.19** |
| plastin 3 (T isoform) (PLS3) | DW521782 | 1 | 0.49** | 0.43** | 0.20** |
| osteonectin (SPARC) | CF613734 | 8 | 0.69* | 0.68* | 0.21** |
| osteopontin (SPP1) | DW521777 | 3 | 0.53** | 0.19** | 0.15** |
| stathmin 1/oncoprotein 18 (STMN1) | DW521774 | 1 | 0.54** | 0.38** | 0.10** |
| tubulin, alpha 1 (TUBA1) | CF613655 | 2 | 0.54** | 0.46** | 0.17** |
| vimentin (VIM) | CF613639 | 7 | 0.51** | 0.49** | 0.16** |
1* indicates a statistically significant decrease in gene expression at P < 0.05 level; ** indicates a statistically significant decrease in gene expression at P < 0.01 level.
2Sequence annotation was carried out using approved gene symbols (human genome nomenclature) via the IBISS4 database [17]. BLAST [15] scores > 110 and e-values < 2 E-23 were accepted for annotation purposes.
3in cases where more than 1 microarray element was used to calculate the expression data, the accession number of 1 representative element is shown.
4Number of microarray elements representing the same gene that were used to calculate the expression data.
5To obtain the ratios shown, averaged absolute microarray signal intensity values for d 135, 195 and birth were divided by the signal intensities measured at d 60.
Table 2.
Genes showing increasing expression in longissimus muscle of bovine fetuses over developmental time1
| Gene2 | GenBank Accession3 | Number of array elements4 | Gene expression ratio5 135 d/60 d | Gene expression ratio5 195 d/60 d | Gene expression ratio5 birth/60 d |
| actin, alpha 1, skeletal muscle (ACTA1) | CF614694 | 6 | 0.80 | 2.49** | 4.09** |
| actinin, alpha 3 (ACTN3) | CF614924 | 3 | 0.62 | 3.79** | 5.63** |
| adenylate kinase 1 (AK1) | CF614671 | 1 | 1.38** | 4.25** | 4.60** |
| aldolase A, fructose-biphosphate (ALDOA) | CF615006 | 1 | 1.28* | 3.27** | 5.82** |
| ankyrin repeat domain 1 (cardiac muscle) (ANKRD1) | CF614403 | 2 | 0.83 | 2.62** | 6.62** |
| mitochondrially encoded ATP synthase 6 (MT-ATP6) | CF613499 | 1 | 1.10 | 2.69** | 5.48** |
| chaperone, ABC1 activity of bc1 complex like (S. pombe) (CABC1) | CF615094 | 1 | 0.51 | 1.18 | 6.43** |
| calsequestrin 1 (fast-twitch, skeletal muscle) (CASQ1) | CO729180 | 1 | 1.08 | 3.93** | 5.00** |
| creatine kinase, muscle (CKM) | CF614901 | 15 | 2.15** | 3.04** | 7.49** |
| creatine kinase, mitochondrial 2 (sarcomeric) (CKMT2) | CF614479 | 1 | 0.60 | 1.76** | 10.08** |
| cardiomyopathy associated 5 (CMYA5) | CF615342 | 4 | 0.77 | 1.63** | 4.79** |
| cold shock domain protein A (CSDA) | CF615315 | 3 | 0.64 | 1.74** | 4.71** |
| cytochrome C (CYC1) | CF614629 | 1 | 2.14** | 2.53** | 6.90** |
| enolase 3 (beta, muscle) (ENO3) | CF614534 | 16 | 1.93** | 3.58** | 5.25** |
| fructose-1,6-bisphosphatase 2 (FBP2) | CF614569 | 2 | 0.53 | 2.39** | 6.73** |
| heat shock 70 kDa protein 1B (HSPA1B) | CF614728 | 1 | 1.03 | 2.17** | 4.02** |
| heat shock 90 kDa protein 1, alpha (HSPCA) | DW521445 | 1 | 1.30 | 3.63** | 6.10** |
| lactate dehydrogenase A (LDHA) | CF614400 | 5 | 0.70 | 2.40** | 4.31** |
| similar to dysferlin interacting protein 1, transcript variant 1 (LOC616223), mRNA | CO729195 | 2 | 1.57** | 3.62** | 4.97** |
| myoglobin (MB) | CF615014 | 18 | 1.79** | 2.51** | 6.68** |
| Myosin binding protein C, fast type (MYBPC2) | CF615254 | 14 | 0.89 | 2.73** | 9.83** |
| myosin, heavy polypeptide 1, skeletal muscle, adult (MYH1) | CF615300 | 3 | 0.59 | 2.70** | 5.85** |
| myosin, heavy polypeptide 2, skeletal muscle, adult (MYH2) | CF614936 | 13 | 1.40** | 2.70** | 5.37** |
| myozenin 1 (MYOZ1) | CF614412 | 18 | 2.22** | 2.89** | 7.66** |
| ncRNA orthologous with NEAT1 | CF614858 | 12 | 0.91 | 1.97** | 6.75** |
| PDZ and LIM domain 3 (PDLIM3) | CF615153 | 10 | 0.92 | 3.88** | 5.05** |
| 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3) | CF615087 | 1 | 0.36 | 0.51 | 6.27** |
| phosphofructokinase, muscle (PFKM) | DW521807 | 1 | 1.01 | 4.12** | 6.71** |
| phosphoglycerate mutase 2 (muscle) (PGAM2) | CF614969 | 6 | 1.12 | 4.03** | 4.65** |
| phosphoglucomutase 1 (PGM1) | CF615240 | 7 | 1.06 | 3.50** | 5.47** |
| Phosphorylase, glycogen; muscle (PYGM) | CF614749 | 6 | 1.06 | 3.17** | 7.27** |
| solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 (SLC25A4) | CF614535 | 3 | 0.80 | 1.93** | 5.14** |
| titin-cap (telethonin) (TCAP) | CF614598 | 5 | 2.81** | 4.15** | 8.78** |
| tropomodulin 4 (muscle) (TMOD4) | CF614407 | 2 | 0.79 | 3.62** | 6.53** |
| Titin immunoglobulin domain protein (myotitilin) (TTID) | CF614674 | 5 | 0.58 | 1.63** | 4.61** |
1* indicates a statistically significant increase in gene expression at P < 0.05 level; ** indicates a statistically significant increase in gene expression at P < 0.01 level.
2Sequence annotation was carried out using approved gene symbols (human genome nomenclature) via the IBISS4 database [17]. BLAST [15] scores > 110 and e-values < 2 E-23 were accepted for annotation purposes.
3In cases where more than 1 microarray element was used to calculate the expression data, the accession number of 1 representative element is shown.
4Number of microarray elements representing the same gene that were used to calculate the expression data.
5To obtain the ratios shown, averaged absolute microarray signal intensity values for d 135, 195 and birth were divided by the signal intensities measured at d 60.
Figure 1.
Annotation of differentially expressed genes in bovine longissimus muscle. The Expression Analysis Systematic Explorer (EASE) tool [41] was used to identify classes of overrepresented gene ontology annotations in the differentially expressed gene lists (Tables 1 and 2). (A) Biological process annotation of genes which showed decreasing expression over the developmental time course. (B) Biological process annotation of genes which showed increasing expression over the developmental time course.
Two genes, FSTL1 and IGFBP5 (Table 1), which are implicated in the regulation of cell growth and differentiation, are significantly downregulated in developing bovine muscle. These genes were chosen for quantitative reverse transcription-PCR (qRT-PCR) validation (see below).
Twelve microarray probes that reported increasing expression levels during development all matched to a single consensus sequence (btcn26022) in the IBISS4 database [17]. Using the NETBLAST function in IBISS4, a match with a non-coding (nc) RNA transcript orthologous with the recently described NEAT1 transcript was identified [18] (Table 2).
Gene expression differences correlated with sire breed
A subset of gene expression differences according to sire breed is summarized in Table 3. In total, 82 genes were differentially expressed according to the breed of the fetus' sire in at least one time point [see Additional file 2]. The temporal changes in gene expression profiles were greater than the expression differences that were correlated to sire breed. Only 3 of these DE genes (ALDOA, FABP5 and MATR3) show a fold-change of more than 2 [see Additional file 2]. ALDOA was around two-fold less expressed in the longissimus muscle of Wagyu-sired fetuses at d195, while FABP5 showed more than two-fold less expression in the longissimus muscle of Piedmontese-sired calves at birth. FABP4 expression is also highlighted in Table 3, as this gene showed less gene expression in the longissimus muscle of Piedmontese-sired fetuses samples at d 135, but at no other time point. MATR3 showed less gene expression in the longissimus muscle of day 60 Wagyu-sired fetuses.
Table 3.
Examples of genes showing differential expression in longissimus muscle of bovine fetuses from different sire breeds in at least one time point1
| Gene2 | GenBank Accession3 | Number of array elements4 | Gene expression ratio5 P60d/W60d | Gene expression ratio5 P135d/W135d | Gene expression ratio5 P195d/W195d | Gene expression ratio5 Pbirth/Wbirth |
| Aldolase A, fructose-biphosphate (ALDOA) | CF615006 | 4 | 0.87 | 1.04 | 2.16** | 0.90 |
| Collagens (COL1A1, COL1A2, COL3A1, COL12A1, COL15A1) | Additional file 2 | 42 | 1.21 | 0.70 | 0.96 | 0.64** |
| Cytochrome c oxidases (COX5B, 6C, 7B, 7C) | Additional file 2 | 9 | 0.93 | 0.99 | 0.84 | 1.74** |
| Fatty acid binding protein 4, adipocyte (FABP4) | CF614083 | 3 | 1.01 | 0.70** | 0.90 | 0.98 |
| Fatty acid binding protein 5 (FABP5) | CF613827 | 1 | 1.29 | 0.93 | 1.16 | 0.44** |
| Matrin 3 (MATR3) | CF614914 | 1 | 2.35** | 1.15 | 0.79 | 0.64** |
| Ribosomal L proteins (RPL) | Additional file 2 | 32 | 0.75 | 0.93 | 1.01 | 1.52** |
| Ribosomal S proteins (RPS) | Additional file 2 | 13 | 0.80 | 0.89 | 0.99 | 1.51** |
1* indicates a statistically significant gene expression difference at P < 0.05 level; ** indicates a statistically significant gene expression difference at P < 0.01 level.
2Sequence annotation was carried out using approved gene symbols (human genome nomenclature) via the IBISS4 database [17]. BLAST [15] scores > 110 and e-values < 2 E-23 were accepted for annotation purposes.
3in cases where more than 1 microarray element was used to calculate the expression data, the accession number of 1 representative element is shown.
4Number of microarray elements representing the same gene that were used to calculate the expression data.
5To obtain the ratios shown, averaged absolute microarray signal intensity values for P (Piedmontese × Hereford) fetuses were divided by the signal intensities measured for W (Wagyu × Hereford) fetuses at d 60, d 135, d 195 and birth.
A number of genes showed smaller, yet still significant differences in gene expression between breeds in at least one time point [see Additional file 2]. Table 3 provides a summary for some of these classes of genes, for example the collagen genes, which were more highly represented in the transcriptome of late-stage Wagyu fetuses. All differentially expressed ribosomal protein genes and cytochrome c oxidase genes were detected as significantly up-regulated in the muscle of Piedmontese crossbred calves at birth (Table 3 and [Additional file 2]). Ribosomal protein and cytochrome c oxidase genes were not detected as differentially expressed over developmental time (Tables 1 and 2).
qRT-PCR validation of microarray gene expression patterns
We conducted qRT-PCR studies on the amplified RNA used in the microarray study. Assays for FABP4, FABP5, FSTL1, IGFBP5, and MATR3 were developed to confirm temporal and sire-breed specific gene expression differences. In order to confirm that breed-specific differences caused by the Piedmontese myostatin (GDF8) mutation could be detected by gene expression measurements, and because GDF8 was not represented on the microarray, GDF8 expression was studied using qRT-PCR. Goodness of fit, as measured by the R2, resulted in the ANOVA model explaining 88.7%, 80.7%, 63.6%, 87.7%, 94.1% and 98.3% of the total variation in the expression of GDF8, FABP4, FABP5, IGFBP5, FSTL1, and MATR3, respectively [see Additional file 3]. Temporal gene expression patterns detected by microarray were confirmed for IGFBP5 and FSTL1 (Table 4). The breed-specific differences in FABP4 and FABP5 expression documented by microarray were accurately confirmed by qRT-PCR (Table 5). FABP4 expression was higher in the longissimus muscle of Wagyu-sired fetuses at d195 and birth. FABP5 expression was higher in the longissimus muscle of Wagyu-sired calves at birth, but no significant differences were detected at any other time point. GDF8 expression was significantly higher in the longissimus muscle of Piedmontese-sired fetuses at d60, d135 and birth (Table 5). No difference could be detected at d195.
Table 4.
Quantitative reverse transcription PCR (qRT-PCR) measurements of gene expression in total RNA from longissimus muscle of bovine fetuses – gene expression ratios over developmental time
| Gene | Fold Ratio1 | ||
| 135 d/60 d | 195 d/60 d | birth/60 d | |
| IGF-binding protein 5 (IGFBP5) | 0.97 | 0.55 ** | 0.08 ** |
| Follistatin-like (FSTL1) | 0.62 ** | 0.24 ** | 0.05 ** |
1To obtain fold ratios, mean normalized (from ANOVA and against 18S RNA) qRT-PCR values at 135 d gestation, 195 d gestation and birth were divided by the values at 60 d gestation.
** Significance at P < 0.01.
Table 5.
Quantitative reverse transcription PCR (qRT-PCR) measurements of gene expression in total RNA from longissimus muscle of bovine fetuses – gene expression ratios between breeds
| Gene | Fold Ratio1 | |||
| P60d/W60d | P135d/W135d | P195d/W195d | Pbirth/Wbirth | |
| Myostatin (GDF8) | 1.52 ** | 1.45 * | 1.15 | 1.51 ** |
| Fatty-acid binding protein 4 (FABP4) | 1.32 | 0.56 | 0.14 ** | 0.20 ** |
| Fatty-acid binding protein 5 (FABP5) | 0.88 | 1.09 | 1.00 | 0.54 ** |
| Matrin 3 (MATR3) | 1.10 | 1.10 | 0.96 | 0.75 |
1To obtain fold ratios, mean normalized (from ANOVA and against 18S RNA) qRT-PCR values from P (Piedmontese × Hereford) samples were divided by those from W (Wagyu × Hereford) samples at 60 d gestation, 135 d gestation, 195 d gestation and birth.
* Significance at P < 0.05; ** Significance at P < 0.01.
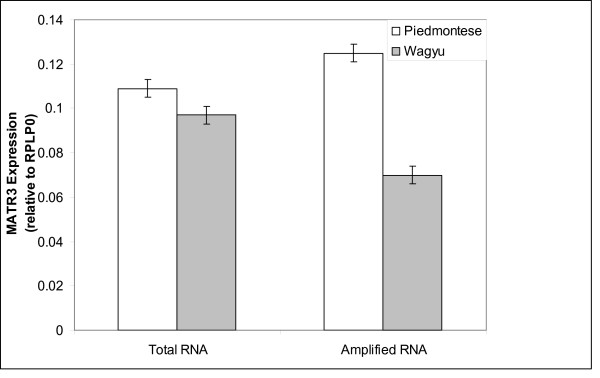
Using the MATR3 qRT-PCR assay, we were unable to confirm the >2 fold higher gene expression detected by microarray in d60 muscle RNA samples from Wagyu-sired fetuses (Table 5). In order to clarify what may have caused this discrepancy, qRT-PCR assays for MATR3 were performed on the amplified RNA samples that had been used for the microarray analysis. This analysis showed that the RNA samples amplified from the d 60 longissimus muscle of Piedmontese-sired fetuses contained higher amounts of MATR3 RNA than the non-amplified RNA sample (Figure 2). Specifically, this elevated expression was due to one individual RNA sample (P663, data not shown). qRT-PCR testing of MATR3 expression therefore confirmed that the microarray analysis, which was carried out with amplified RNA, had accurately identified the elevated MATR3 expression levels in the d 60 sample. The results did, however uncover a discrepancy between amplified and total RNA for at least one particular individual and one gene.
Figure 2.
Quantitative reverse transcription (qRT-PCR) measurements of matrin 3 (MATR3) gene expression in bovine d 60 longissimus muscle. qRT-PCR was used to measure MATR3 expression from Hereford × Piedmontese (clear) or Hereford × Wagyu (shaded) fetuses, comparing total and amplified RNA. The data represent average values and standard error measurements from 3 individual animal samples, normalized against ribosomal protein, large PO (RPLPO) expression measured in the same sample.
Discussion
This study was designed to map out gene expression changes that accompany four discrete phases of muscle development: the first wave of myogenesis (d 60), the second generation myoblast formation (d 135) and the beginning (d 195) and end (birth) stages of contractile and metabolic differentiation of muscle fibers. As two different breeds of sire (Wagyu and Piedmontese) were used to generate the fetuses in this study, we also attempted to detect gene expression differences that may be associated with the genetic background of the fetus. The fact that Hereford crossbreeds rather than purebred fetuses were used in this comparison is a limitation imposed by resource constraints. However, the postnatal phenotypes of offspring from Hereford dams sired by these breeds have been studied in a separate investigation and were found to be highly divergent for muscle growth and intramuscular fat content [13].
At d 60, the longissimus muscle was not identifiable as a distinct anatomical entity. An area of tissue corresponding to the future position of the longissimus muscle was therefore collected at this time point. At the d 60 time point it was more problematic to accurately dissect the overlying skin tissue away from the muscle. Reliable and consistent sampling of the longissimus muscle, using position along the vertebral column as a positional marker, was achieved from d 135 onwards. Due to limiting sample amounts, particularly at the d 60 time point, all RNA samples were subjected to a linear amplification procedure before labelling for microarray hybridization.
The gene expression analysis of longissimus muscle RNA samples provided evidence that the linear amplification procedure may have introduced bias into the representation of mRNA sequences in the d 60 microarray sample from one individual. While numerous studies have shown that linear amplification will conserve transcript abundance to an acceptable level, the potential to introduce significant bias to gene expression studies does exist [19]. For the above reasons, the discussion of differential gene expression will focus on the time points d 135, d 195 and birth.
Temporal changes in gene expression
This microarray study of fetal bovine muscle development detected a large number of differentially expressed genes across developmental time. The expression pattern of muscle fiber structural and metabolism genes observed in this study is correlated to the timing of contractile differentiation of muscle fibers during fetal development. The overall trend of decreasing representation of extracellular matrix gene expression in fetal bovine muscle over time is in accordance with histological observations that show the proportion of extracellular matrix material on a muscle section decreasing from 70% to 27% over the time course studied here [7]. The increasing representation of muscle fiber-specific gene expression over time may therefore be a reflection of the proliferation and differentiation of muscle fibers within a connective tissue "scaffold" established early during development [20].
An earlier transcriptome study of bovine muscles during ontogenesis [10] did not identify connective tissue gene expression as a significant contributor to differential gene expression across developmental time. This may be a consequence of the skeletal muscle focused macroarray that was used in their study. The bovine fat/muscle microarray used in this study was constructed from muscle and subcutaneous fat cDNA libraries [21]. The differentially expressed microarray elements that showed decreasing gene expression over time, mainly had their origin in the subcutaneous fat cDNA library.
The observation that the most marked gene expression changes occur between d 195 and birth is in accordance with other studies that have shown the dramatic adaptations of skeletal muscle in preparation for birth [10]. As the postnatal muscle samples studied here were collected within 24 hours after birth, our study may, in addition, detect gene expression differences that reflect birth trauma.
We were unable to detect transcriptional changes that reflect the replacement of fetal with adult isoforms of contractile protein transcripts during the final trimester of pregnancy, as we are using a microarray based on adult skeletal muscle gene expression [21].
The vast majority of temporal changes in gene expression in fetal longissimus muscle comprise structural and metabolic components of extracellular matrix and muscle fibers. Two exceptions are FSTL1and IGFBP5, which have been implicated in muscle cell growth and differentiation. The expression patterns for these two genes were confirmed by qRT-PCR and their reduced expression over the developmental time course is in accord with postulated roles in the inhibition of muscle cell differentiation [22,23]. Both genes have been reported as early targets of MyoD expression in a mouse model of MyoD-induced differentiation [24]. In this model of myogenesis, Fst1 was down-regulated, whereas Igfbp5 was strongly up-regulated. This discrepancy with our results may indicate species-specific differences in IGFBP5 action in bovine muscle development, be due to the fact that our results are derived from an in vivo, not an in vitro model of myogenesis, or hint at the proposed dual role of this gene in the regulation of muscle cell development [25]. Our results lend further support to the notion that FSTL1, while its precise function is still unknown, has a crucial role in muscle development.
The NEAT1orthologue, a ncRNA species, is found at increasing levels during development of cattle muscle. NEAT1 was one of only three large abundant ncRNAs found enriched in the nucleus and highly conserved across mammalian species [18]. This, together with its association with SC35 splicing domains, suggests a fundamental function in mRNA metabolism for the NEAT1 transcript. The increasing representation of this ncRNA species in developing cattle muscle may be a reflection of the increasing number of nuclei in developing myofibres. Myonuclei accumulate in muscle cross-sections at an increasing rate up to about 180 days post conception [26]. The 6.8 fold higher representation of the NEAT1 orthologue transcript in the muscle of newborn cattle, compared with the muscle at d60, may therefore indicate a correspondingly higher proportion of nuclei per cell volume sampled. The observations of Ansay [27] on the DNA content of developing cattle muscle lend support to this view.
Individual gene expression differences correlated with sire breed
While the microarray analysis detected a large number of genes that were significantly differentially expressed in at least one of the time points studied, only 2 of those genes showed a more than two-fold difference between the two sire breeds. We therefore examined the expression of myostatin (GDF8) itself by qRT-PCR. GDF8 RNA was expressed at significantly higher levels in the muscle of Piedmontese-sired fetuses at d 60 and d 135, in accordance with the findings of Forbes et al. [28] that the non-functional GDF8 molecule of the Piedmontese breed failed to repress the GDF8 promoter and therefore led to higher GDF8 transcript levels in the fetal muscle. Our observations therefore confirm that the GDF8 mutation did affect gene regulation in the fetal longissimus muscle as predicted.
One of the genes showing more than two-fold elevated expression in the muscle of newborn Wagyu-sired calves, FABP5, may indicate a higher level of adipogenic differentiation in the longissimus muscle of these animals, even at this early stage of their development. FABP4 showed a similar pattern of breed-specific differences, and the differential expression of both genes was confirmed by qRT-PCR (Tables 3 and 5). Fatty acid binding proteins are expressed at very high levels by adipocytes and are themselves, by interaction with peroxisome proliferator-activated receptors (PPAR) involved in mediating the effects of fatty acids on gene regulation [29]. Genetic variation in the bovine FABP4 gene has been found to be associated with intramuscular fat content and subcutaneous fat depth [30]. The possibility that FABP5 and FABP4 may be involved in defining the early onset of intramuscular fat deposition in the Wagyu breed remains to be explored. On the other hand, this finding may be an indication of relatively lower expression of FABP4 and FABP5 in Piedmontese-sired fetuses and therefore a consequence of the biochemical changes initiated by the myostatin mutation. In support of this latter possibility, a recent study of bovine fetuses carrying a myostatin mutation found a gene marker of adipocyte differentiation down-regulated in semitendinosus muscle at d 260 of gestation [11].
Global gene expression differences correlated with sire breed
The microarray used in these experiments was based on adult muscle gene expression and is therefore less likely to detect expression differences in developmental regulatory genes involved with determining the double-muscled phenotype. However, the comprehensive view of developing muscle biochemistry that these experiments describes, points to subtle differences in timing of gene expression, as well as differences in the translational capacity of the Piedmontese muscle and an altered biochemical profile as the main consequences of the GDF8 mutation. Our study provides evidence for gene expression timing differences by the large number of ribosomal genes, which tend to be expressed at a relatively lower level in the longissimus muscle of Piedmontese-sired fetuses at the earlier time points and are then relatively more highly expressed at birth. This confirms the findings by Steelman et al. [31], who found genes involved in translation overrepresented among the up-regulated genes in 5-week-old myostatin null mice. A study of d 30- d 32 bovine embryos had identified a number of ribosomal protein genes as differentially expressed in embryos carrying the GDF8 mutation [32]. Cassar-Malek et al. [11], in a gene expression study in bovine semitendinosus muscle from d 260 fetuses carrying myostatin mutations also described higher levels of expression of ribosomal protein genes at this time point. Support for the view that timing differences characterize the development of GDF8 mutants comes from the work of Deveaux et al[33], which showed evidence for a delay in contractile differentiation of muscle from Belgian Blue fetuses, and Cagnazzo et al. [9], who showed that the fetuses from a more highly muscled breed of pigs showed a later peak of myogenesis-related gene expression during fetal development.
Our study confirms the findings by Cassar-Malek et al. [11] of decreased collagen gene expression in the muscle of late stage bovine fetuses carrying a myostatin mutation. This finding lends support to our interpretation of the temporal changes of collagen gene expression (above). As the GDF8 mutation results in larger numbers of muscle fibers that have larger cross-sectional areas on average, connective tissue fibroblasts (with collagen as the main part of their expression signature) should contribute relatively less to the overall gene expression profile.
GDF8 mutants are known to have a more glycolytic adult muscle phenotype than wild type animals [33] and this is clearly reflected in the gene expression comparisons between breeds at this time point. For example, the Piedmontese-sired fetuses show higher expression of genes that reflect fast, glycolytic fiber structural differentiation such as ACTN3, MYH1 and MYH2, and MYBPC2, coupled with higher expression of genes for glycolytic metabolic enzymes such as ALDOA and LDHA. These findings confirm the observations of Cassar-Malek et al[11] on bovine muscle and also of Steelman et al. [31], who documented a shift away from slow muscle fiber isoforms in myostatin null mice.
At birth, a suite of gene expression changes suggesting an increased ATP supply to the muscle of the Piedmontese-sired fetuses is observed. In fact, there is a coordinated up-regulation of genes encoding enzymes that determine mitochondrial activity at multiple steps in cellular respiration. These include key oxidative enzymes, in particular isoforms of cytochrome c oxidase (COX5B, 6C, 7B, 7C), the terminal enzyme in the citric acid cycle. Cagnazzo et al[9] made a similar observation in their comparison of transcription profiles of developing muscles from two contrasting breeds of pigs. They found that a highly muscled breed of pigs (Pietrain) showed more active transcription of ATP metabolism-related genes during late fetal development.
This surge in ATP supply may reflect increased energetic demand in the immediate postnatal period in animals that experience very rapid postnatal growth, such as Piedmontese-sired calves or Pietrain piglets. Two major ATP consuming cellular processes are the maintenance of transmembrane ion gradients and protein turnover [34]. The demand for sarcolemmal ion gradients is unlikely to increase at this time. In fact, the expression of ATPase genes involved in this process (ATP1A2, ATP2A1, ATP2A2) is relatively lower in the muscle of Piedmontese-sired calves.
Therefore, taken together with the observation that a suite of ribosomal proteins is upregulated in Piedmontese-sired calves, we suggest that the extra ATP might be supporting more active ribosomal synthetic machinery. Consequently, the gene expression changes captured by this microarray appear to describe the energetic modifications necessary for the rapid muscular growth of newborn GDF8 mutants.
Conclusion
This study describes the molecular events accompanying skeletal muscle differentiation in the bovine, namely a coordinate down-regulation of extracellular matrix-related gene expression at the same time as increasing gene expression levels of structural and metabolic constituents of muscle fibers. This study also highlights the developmental expression pattern of FSTL1 and IGFBP5, which have previously been implicated in myogenesis regulation, as well as describing the changing representation of a recently-described ncRNA (NEAT1 orthologue) in developing cattle muscle. A large number of breed-related gene expression differences were detected by contrasting the development of Wagyu-sired and Piedmontese-sired fetuses. While no breed-related differences in regulatory pathways were detected in this study, it documents the differential timing and magnitude of gene expression that result in the more glycolytic muscle fiber profile as well as the lower level of intramuscular fat observed in animals carrying myostatin mutations.
Methods
Animals
All animal experimentation complied with the Animal Ethics requirements of NSW Agriculture/GARAS. Hereford cows were artificially inseminated or mated to one of 5 different Wagyu sires or one of 6 different Piedmontese sires. All Piedmontese sires were homozygous for the GDF8 missense mutation in exon 3 and none of the Wagyu sires carried the mutation. 3 Fetuses from each sire breed were recovered by caesarean section at around 60, 130 and 195 of gestation. 3 newborn calves per sire breed were euthanased by lethal injection within 24 hours of birth. Details of the sampled fetuses and calves were recorded [see Additional file 4]. Longissimus muscle was dissected immediately after death of the fetuses or calves and tissue samples snap frozen in liquid nitrogen.
Microarray methods
The process for RNA extraction and purification was performed as described in Lehnert et al. [21]. Anti-sense RNA (aRNA) amplification was performed using the MessageAmp aRNA Kit (Ambion, Austin, TX). Indirect labeling procedures and hybridizations were performed as described in Lehnert et al. [21] with the following modifications. The cDNA was generated using a modified amino C6 dT random primer [35] (Geneworks, Adelaide, SA) and the reverse transcriptase Superscript III (Invitrogen, Carlsbad, CA). The cDNA (pre-labeling) and the post-labeled cDNA were purified using the QIAquick PCR purification columns (Qiagen, Valencia, CA). The hybridization mixture used a reduced detergent concentration of 0.2%SDS.
We utilized a bovine cDNA microarray constructed from two cattle cDNA libraries; longissimus muscle and subcutaneous fat tissue derived from a 24 mo-old grass-fed Angus steer [21]. The array contained 9,600 bovine cDNA probes, printed in duplicate.
Experimental design and data acquisition criteria
The weight of the individual fetuses [see Additional file 4], was analyzed by fitting an ANOVA model that contained the effects of breed, time, sex, and breed by time interaction. The ANOVA model accounted for 97.8% of the total variation in fetus weight and significant effects included time (P < 0.001), breed by time (P < 0.05) and breed (P < 0.10). The design of the microarray experiment therefore focused on the developmental aspect of the study, as well as exploring sire breed.
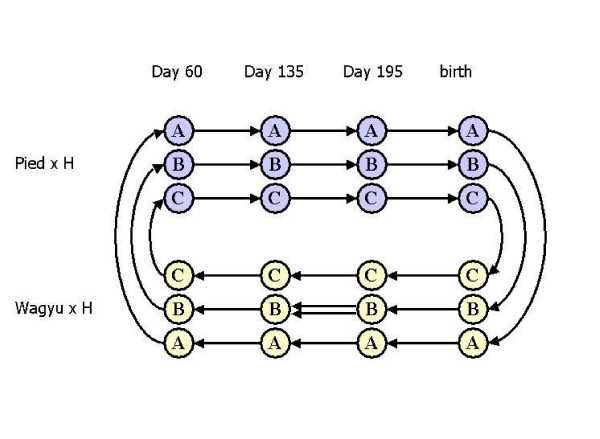
Factors considered in designing the experiment included the availability of RNA for each sample of interest as well as the cost of the arrays themselves. Figure 3 illustrates the configuration layout for the experimental design that was developed with a total of three biological replicates, two breeds and four time points that were compared using 25 microarray hybridizations. The design was chosen to allow a focus on the developmental aspects of the study, but to also permit a breed comparison to be carried out. The entire set of expression data was deposited on the GEO database [14] and can be accessed using accession numbers GPL4196 and GPL4197.
Figure 3.
Microarray experimental design. Each microarray assay is symbolized by an arrow connecting the two longissimus muscle RNA samples that were hybridized to the array. The direction of the arrow indicates which sample was labeled with Cy3 (arrowhead) and Cy5 (origin).
We used the GenePix 4000A optical scanner (Molecular Devices, Sunnyvale, CA) and the GenePixPro 5.1 image analysis software (Molecular Devices) to quantify the gene expression level intensities, employing editing criteria detailed in [36]. Editing criteria were applied separately for the red (Cy5) and for the green (Cy3) intensity channels so that a different number of observations for each channel were obtained. These resulted in a total of 887,706 gene expression intensity readings (443,292 red and 444,414 green) that were background corrected and base-2 log transformed. The arithmetic mean and standard deviation (in brackets) for the red and green intensities were 11.93 (2.02) and 11.97 (1.95), respectively.
Mixed-model equations and identification of differentially expressed genes
The following linear mixed-effect model was fitted to the data:
Yijktmn = μ + Cijk + Gm + AGijm + DGkm + TGtm + εijktmn
where Yijktmn represents the n-th background-adjusted, normalized base-2 log-intensity from the m-th gene (probe) at the t-th treatment (fetal age and breed sample) from the i-th array, j-th printing block and k-th dye channel; C represents a comparison group fixed effect defined as those intensity measurements from the same array slide, printing block and dye channel; G represent the random gene (probe) effects with 8,845 levels; AG, DG, and TG are the random interaction effects of array × gene, dye × gene, and treatment × gene, respectively. Finally, ε is the random error term.
Variance components for random effects were estimated using restricted maximum likelihood. Differentially expressed (DE) genes were identified after processing the appropriate linear combination of the solutions (best linear unbiased predictions) of TG via model-based clustering, addressing the developmental as well as the breed comparison contrasts of interest [37,38].
A total of six contrasts were considered in the identification of DE genes. These included the two within-breed over-time contrasts and the four within-time across-breed contrasts. For each contrast, a two-component normal mixture model was fitted and posterior probabilities of belonging to the non-null component used to identify DE genes for an estimated experiment-wise false discovery rate of < 1% were computed as described by McLachlan et al. [39].
Synthesis of complementary DNA (cDNA) for quantitative reverse transcription PCR (qRT-PCR) validation
TRIzol-extracted (Invitrogen) total RNA from longissimus muscle samples was further purified on RNeasy mini columns and treated on-column with DNase I (Qiagen) according to the manufacturer's instructions. Total RNA concentrations and relative purity were determined spectrophotometrically by measurement of UV absorbance at 260 and 280 nm (GeneQuant; GE Healthcare, Little Chalfont, UK). Reverse transcription of 1.5 μg of total RNA was performed with the Omniscript cDNA synthesis kit (Qiagen) using a mixture of oligonucleotides [100 μM OligodTVN (5'-TTTTTTTTTTTTTTVN-3' where V = A,C,G and N = A,C,G,T) and 1 μM 18Sprimer (5'-CACACGCTGAGCCAGTCAGT-3')]. The cDNA was diluted 1:10 in 10 mM Tris-HCl (pH8.0) for target gene measurements, whereas the 18S rRNA reference gene measurements were performed on a 1:200 dilution of cDNA.
Total RNA from d 60 fetal muscle samples and amplified RNA from the d 60 samples that were used to probe the DNA microarray were separately reverse transcribed. 1.5 μg of total or amplified RNA was reverse transcribed with the Superscript III first strand cDNA synthesis kit (Invitrogen). The cDNA synthesis used random hexamers and was performed at 37°C for 1 hr in the presence of RNase inhibitor (RNaseOUT, Invitrogen). The cDNA was diluted 1:10 in 10 mM Tris-HCl (pH8.0) and was used to measure the mRNA levels of MATR3 and RPLP0.
qRT-PCR analyses
qRT-PCR to measure mRNA transcript levels was performed as described in Nattrass et al. [40], using the primer sequences listed in Table 6. qRT-PCR measurements were performed in triplicate on each cDNA sample (n = 24) on two separate occasions or once on the d 60 amplified and unamplifed samples (n = 12). Relative transcript quantitation was performed using standard curves generated for each gene from a 10-fold serial dilution of cDNA. Pooled cDNA from a subset of the longissimus muscle samples examined in this study was used to generate the standard curves. The standard curve dilutions, preparation and dispensing of the SYBR green I reagent and addition of the cDNA standards, reference cDNAs and cDNA samples were performed with a CAS1200 liquid handling robotics system (Corbett Life Science, Sydney, Australia). Target gene expression data from samples collected at day 60, day 135, day 195 and birth were determined by normalisizing against the measurements for 18SrRNA. Matrin 3 expression dataon the unamplified and amplified day 60 samples were determined by normalisizing against the measurements for RPLP0.
Table 6.
Oligonucleotides used in quantitative reverse transcription PCR (qRT-PCR)
| Gene | Symbol | sense | sequence | Genbank accession number |
| Myostatin (growth differentiation factor 8) | GDF8 | FOR1 | ACCTTCCCAGAACCAGGAGAA | AF019620 |
| REV | TCACAATCAAGCCCAAAATCTCT | |||
| Fatty acid binding protein 4 (adipocyte) | FABP4 | FOR | TGGAAACTTGTCTCCAGTGAAA | X89244 |
| REV | ACCCCCATTCAAACTGATGA | |||
| Fatty acid binding protein 5 (psoriasis-associated) | FABP5 | FOR | TGGGAGAGAAGTTTGAAGAGA | BT020981 |
| REV | TTCCTGATGTTGAACCAATGC | |||
| Follistatin-like 1 | FSTL1 | FOR | TGCAGACCAGGAGAACAACA | BC114758 |
| REV | GGTTGAGGCACTTGAGGAAC | |||
| Insulin-like growth factor binding protein 5 | IGFBP5 | FOR | GGTTTGCCTGAACGAAAAGA | S52657 |
| REV | CTTGGGCGAGTAGGTCTCC | |||
| Matrin 3 | MATR3 | FOR (1) | GGAAAAAAGAACCTTCAGACA | CB434458 |
| FOR (2) | GACAAAGCTGTGAAAAAAGAT | |||
| REV | CCTCGATCTTGTCCACCTTT | |||
| 18S ribosomal RNA | 18SrRNA | CGGTCGGCGTCCCCCAACTT | AY779625 | |
| GCGTGCAGCCCCGGACATCTAA | ||||
| Ribosomal protein large, P0 | RPLP0 | FOR | CAACCCTGAAGTGCTTGACAT | NM001012682 |
| REV | GCAAGTGGGAAGGTGTAATCA |
FOR = forward primer; REV = reverse primer
Statistical assessment of qRT-PCR gene expression data
Statistical analyses were performed using the Procedure GLM of SAS (SAS v9.1, SAS Institute Inc., Cary NC, USA) with an ANOVA model that included the expression of the genes (both target and reference genes), as measured by their threshold cycle (Ct) of the PCR reaction, as dependent (response) variable and biological replicate (with two levels), breed (with two levels), age (with four levels), breed by age interaction (with eight levels) and residual as independent variables. Biological replicates were fitted as nested within breed by age interaction. Least square means of Ct for each gene and at each level of breed, age, and breed by age interaction were compared for differential expression. To ensure overall protection level, only probabilities associated with pre-planned comparisons were used.
Authors' contributions
SAL participated in the study design, sample collections and data analysis and drafted the manuscript. AR participated in the microarray experimental design and carried out the statistical analyses. KAB participated in the microarray experimental design and analysis, and carried out RNA extractions and microarray experimentation. YHW participated in the study design and coordination. GSN carried out quantitative PCR validation. NJH helped to draft the manuscript. PLG was instrumental in developing the concepts for this study, and participated in the sample collections and helped to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Statistical thresholds for t-statistics and fold changes at two levels of significance and for down- and up-regulated genes. Statistical cut-off values for inclusion in differentially expressed gene lists
Genes showing differential gene expression in LM of bovine fetuses from different sire breeds in at least one time point. Full listing of all genes that showed differential expression between the two sire breeds.
Goodness of fit of the ANOVA model and significance of design effects on the expression of target and housekeeping genes as measured by the Ct of the qRT-PCR. Background detail to the analysis of qRT-PCR data.
Bovine fetuses sampled for microarray study. Listing of genetic background, weight, sex and accurate ages of the fetuses used in this study.
Acknowledgments
Acknowledgements
This research was supported by the Cooperative Research Center for Cattle and Beef Quality and its core partners: The University of New England, NSW Agriculture, CSIRO Livestock Industries and Queensland Department of Primary Industries. Meat and Livestock Australia also supported this project (BSC.010). NJH was a recipient of a CSIRO postdoctoral fellowship. The authors thank H Hearnshaw, L Cafe and K Newby at NSWDPI's Grafton Agricultural Research Station for their invaluable support, as well as S McClelland, S McWilliam, N Bower, S H Tan and G Kelly for their assistance. The authors also acknowledge the support of many other colleagues in the Beef CRC network, notably G Harper and D Ferguson of CSIRO Livestock Industries. R Moore provided microarray printing services and B Dalrymple and R Tellam contributed valuable comments on the manuscript.
Contributor Information
Sigrid A Lehnert, Email: sigrid.lehnert@csiro.au.
Antonio Reverter, Email: tony.reverter-gomez@csiro.au.
Keren A Byrne, Email: keren.byrne@csiro.au.
Yonghong Wang, Email: yonghong.wang@csiro.au.
Greg S Nattrass, Email: nattrass.greg@saugov.sa.gov.au.
Nicholas J Hudson, Email: n.hudson1@uq.edu.au.
Paul L Greenwood, Email: paul.greenwood@dpi.nsw.gov.au.
References
- Grobet L, Martin LJ, Poncelet D, Pirottin D, Brouwers B, Riquet J, Schoeberlein A, Dunner S, Menissier F, Massabanda J, et al. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet. 1997;17:71–4. doi: 10.1038/ng0997-71. [DOI] [PubMed] [Google Scholar]
- Maxfield EK, Sinclair KD, Dunne LD, Broadbent PJ, Robinson JJ, Stewart E, Kyle DG, Maltin CA. Temporary exposure of ovine embryos to an advanced uterine environment does not affect fetal weight but alters fetal muscle development. Biol Reprod. 1998;59:321–5. doi: 10.1095/biolreprod59.2.321. [DOI] [PubMed] [Google Scholar]
- Picard B, Lefaucheur L, Berri C, Duclos MJ. Muscle fibre ontogenesis in farm animal species. Reprod Nutr Dev. 2002;42:415–31. doi: 10.1051/rnd:2002035. [DOI] [PubMed] [Google Scholar]
- Russell RG, Oteruelo FT. An ultrastructural study of the differentiation of skeletal muscle in the bovine fetus. Anat Embryol (Berl) 1981;162:403–17. doi: 10.1007/BF00301866. [DOI] [PubMed] [Google Scholar]
- Robelin J, Picard B, Listrat A, Jurie C, Barboiron C, Pons F, Geay Y. Myosin expression in semitendinosus muscle during fetal development of cattle: immunocytochemical and electrophoretic analyses. Reprod Nutr Dev. 1993;33:25–41. doi: 10.1051/rnd:19930103. [DOI] [PubMed] [Google Scholar]
- Gagniere H, Picard B, Geay Y. Contractile differentiation of foetal cattle muscles: intermuscular variability. Reprod Nutr Dev. 1999;39:637–55. doi: 10.1051/rnd:19990510. [DOI] [PubMed] [Google Scholar]
- Robelin J, Lacourt A, Bechet D, Ferrara M, Briand Y, Geay Y. Muscle differentiation in the bovine fetus: a histological and histochemical approach. Growth Dev Aging. 1991;55:151–60. [PubMed] [Google Scholar]
- Te Pas MF, De Wit AA, Priem J, Cagnazzo M, Davoli R, Russo V, Pool MH. Transcriptome expression profiles in prenatal pigs in relation to myogenesis. J Muscle Res Cell Motil. 2005;26:157–65. doi: 10.1007/s10974-005-7004-6. [DOI] [PubMed] [Google Scholar]
- Cagnazzo M, te Pas MF, Priem J, de Wit AA, Pool MH, Davoli R, Russo V. Comparison of prenatal muscle tissue expression profiles of two pig breeds differing in muscle characteristics. J Anim Sci. 2006;84:1–10. doi: 10.2527/2006.8411. [DOI] [PubMed] [Google Scholar]
- Sudre K, Leroux C, Pietu G, Cassar-Malek I, Petit E, Listrat A, Auffray C, Picard B, Martin P, Hocquette JF. Transcriptome analysis of two bovine muscles during ontogenesis. J Biochem (Tokyo) 2003;133:745–56. doi: 10.1093/jb/mvg096. [DOI] [PubMed] [Google Scholar]
- Cassar-Malek I, Passelaigue F, Bernard C, Leger J, Hocquette JF. Target genes of myostatin loss-of-function in muscles of late bovine fetuses. BMC Genomics. 2007;8:63. doi: 10.1186/1471-2164-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zembayashi M, Nishimura K, Lunt DK, Smith SB. Effect of breed type and sex on the fatty acid composition of subcutaneous and intramuscular lipids of finishing steers and heifers. J Anim Sci. 1995;73:3325–3332. doi: 10.2527/1995.73113325x. [DOI] [PubMed] [Google Scholar]
- Greenwood PL, Cafe LM, Hearnshaw H, Hennessy DW, Thompson JM, Morris SG. Long-term consequences of birth weight and growth to weaning for carcass, yield and beef quality characteristics of Piedmontese- and Wagyu-sired cattle. Australian Journal of Experimental Agriculture. 2006;46:257–269. doi: 10.1071/EA05240. [DOI] [Google Scholar]
- Gene Expression Omnibus (GEO) database http://www.ncbi.nlm.nih.gov/geo/
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawken RJ, Barris WC, McWilliam SM, Dalrymple BP. An Interactive Bovine in Silico SNP Database (IBISS) Mammalian Genome. 2004;15:819–27. doi: 10.1007/s00335-004-2382-4. [DOI] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard V, Hovig E. Options available for profiling small samples: a review of sample amplification technology when combined with microarray profiling. Nucleic Acids Res. 2006;34:996–1014. doi: 10.1093/nar/gkj499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Futami E, Taneichi A, Mori T, Hattori A. Decorin Expression During Development of Bovine Skeletal Muscle and Its Role in Morphogenesis of the Intramuscular Connective Tissue. Cells Tissues Organs. 2002;171:199–214. doi: 10.1159/000063713. [DOI] [PubMed] [Google Scholar]
- Lehnert SA, Wang YH, Byrne KA. Development and application of a bovine cDNA microarray for expression profiling of muscle and adipose tissue. Australian Journal of Experimental Agriculture. 2004;44:1127–1133. doi: 10.1071/EA03238. [DOI] [Google Scholar]
- Amthor H, Connolly D, Patel K, Brand-Saberi B, Wilkinson DG, Cooke JCB. The Expression and Regulation of Follistatin and a Follistatin-like Gene during Avian Somite Compartmentalization and Myogenesis. Developmental Biology. 1996;178:343–362. doi: 10.1006/dbio.1996.0223. [DOI] [PubMed] [Google Scholar]
- James PL, Stewart CE, Rotwein P. Insulin-like growth factor binding protein-5 modulates muscle differentiation through an insulin-like growth factor-dependent mechanism. J Cell Biol. 1996;133:683–93. doi: 10.1083/jcb.133.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom DA, Penn BH, Strand A, Perry RL, Rudnicki MA, Tapscott SJ. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell. 2002;9:587–600. doi: 10.1016/S1097-2765(02)00481-1. [DOI] [PubMed] [Google Scholar]
- Cobb LJ, Salih DA, Gonzalez I, Tripathi G, Carter EJ, Lovett F, Holding C, Pell JM. Partitioning of IGFBP-5 actions in myogenesis: IGF-independent anti-apoptotic function. J Cell Sci. 2004;117:1737–46. doi: 10.1242/jcs.01028. [DOI] [PubMed] [Google Scholar]
- Stickland NC. A quantitative study of muscle development in the bovine foetus (Bos indicus) Anat Histol Embryol. 1978;7:193–205. doi: 10.1111/j.1439-0264.1978.tb00795.x. [DOI] [PubMed] [Google Scholar]
- Ansay M. Developmental biochemistry of bovine foetal muscle. I. DNA, RNA, dry matter and total nitrogen. Zentralbl Veterinarmed A. 1974;21:603–10. doi: 10.1111/j.1439-0442.1974.tb01155.x. [DOI] [PubMed] [Google Scholar]
- Forbes D, Jackman M, Bishop A, Thomas M, Kambadur R, Sharma M. Myostatin auto-regulates its expression by feedback loop through Smad7 dependent mechanism. J Cell Physiol. 2006;206:264–72. doi: 10.1002/jcp.20477. [DOI] [PubMed] [Google Scholar]
- Tan NS, Shaw NS, Vinckenbosch N, Liu P, Yasmin R, Desvergne B, Wahli W, Noy N. Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription. Mol Cell Biol. 2002;22:5114–27. doi: 10.1128/MCB.22.14.5114-5127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michal JJ, Zhang ZW, Gaskins CT, Jiang Z. The Bovine Fatty Acid Binding Protein 4 Gene Is Significantly Associated With Marbling and Subcutaneous Fat Depth in Wagyu × Limousin F2 Crosses. Animal Genetics. 2006;37:400–402. doi: 10.1111/j.1365-2052.2006.01464.x. [DOI] [PubMed] [Google Scholar]
- Steelman CA, Recknor JC, Nettleton D, Reecy JM. Transcriptional profiling of myostatin-knockout mice implicates Wnt signaling in postnatal skeletal muscle growth and hypertrophy. FASEB J. 2006 doi: 10.1096/fj.05-5125fje. [DOI] [PubMed] [Google Scholar]
- Potts JK, Echternkamp SE, Smith TPL, Reecy JM. Characterization of Gene Expression in Double-Muscled and Normal-Muscled Bovine Embryos. Animal Genetics. 2003;34:438–444. doi: 10.1046/j.0268-9146.2003.01055.x. [DOI] [PubMed] [Google Scholar]
- Deveaux V, Cassar-Malek I, Picard B. Comparison of contractile characteristics of muscle from Holstein and double-muscled Belgian Blue foetuses. Comp Biochem Physiol A Mol Integr Physiol. 2001;131:21–9. doi: 10.1016/S1095-6433(01)00459-7. [DOI] [PubMed] [Google Scholar]
- Schrauwen P, Hesselink M. UCP2 and UCP3 in muscle controlling body metabolism. J Exp Biol. 2002;205:2275–85. doi: 10.1242/jeb.205.15.2275. [DOI] [PubMed] [Google Scholar]
- Xiang CC, Kozhich OA, Chen M, Inman JM, Phan QN, Chen Y, Brownstein MJ. Amine-modified random primers to label probes for DNA microarrays. Nat Biotechnol. 2002;20:738–42. doi: 10.1038/nb0702-738. [DOI] [PubMed] [Google Scholar]
- Tan SH, Reverter A, Wang YH, Byrne K, McWilliam S, Lehnert S. Gene expression profiling of bovine in vitro adipogenesis using a cDNA microarray. Functional and Integrative Genomics. 2006;6:235–249. doi: 10.1007/s10142-005-0016-x. [DOI] [PubMed] [Google Scholar]
- Reverter A, Wang YH, Byrne KA, Tan SH, Harper GS, Lehnert SA. Joint Analysis of Multiple cDNA Microarray Studies Via Multivariate Mixed Models Applied to Genetic Improvement of Beef Cattle. Journal of Animal Science. 2004;82:3430–3439. doi: 10.2527/2004.82123430x. [DOI] [PubMed] [Google Scholar]
- Reverter A, Ingham A, Lehnert SA, Tan SH, Wang Y, Ratnakumar A, Dalrymple BP. Simultaneous identification of differential gene expression and connectivity in inflammation, adipogenesis and cancer. Bioinformatics. 2006;22:2396–404. doi: 10.1093/bioinformatics/btl392. [DOI] [PubMed] [Google Scholar]
- McLachlan GJ, Bean RW, Jones LB. A simple implementation of a normal mixture approach to differential gene expression in multiclass microarrays. Bioinformatics. 2006;22:1608–15. doi: 10.1093/bioinformatics/btl148. [DOI] [PubMed] [Google Scholar]
- Nattrass GS, Quigley SP, Gardner GE, Bawden CS, McLaughlan CJ, Hegarty RS, Greenwood PL. Genotypic and nutritional regulation of gene expression in two sheep hindlimb muscles with distinct myofibre and metabolic characteristics. Australian Journal of Agricultural Research. 2006;57:691–698. doi: 10.1071/AR05101. [DOI] [Google Scholar]
- Expression Analysis Systematic Explorer (EASE) http://david.abcc.ncifcrf.gov/ease/ease.jsp
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistical thresholds for t-statistics and fold changes at two levels of significance and for down- and up-regulated genes. Statistical cut-off values for inclusion in differentially expressed gene lists
Genes showing differential gene expression in LM of bovine fetuses from different sire breeds in at least one time point. Full listing of all genes that showed differential expression between the two sire breeds.
Goodness of fit of the ANOVA model and significance of design effects on the expression of target and housekeeping genes as measured by the Ct of the qRT-PCR. Background detail to the analysis of qRT-PCR data.
Bovine fetuses sampled for microarray study. Listing of genetic background, weight, sex and accurate ages of the fetuses used in this study.