Abstract
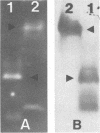
Polyclonal antibodies were used to identify heme or copper nitrite reductases in the following groups: 23 taxonomically diverse denitrifiers from culture collections, 100 numerically dominant denitrifiers from geographically diverse environments, and 51 denitrifiers from a culture collection not selected for denitrification. Antisera were raised against heme nitrite reductases from Pseudomonas aeruginosa and Pseudomonas stutzeri and against copper nitrite reductase from Achromobacter cycloclastes. Nitrite reductases were identified by Western immunoblot. Diethyldithiocarbamate, which specifically inhibits copper nitrite reductases, was used to confirm the immunological characterization and determine which type was present in strains nonreactive with any antiserum. For groups in which the type of nitrite reductase has not been previously described, we found that Alcaligenes eutrophus, Bacillus azotoformans, Bradyrhizobium japonicum, Corynebacterium nephridii, and Rhizobium spp. contained copper nitrite reductase, while Aquaspirillum itersonii, Flavobacterium spp., and Pseudomonas fluorescens contained heme nitrite reductase. Heme nitrite reductases dominated, regardless of soil type or geographic origin. They occurred in 64 and 92%, respectively, of denitrifiers in the numerically dominant and nonselected collections. The two nitrite reductase types were mutually exclusive in individual bacteria, but both appeared in different strains from the Alcaligenes and Pseudomonas genera. The heme type predominated in Pseudomonas strains. The heme-type nitrite reductase appeared more conserved if judged by similarities in molecular weights and immunological reactions. The Cu type was found in more taxonomically unrelated strains and varied in molecular weight and antiserum recognition.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abd-el-Malek Y., Hosny I., Emam N. F. Evaluation of media, used for enumeration of denitrifying bacteria. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg. 1974;129(5):415–421. doi: 10.1016/s0044-4057(74)80003-1. [DOI] [PubMed] [Google Scholar]
- Betlach M. R. Evolution of bacterial denitrification and denitrifier diversity. Antonie Van Leeuwenhoek. 1982;48(6):585–607. doi: 10.1007/BF00399543. [DOI] [PubMed] [Google Scholar]
- Gamble T. N., Betlach M. R., Tiedje J. M. Numerically dominant denitrifying bacteria from world soils. Appl Environ Microbiol. 1977 Apr;33(4):926–939. doi: 10.1128/aem.33.4.926-939.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M. A., Hochstein L. I. A dissimilatory nitrite reductase in Paracoccus halodenitrificans. Arch Microbiol. 1984 Jan;137(1):79–84. doi: 10.1007/BF00425812. [DOI] [PubMed] [Google Scholar]
- Hochstein L. I., Tomlinson G. A. The enzymes associated with denitrification. Annu Rev Microbiol. 1988;42:231–261. doi: 10.1146/annurev.mi.42.100188.001311. [DOI] [PubMed] [Google Scholar]
- Hulse C. L., Tiedje J. M., Averill B. A. A spectrophotometric assay for dissimilatory nitrite reductases. Anal Biochem. 1988 Aug 1;172(2):420–426. doi: 10.1016/0003-2697(88)90464-2. [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Matsubara T. A nitrite reductase from Achromobacter cycloclastes. J Biochem. 1972 Apr;71(4):645–652. [PubMed] [Google Scholar]
- Kakutani T., Watanabe H., Arima K., Beppu T. Purification and properties of a copper-containing nitrite reductase from a denitrifying bacterium, Alcaligenes faecalis strain S-6. J Biochem. 1981 Feb;89(2):453–461. doi: 10.1093/oxfordjournals.jbchem.a133220. [DOI] [PubMed] [Google Scholar]
- Körner H., Frunzke K., Döhler K., Zumft W. G. Immunochemical patterns of distribution of nitrous oxide reductase and nitrite reductase (cytochrome cd1) among denitrifying pseudomonads. Arch Microbiol. 1987 Jun;148(1):20–24. doi: 10.1007/BF00429641. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- LeGall J., Payne W. J., Morgan T. V., DerVartanian D. On the purification of nitrite reductase from Thiobacillus denitrificans and its reaction with nitrite under reducing conditions. Biochem Biophys Res Commun. 1979 Mar 30;87(2):355–362. doi: 10.1016/0006-291x(79)91804-7. [DOI] [PubMed] [Google Scholar]
- Liu M. Y., Liu M. C., Payne W. J., Legall J. Properties and electron transfer specificity of copper proteins from the denitrifier "Achromobacter cycloclastes". J Bacteriol. 1986 May;166(2):604–608. doi: 10.1128/jb.166.2.604-608.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuko M., Iwasaki H., Sakurai T., Suzuki S., Nakahara A. Characterization of nitrite reductase from a denitrifier, Alcaligenes sp. NCIB 11015. A novel copper protein. J Biochem. 1984 Aug;96(2):447–454. doi: 10.1093/oxfordjournals.jbchem.a134856. [DOI] [PubMed] [Google Scholar]
- Matsubara T., Iwasaki H. Enzymatic steps of dissimilatory nitrite reduction in Alcaligenes faecalis. J Biochem. 1971 May;69(5):859–868. doi: 10.1093/oxfordjournals.jbchem.a129537. [DOI] [PubMed] [Google Scholar]
- Newton N. The two-haem nitrite reductase of Micrococcus denitrificans. Biochim Biophys Acta. 1969;185(2):316–331. doi: 10.1016/0005-2744(69)90425-2. [DOI] [PubMed] [Google Scholar]
- Ohkubo S., Iwasaki H., Hori H., Osawa S. Evolutionary relationship of denitrifying bacteria as deduced from 5S rRNA sequences. J Biochem. 1986 Nov;100(5):1261–1267. doi: 10.1093/oxfordjournals.jbchem.a121832. [DOI] [PubMed] [Google Scholar]
- Ritchie G. A., Nicholas D. J. The partial characterization of purified nitrite reductase and hydroxylamine oxidase from Nitrosomonas europaea. Biochem J. 1974 Mar;138(3):471–480. doi: 10.1042/bj1380471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römermann D., Friedrich B. Denitrification by Alcaligenes eutrophus is plasmid dependent. J Bacteriol. 1985 May;162(2):852–854. doi: 10.1128/jb.162.2.852-854.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton D. C., Weintraub S. T. Identification of nitric oxide and nitrous oxide as products of nitrite reduction by Pseudomonas cytochrome oxidase (nitrate reductase). Biochem Biophys Res Commun. 1980 Nov 17;97(1):236–242. doi: 10.1016/s0006-291x(80)80159-8. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAMANAKA T., OKUNUKI K. Crystalline Pseudomonas cytochrome oxidase. I. Enzymic properties with special reference to the biological specificity. Biochim Biophys Acta. 1963 Mar 12;67:379–393. doi: 10.1016/0006-3002(63)91844-4. [DOI] [PubMed] [Google Scholar]
- Zablotowicz R. M., Focht D. D. Physiological Characteristics of Cowpea Rhizobia: Evaluation of Symbiotic Efficiency in Vigna unguiculata. Appl Environ Microbiol. 1981 Mar;41(3):679–685. doi: 10.1128/aem.41.3.679-685.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft W. G., Gotzmann D. J., Kroneck P. M. Type 1, blue copper proteins constitute a respiratory nitrite-reducing system in Pseudomonas aureofaciens. Eur J Biochem. 1987 Oct 15;168(2):301–307. doi: 10.1111/j.1432-1033.1987.tb13421.x. [DOI] [PubMed] [Google Scholar]