Summary
The vertebrate brain is anatomically and functionally asymmetric, however, the molecular mechanisms that establish left-right brain patterning are largely unknown. In zebrafish, asymmetric left-sided Nodal signaling within the developing dorsal diencephalon is required for determining the direction of epithalamic asymmetries. How diencephalic Nodal activity is regulated is not well understood. Here we show that the transcription factor Six3, which is essential for forebrain formation and is associated with holoprosencephaly in humans, regulates diencephalic Nodal expression during initial establishment of brain asymmetry. Reduction of Six3 function causes brain-specific de-regulation of Nodal pathway activity, resulting in epithalamic laterality defects. Based on misexpression and genetic epistasis experiments, we propose that Six3 acts in the neuroectoderm, where it establishes an early prepattern of bilateral repression of Nodal activity. Subsequently, Nodal signaling from the left lateral plate mesoderm alleviates this repression ipsilaterally. Our data reveal a Six3-dependent mechanism for establishment of correct brain laterality, which does not require asymmetric expression of six3, and provide an entry point to understanding the molecular basis of the regulation of Nodal signaling in the brain.
Introduction
Brain asymmetry is evident at anatomical and molecular levels, and has implications for behavior and cognitive functions. To date, our only insights into the molecular mechanisms underlying development of brain laterality come from studies of asymmetry in the epithalamus of zebrafish (reviewed in Halpern et al., 2003). The epithalamus contains the centrally positioned pineal organ (a component of the photoneuroendocrine system) and, in many vertebrates, an accessory organ (named parapineal in fish). Additionally, the epithalamus contains the bilateral habenular nuclei, which are part of a conserved conduction system connecting the forebrain and ventral midbrain. Epithalamic asymmetries are conserved throughout many vertebrate species (reviewed in Concha and Wilson, 2001; Harris et al., 1996). In larval zebrafish they include localization of the parapineal to the left side (Concha et al., 2000; Gamse et al., 2002) as well as differences in size, anatomy, gene expression and pattern of target innervation between the left and right habenular nuclei (Aizawa et al., 2005; Concha et al., 2000; Concha et al., 2003; Gamse et al., 2005; Gamse et al., 2003).
Transient left-sided activation of the Nodal signaling pathway in the developing dorsal diencephalon during mid-late segmentation stages is required to determine the direction of epithalamic lateralities (Concha et al., 2000; Concha et al., 2003; Liang et al., 2000). It has been proposed that diencephalic expression of Nodal pathway genes is first repressed altogether by early Nodal signaling, likely emanating from midline tissues during gastrulation (Concha et al., 2000; Liang et al., 2000). Subsequently, diencephalic Nodal pathway gene expression is activated on the left side during late segmentation presumably by Nodal signaling from the left lateral plate mesoderm (LPM) (Long et al., 2003). However, what factors function within the developing brain itself to regulate diencephalic Nodal pathway activity remains unknown.
Six3 is a homeodomain transcription factor essential for forebrain and eye development (Carl et al., 2002; Lagutin et al., 2003). Mutations in human SIX3 can cause holoprosencephaly, the most common congenital malformation of the forebrain (reviewed in Cohen, 2006). We have generated a zebrafish model of Six3 loss of function, which contrary to previously described Six3-deficient murine and Medaka embryos lacking forebrain tissue, enables studying the roles of Six3 in forebrain patterning. We find that Six3 is also essential for correct left-right epithalamic patterning in zebrafish. The combined inactivation of Six3b and Six7, two zebrafish homologs of Six3 (Kobayashi et al., 1998; Seo et al., 1998a; Seo et al., 1998b), causes brain-specific spatial and temporal de-regulation of Nodal pathway activity, as evidenced by excessive, symmetric and precocious Nodal pathway gene expression during late segmentation, and subsequently results in randomization of epithalamic asymmetries. Rescue experiments, in which Six3 function is restored in a tissue- or time-specific manner, and epistasis experiments, support a model whereby Six3 functions in the neuroectoderm no later than early segmentation to bilaterally repress diencephalic Nodal activity. Subsequently, at late segmentation, Nodal signaling from the left LPM is required to alleviate this repression ipsilaterally. Our results uncover a Six3-dependent regulatory mechanism of Nodal pathway activity in the developing brain that is essential for establishment of correct brain laterality, without requiring asymmetric expression of six3. Identification of Six3 targets in this process should help delineate the molecular genetic hierarchy that governs left-right brain patterning.
Results and Discussion
six3b/six7-Deficient Embryos
Vertebrate Six3 is expressed in the anterior neuroectoderm during early embryogenesis and afterward in specific forebrain territories. Its loss of function in mice or Medaka fish leads to severe forebrain deficiencies by early segmentation (Carl et al., 2002; Lagutin et al., 2003), thereby leaving open the functional significance of Six3 in later forebrain development. In zebrafish, three six3-related genes, six3a, six3b and six7, are similarly expressed in the prechordal plate throughout gastrulation and in the anterior neuroectoderm during late gastrulation and early segmentation (Kobayashi et al., 1998; Seo et al., 1998a; Seo et al., 1998b). We reasoned that loss of function of one or two of these genes may lead to specific forebrain malformations without causing severe forebrain deficiencies. Using TILLING (Targeting Induced Local Lesions IN Genomes) (Draper et al., 2004; Wienholds and Plasterk, 2004) we identified a nonsense mutation in six3b (E109->stop), which is expected to result in a truncated protein lacking part of the Six domain and the entire homeodomain (Figure 1A). Whereas misexpression assays suggest that six3bvu87 is a null allele (Figure S1), six3bvu87/vu87 embryos appear normal and develop to adulthood, most likely due to redundancy between six3-related genes. We therefore used antisense morpholino oligonucleotide translation interference to inhibit specifically Six7 function (MOsix7) (see Experimental Procedures). Injection of MOsix7 into wild-type or six3bvu87/+ embryos did not produce any specific defects. However, six3bvu87/vu87 embryos injected with MOsix7 exhibited strongly reduced or no eyes (Figure 1C), a phenotype that could be rescued by co-injection of synthetic RNA encoding Six3b or Six7 (insensitive to MOsix7) (Figure S1; not shown). Morphological and molecular marker analyses revealed that gross regionalization of the brain in these embryos was relatively normal, and the forebrain was not reduced significantly (Figure 1E).
Figure 1. Combined Loss of Six3b/Six7 Function Results in Lack of Eye Tissue and Abnormal Brain Laterality.
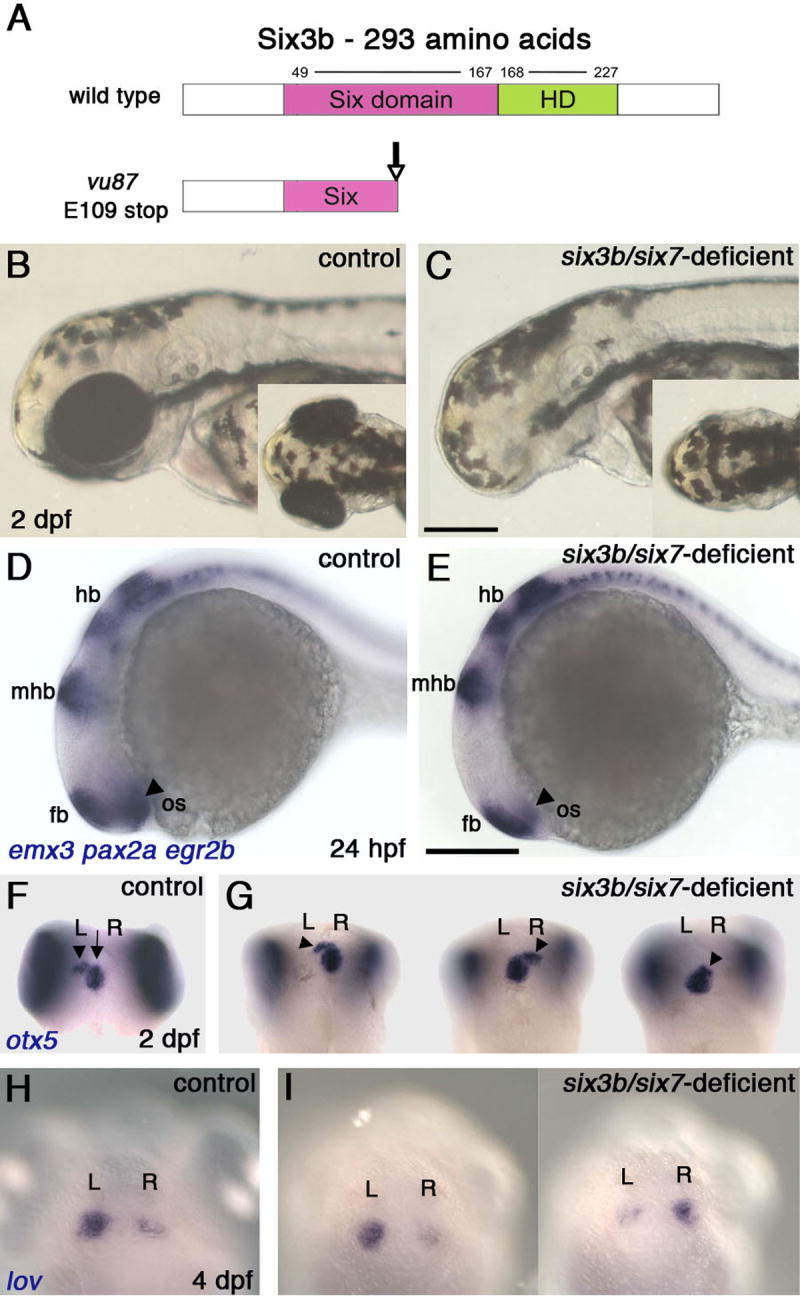
(A) Schematic presentation of normal Six3b and the predicted truncated Six3b protein, encoded by the six3bvu87 allele. (B, C) Live control embryo (B) and six3b/six7-deficient embryo (B), demonstrating lack of eye tissue. (D, E) Comparable brain patterning in 24 hpf control (D) and six3b/six7-deficient (E) embryos. Forebrain (fb), midbrain-hindbrain boundary (mhb) and hindbrain (hb) are labeled with emx3, pax2a, and egr2b, respectively. The optic stalk (os, arrowheads, pax2a) is reduced in the six3b/six7-deficient embryo. (F, G) Pineal (arrow) and parapineal localization (arrowheads) in control (F), and six3b/six7-deficient (G) embryos. (H, I) Habenular nuclei labeled with lov, in control (H) and six3b/six7-deficient (I) embryos. Control embryos are six3bvu87/+ or six3bvu87/vu87 and present normal phenotypes. R, right; L, left. Anterior is to the left in B-E and up in F-I. B-E, Lateral views and F-I and insets in B and C are dorsal views. Scale bars are 200 μm.
Brain-Specific Randomization of Asymmetry in six3b/six7-Deficient Embryos
Detailed analysis of the forebrain of six3b/six7-deficient embryos revealed laterality defects. We used molecular markers to examine the developing dorsal diencephalon (epithalamus). At 1 day post-fertilization (dpf), expression of floating head (flh) (Talbot et al., 1995), marking the pineal, was comparable between control and six3b/six7-deficient embryos (Figure 2G-J). At 2 dpf, otx5 staining, which labels both the pineal and parapineal (Gamse et al., 2002), revealed a normal-looking pineal organ, whereas the parapineal was often found more anteriorly than normal, and was sometimes positioned to the right of the pineal (Figure 1G). By 4 dpf, the parapineal was localized more posteriorly, on the left or the right in similar numbers of embryos (not shown, nleft=29 (54%), nright=25 (46%), Figure S2A). Similar defects were observed with gfi1, a parapineal-specific marker (Dufourcq et al., 2004) (not shown, nleft=22 (60%), nright=15 (40%), Figure S2B). Parapineal laterality is correlated with and affects the asymmetry of the habenular nuclei (Concha et al., 2000; Concha et al., 2003; Gamse et al., 2003). Indeed, consistent with randomization of parapineal localization, habenular nuclei asymmetry was also randomized in six3b/six7-deficient embryos, as demonstrated by expression of leftover (lov), an asymmetric marker of the habenula (Gamse et al., 2003) (Figure 1I, nleft=13 (50%), nright=13 (50%), Figure S2C). The randomization of parapineal sidedness and habenular asymmetry is indicative of a defect in establishment of correct brain laterality. By contrast, heart laterality was normal in six3b/six7-deficient embryos (not shown, nleft=21, nright=0, Figure S2D), suggesting that laterality defects in these embryos were brain-specific (also see below).
Figure 2. Brain-Specific Loss of Asymmetry and Excessive Diencephalic Nodal Activation in six3b/six7-Deficient Embryos.
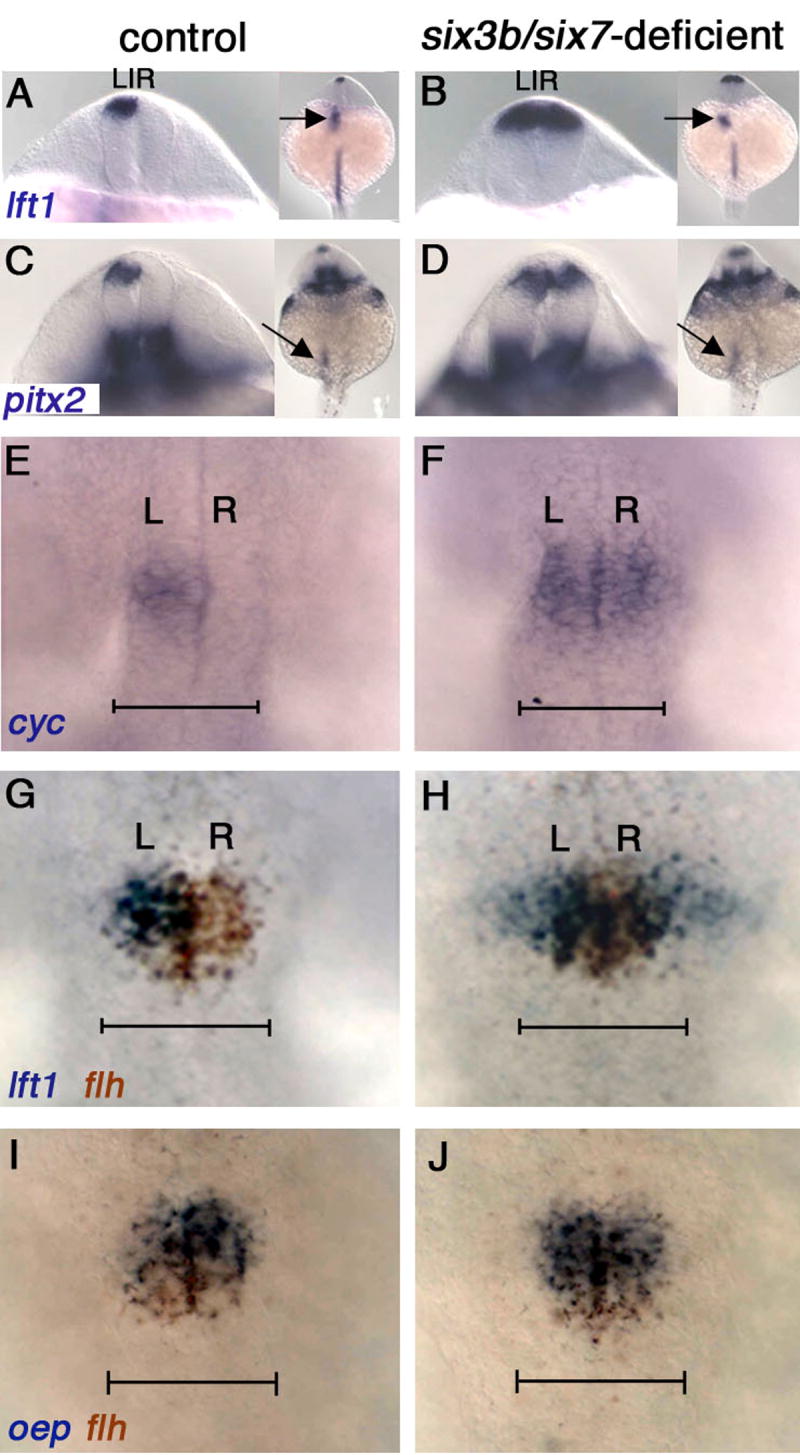
(A-F) Left-sided expression in control (A, C, E) and bilateral expression in six3b/six7-deficient embryos (B, D, F), of lft1 (A, B), pitx2 (C, D) and cyc (E, F). In the LPM, expression remains left-sided (arrows in insets, A-D). (E-H) The diencephalic domain of Nodal expression and activity is expanded in six3b/six7-deficient embryos (compare F to E and H to G). lft1 expression extends outside the flh-positive pineal anlage (H). (I, J) oep expression continues to coincide with the pineal anlage in six3b/six7-deficient embryos (J). Control embryos in all panels are six3bvu87/+ or six3bvu87/vu87 and present normal phenotypes. R, right; L, left. (A-D) Dorsoposterior views; insets show smaller magnification of the same embryo in each panel. (E-J) Dorsal views, anterior is up. All embryos are 21-24s stage. Scale bars are 100 μm.
Loss of Early Brain Asymmetry in six3b/six7-Deficient Embryos
Next we wished to determine when during embryogenesis the brain asymmetry defects arose in six3b/six7-deficient embryos. Brain asymmetry can be first detected at 18-somite stage (18s, 18 hpf - hours post fertilization), by left-sided expression of cyclops (cyc), a zebrafish Nodal homolog (Rebagliati et al., 1998a; Sampath et al., 1998), followed by left-sided expression of lefty1 (lft1), encoding a Nodal pathway feedback inhibitor (Bisgrove et al., 1999; Thisse and Thisse, 1999) and pitx2, encoding a Nodal effector (Campione et al., 1999; Essner et al., 2000), largely within the presumptive pineal (Bisgrove et al., 2000; Concha et al., 2000; Concha et al., 2003; Liang et al., 2000) (Figure 2G). This asymmetric expression is essential for establishment of correct laterality in the epithalamus: both lack of expression of these genes or their bilateral expression lead to randomization of parapineal and habenular laterality (Concha et al., 2000; Gamse et al., 2003). We found that in six3b/six7-deficient embryos, cyc, lft1 and pitx2 are expressed bilaterally in the dorsal diencephalon (Figure 2B,D,F). In accordance with laterality defects being confined to the brain, expression of lft1, pitx2 and the Nodal homolog southpaw (spaw) (Long et al., 2003) remained asymmetric in the left lateral plate mesoderm (LPM), as in wild type (Figure 2B,D; not shown).
In addition to being bilateral, Nodal pathway gene expression is broader and precocious in six3b/six7-deficient embryos. Nodal pathway activation, as marked by expression of lft1, was also detected well outside the presumptive pineal, marked by flh expression (Figure 2H), suggesting that Six3 is required to limit the domain of Nodal pathway activity within the dorsal diencephalon. Interestingly, the expression domain of the one-eyed-pinhead (oep) gene, encoding an obligatory Nodal co-receptor (Gritsman et al., 1999), was only mildly enlarged and continued to coincide with flh (Figure 2J). In addition, loss of Six3b/Six7 function caused precocious diencephalic Nodal activity. Similar to wild-type embryos, at 18-20s Nodal pathway activation was observed in only a small fraction of six3bvu87/+ or six3bvu87/vu87 embryos; but in six3b/six7-deficient sibling embryos, it was significantly more frequent (Figure S3).
Altogether, these data demonstrate that the concerted action of Six3b and Six7 is necessary for spatial and temporal regulation of Nodal activity in the developing diencephalon, so that early brain asymmetry and afterward, correct brain laterality, are achieved. The penetrance of the excessive, symmetric Nodal expression phenotype observed in six3b/six7-deficient embryos varied depending on the parents. Incomplete penetrance is also frequently seen in other laterality mutants (Bisgrove et al., 2000), suggesting the phenotype is influenced by the presence of modifier/s, which are yet to be identified.
Six3 Function is Required in the Neuroectoderm to Represses Diencephalic Nodal Activity
Diencephalic Nodal pathway gene expression is thought to be controlled by Nodal pathway activity in two phases. Loss of Nodal signaling during both gastrulation and segmentation results in bilateral diencephalic Nodal expression (Concha et al., 2000; Liang et al., 2000). However, loss of Nodal signaling only during segmentation results in absence of diencephalic Nodal expression (Concha et al., 2000; Liang et al., 2000; Long et al., 2003). Thus, the current model proposes that early Nodal pathway activity is required to repress Nodal pathway gene expression in the brain bilaterally, whereas later Nodal signaling, likely initiated by expression of spaw in the left LPM, is required to activate it ipsilaterally (Concha et al., 2000; Liang et al., 2000; Long et al., 2003).
To determine whether and how Six3b/Six7 affect the hierarchy of Nodal signaling in left-right axis formation, we addressed the spatiotemporal requirements for their function in establishment of early brain asymmetry. six3b and six7 are expressed in the prechordal plate throughout gastrulation and early segmentation, and in the anterior neuroectoderm from late gastrulation. By midsegmentation, six7 expression ceases, whereas six3b transcripts become localized mostly to the presumptive telencephalon, eyes and rostral diencephalon (Kobayashi et al., 1998; Seo et al., 1998a; Seo et al., 1998b). During early segmentation, six3b and six7 expression partially overlaps with the flh-positive domain, shown to contribute to the epithalamus and dorsal telencephalon (Masai et al., 1997; Staudt and Houart, 2007) (Figure 3A-C; not shown). Importantly, there is no apparent asymmetry in six3b or six7 gene expression. Since deficiencies in axial mesodermal tissues result in bilateral diencephalic Nodal expression (Bisgrove et al., 2000; Concha et al., 2000; Liang et al., 2000; Rebagliati et al., 1998a; Rebagliati et al., 1998b; Sampath et al., 1998), loss of Six3b/Six7 function in the prechordal plate could underlie the loss of early brain asymmetry. Alternatively, asymmetry defects could result from Six3b/Six7 loss of function in the brain/neuroectoderm itself.
Figure 3. Six3 Function is Required in the Neuroectoderm to Repress Diencephalic Nodal Activity.
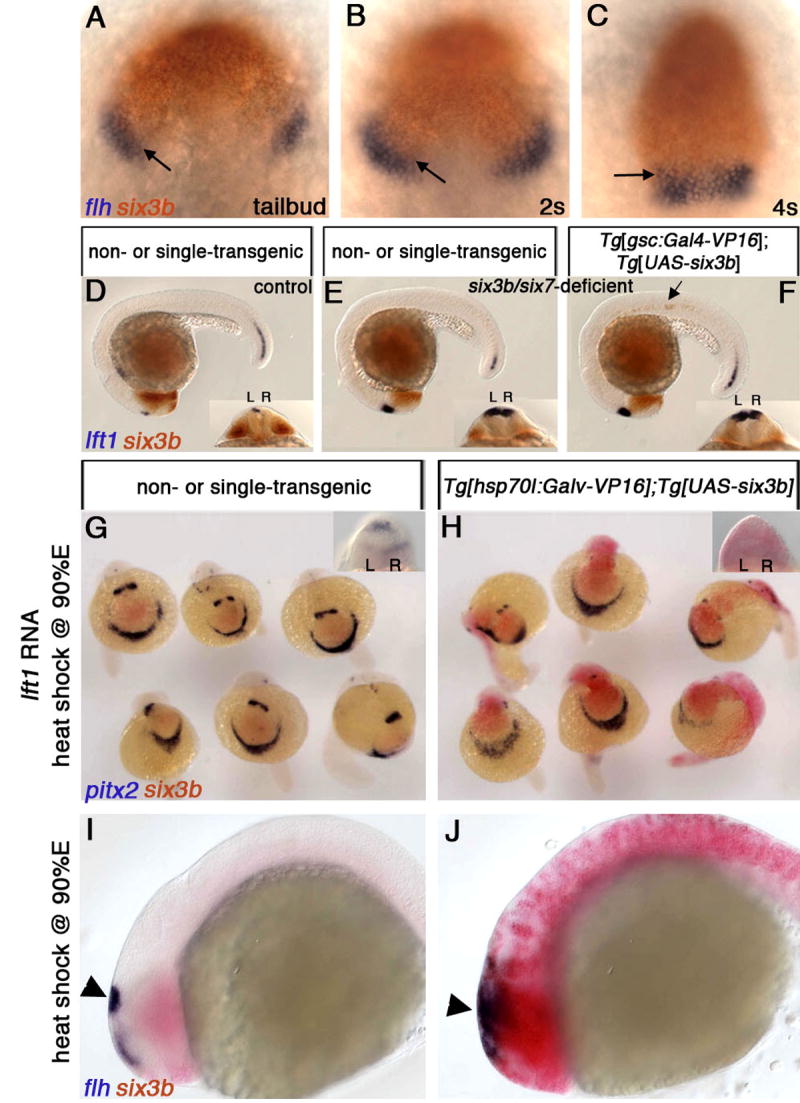
(A-C) Wild-type embryos at tailbud (10 hpf) (A), 2s (B) and 4s (C) stages, labeled for six3b and flh expression, which marks presumptive epithalamic and dorsal telencephalic tissues. Overlap between six3b and the flh-positive domain is indicated by arrows. (D-F) Embryos from a cross between six3bvu87/+; Tg[gsc:Gal4-VP16]vu160 heterozygous and six3bvu87/+; Tg[UAS-six3b]vu156 heterozygous fish. Control embryo (D) not injected with MOsix7. (E, F) six3b/six7-deficient embryos. Only the embryo in (F) carries both transgenes as evidenced by ectopic six3b in the notochord (arrow). (G, H) Embryos from a cross between Tg[hsp70l:Gal4-VP16]vu22 and Tg[UAS-six3b]vu156 heterozygous fish that were injected with lft1 RNA and heat-shocked at late gastrulation (H). (I, J) Ubiquitous six3b does not eliminate presumptive pineal tissue (flh-positive domain, arrowheads in I and J). R, right; L, left. A-C, dorsal views, anterior up. E-F,I,J, Lateral views, anterior to the left. Insets in D-F are dorsoposterior views.
Arguing against the first possibility, several lines of evidence suggest that Six3 activity in the prechordal plate does not influence brain asymmetry. We first examined the prechordal plate in six3b/six7-deficient embryos using several molecular markers, and found no abnormalities in expression of gsc, cyc, lft1 and pitx2 (Figure S4), suggesting that structural integrity and Nodal signaling in the prechordal plate are not impaired. Next, we expressed six3b from early gastrulation specifically in the anterior axial mesoderm of six3b/six7-deficient embryos, harboring Tg[gsc:Gal4-VP16]vu160 and Tg[UAS-six3b]vu156 transgenes. The gsc:Gal4-VP16 transgene drives expression of UAS-regulated genes in the anterior midline mesoderm, including the prechordal plate (Inbal et al., 2006). However, restoring Six3b function in the prechordal plate failed to suppress the excessive diencephalic Nodal pathway gene expression (Figure 3F).
By contrast, when ubiquitous six3b expression was induced by heat shock in six3b/six7-deficient embryos harboring Tg[hsp70l:Gal4-VP16]vu22 and Tg[UAS-six3b]vu156 transgenes, diencephalic Nodal expression was repressed bilaterally (Figure 4H). Therefore, Six3b/Six7 function is likely required in the neuroectoderm to regulate diencephalic Nodal expression. To test this hypothesis further, we asked if Six3b misexpression could repress diencephalic Nodal expression in the absence of axial mesodermal tissues. In embryos injected with synthetic lft1 RNA, early Nodal signaling is inhibited, resulting in deficiencies of axial mesodermal structures including prechordal plate (Bisgrove et al., 1999; Thisse and Thisse, 1999), and later, in bilateral diencephalic Nodal activity (Figure 3G). While ubiquitous six3b misexpression did not restore axial mesodermal tissues, it efficiently down-regulated diencephalic Nodal expression in these embryos (Figure 3H). This result supports the notion that misexpression of Six3b in the neuroectoderm is sufficient to suppress diencephalic Nodal pathway gene expression. The downregulation of diencephalic Nodal expression by ubiquitous Six3b expression was specific, and did not result from loss of presumptive pineal tissue (Figure 3J) or from defects in the left LPM spaw expression, which is essential for Nodal pathway gene expression in the diencephalon (Long et al., 2003) (not shown). Taken together, the data are consistent with a model whereby Six3b/Six7 function in the neuroectoderm to repress bilaterally diencephalic Nodal gene expression.
Figure 4. Six3 Misexpression Can Repress Diencephalic Nodal Signaling if Induced by the Beginning of Somitogenesis.
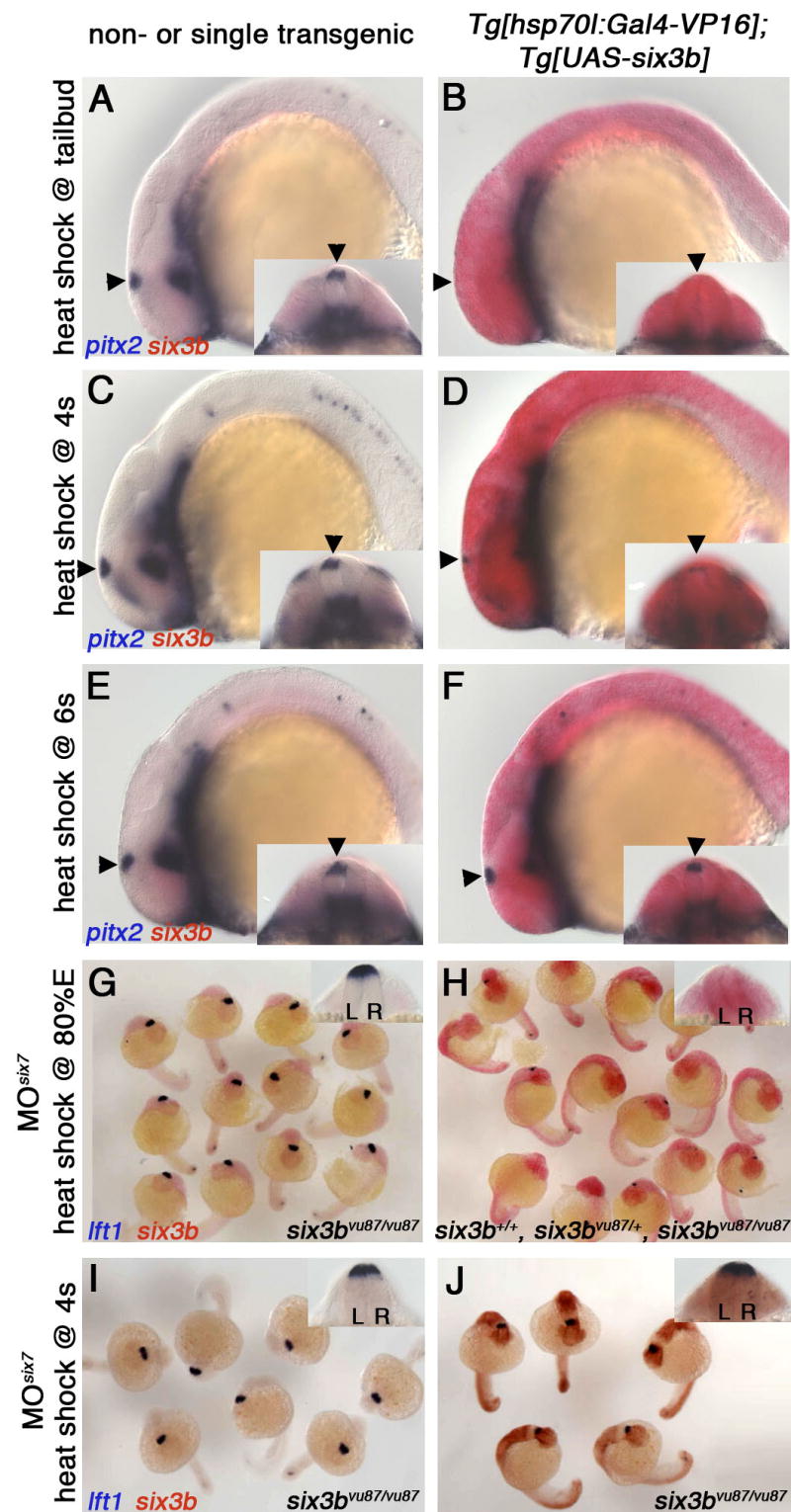
Tg[hsp70l:Gal4-VP16]vu22; Tg[UAS-six3b]vu156 (B, D, F) and single or non-transgenic siblings (A, C, E), heat-shocked at tailbud (10 hpf) (A, B), 4s stage (C, D), or 6s stage (E, F). Arrowheads point at the left-sided dorsal diencephalic domain of Nodal activity represented by pitx2 expression. (G-J) Embryos from a cross between six3bvu87/+; Tg[hsp70l:Gal4-VP16]vu22 heterozygous and six3bvu87/+; Tg[UAS-six3b]vu156 heterozygous fish, that were injected with MOsix7 and heat-shocked at 80% epiboly (80%E) (G, H) or 4s stage (I, J). Insets are dorsoposterior view of the same embryo (A-F) or a representative embryo (G-I). L, left; R, right. Embryos are 22-24s stage. Genotypes in (G,I,J) were inferred from clearly reduced eye size demonstrating embryos are six3b/six7-deficient. Because inducing six3b expression by heat shock at 80%E rescues the reduced eye size in six3b/six7-deficient embryos, embryos in H were PCR-genotyped and the presence of all genotypes at expected ratios was confirmed.
Repression of Nodal Activity by Six3 Occurs by Early Segmentation
Six3 could repress diencephalic Nodal expression directly, by acting in epithalamic cells during midsegmentation stages just prior to initiation of Nodal expression. Alternatively, Six3 functions at earlier stages, given that the proposed bilateral repression of diencephalic Nodal is induced already during gastrulation (Concha et al., 2000). To determine when Six3 can repress diencephalic Nodal expression, we induced ubiquitous six3b expression by heat shock in otherwise wild-type transgenic embryos at different developmental stages. Efficient repression was achieved when heat shock was applied at mid-late gastrulation (8-10 hpf), whereas heat-shocking at early segmentation stages (2s or 4s; 10.6 or 11.3 hpf respectively), caused only partial repression, and heat-shocking after 6s stage (12 hpf) no longer repressed diencephalic Nodal expression (Figure 4B,D,F; not shown). Similarly, ubiquitous six3b misexpression efficiently suppressed the excessive Nodal expression phenotype of six3b/six7-deficient embryos when applied by heat shock at late gastrulation (Figure 4G,H), but not when applied at 4s stage (Figure 4I,J). Given that the effector gene is expressed robustly about an hour after heat shock (not shown), together these results suggest that Six3 functions by 10s stage to efficiently repress diencephalic Nodal expression, which begins at 18s stage. Thus, Six3 likely functions as an early bilateral repressor of diencephalic Nodal expression, similar to early Nodal signaling.
We have shown that Six3 can repress diencephalic Nodal expression bilaterally, and can only do so by early segmentation. This temporary ability of Six3 to repress diencephalic Nodal expression suggests there is transient competence of prospective diencephalic tissue for this function, perhaps due to requirement for another factor, no longer available after early segmentation. Another question is how Nodal repression is maintained during later segmentation. six3b and six7 expression domains partially overlap with presumptive pineal domain during early segmentation (Figure 3A-C). Hence, in one scenario, Six3b/Six7 induce another factor in presumptive dorsal diencephalic cells, which, in turn, represses Nodal expression at later segmentation (Figure 5F). Interestingly, the levels of Six3 activity appear also to be important, since overexpression of Six3 during the limited competence period causes repression that cannot be overcome.
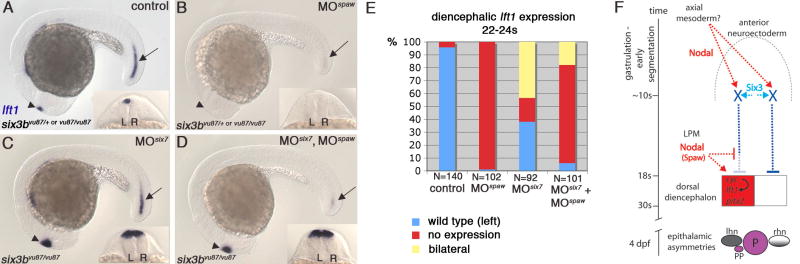
Figure 5. Loss of Six3b/Six7 is Epistatic to Loss of Spaw Function, and a Model for Six3 Function in Regulation of Diencephalic Nodal Activity.
(A) Control, uninjected embryo. (B) six3bvu87/+ or six3bvu87/vu87 injected with Spaw-MO1 (Long et al., 2003) (MOspaw). lft1 expression is abolished in the diencephalon (arrowhead) and very strongly reduced in the posterior notochord (arrow). (C) six3b/six7-deficient embryo. (D) six3b/six7-deficient embryo injected with Spaw-MO1. Excessive, bilateral diencephalic lft1 expression is not abolished, but lft1 posterior notochord expression is strongly reduced. (E) In clutches comprising approximately 1:1 ratio of six3bvu87/+ and six3bvu87/vu87 embryos, inhibiting Spaw function abolishes lft1 diencephalic expression in 99% of embryos, inhibiting Six7 function causes bilateral lft1 expression in more than 40% of embryos (all six3bvu87/vu87), and inhibiting both Spaw and Six7 results in restoring lft1 expression in 24% of embryos (all six3bvu87/vu87). (F) A model of Six3 function and interaction with Nodal signaling during establishment of early brain asymmetry. Six3 activity in the anterior neuroectoderm (light blue) and early Nodal signaling, presumably from the axial mesoderm (red), induce by early segmentation repressor/s of Nodal expression (X, dark blue) in the prospective dorsal diencephalon. Later, diencephalic left sided expression of Nodal is achieved by ipsilateral Spaw, repressing X and activating Nodal. This results in correct epithalamic asymmetries (left sided parapineal; higher lov levels (grey) in left habenular nucleus). P, pineal; pp, parapineal; lhn, left habenular nucleus; rhn, right habenular nucleus.
Loss of Six3 Function is Epistatic to Loss of Spaw Function
Based on our results, we propose that a Six3-dependent prepattern of bilateral Nodal repression in the prospective dorsal diencephalon is established by early segmentation. The question remains, however, whether later activation of diencephalic Nodal pathway gene expression by Spaw signal from the left LPM is achieved through alleviating the Six3-dependent repression, or by directly activating Nodal expression. To distinguish between these two possible models of Spaw action, we inhibited its function using antisense morpholino oligonucleotides in six3b/six7-deficient embryos. In this experiment, if Spaw functions by directly activating Nodal expression, then even in the absence of Six3b/Six7 function diencephalic Nodal pathway gene expression would be absent. Alternatively, if Spaw functions by alleviating the Six3-dependent repression, then even without Spaw function diencephalic Nodal expression should be activated. Consistent with a previous report (Long et al., 2003), inhibiting Spaw function abolished diencephalic Nodal activity, as inferred from lft1 expression, in 99% of progeny from a cross between six3bvu87/+ and six3bvu87/vu87 fish (n=102, Figure 5B,E). However, when function of both Spaw and Six7 was inhibited in sibling embryos, diencephalic lft1 expression was observed, mostly bilaterally, in 24% of the embryos (n=101, Figure 5D,E). Genotyping confirmed that all embryos showing lft1 diencephalic expression were six3bvu87/vu87. Thus, loss of Six3b/Six7 activity can alleviate the need for Spaw-mediated activation of diencephalic Nodal expression. This result supports the notion that Spaw signal from the left LPM promotes ipsilateral diencephalic Nodal expression largely by negatively regulating the bilateral repression set up by Six3. However, because the fraction of six3b/six7-deficient embryos exhibiting diencephalic Nodal pathway activation was reduced when Spaw function was inhibited (24% observed compared to almost 50% expected; Figure 5E), then loss of Six3b/Six7 function is not completely epistatic to loss of Spaw function. We interpret this incomplete epistasis to mean that Spaw has additional functions, possibly directly activating diencephalic Nodal expression (Figure 5F).
Conclusions
We have described a new, essential role for Six3 in left-right brain patterning, which provides a mechanism for regulation of diencephalic Nodal activity within the neuroectoderm. Despite being expressed symmetrically, Six3 is required for brain asymmetry by ensuring repression of diencephalic Nodal activity on both sides of the brain, thus allowing the setting of asymmetric activation later on. That Six3 functions much earlier than the appearance of brain asymmetry is surprising, yet consistent with a similar role proposed for early Nodal signaling (Concha et al., 2000). In agreement is also a recent finding that excessive Wnt activity in the anterior neuroectoderm during a limited time window at late gastrulation, leads to brain-specific bilateral Nodal expression, as seen in Six3 loss of function (M. Carl and S. Wilson, personal communication). Because high levels of canonical Wnt signaling in the neuroectoderm have been shown to repress six3 expression (Braun et al., 2003; Lagutin et al., 2003; Wilson and Houart, 2004), Six3 may be the main target through which Wnt signaling influences left-right brain asymmetry.
Interestingly, unlike mice or Medaka embryos lacking Six3 function, six3b/six7-deficient embryos have a relatively intact forebrain. A possible explanation for this difference is the activity of the six3a gene, which is highly similar to six3b and six7 in both sequence and expression pattern during early development. In the future, it will be interesting to test whether loss of six3a function, alone or in combination with other six3-related genes, affects brain asymmetry as well.
The finding that Six3, a transcription factor, is a neuroectodermal repressor of diencephalic Nodal activity should help to delineate the molecular genetic hierarchy that establishes left-right brain asymmetry by identification of Six3 target genes, and testing its functional interactions with other signaling pathways in this process. It is also intriguing that both left-right asymmetry defects and holoprosencephaly are associated with anomalies in Nodal, Hedgehog and Six3 function (Roessler and Muenke, 2001). Hence, future studies of functional interactions between these genes might be of particular importance to our understanding of normal forebrain development and its pathologies.
Experimental Procedures
Fish Lines and Genotyping
six3bvu87 mutation was identified using TILLING as previously described (Draper et al., 2004; Wienholds and Plasterk, 2004), and maintained in AB background. The mutation introduces a g325>t transversion resulting in gaa (Glutamic Acid, E109) to taa (stop) change. vu87 genotypes were identified using the following protocol: a 590 bp fragment from the six3b locus was amplified from genomic DNA by PCR using the forward primer: 5′ ACGACGTCGGGTTTTCCGTCTTT 3′ and the reverse primer: 5′ GTTCGTTCCTTGAAACAGTGC 3′. Because the mutation introduces an MseI restriction site, the PCR product of the six3bvu87 allele, but not the wild-type allele, is cut to 378 bp and 212 bp fragments.
Tg[UAS-six3b]vu156 fish: six3b coding sequence was cloned into pT2-UAS-pA-γCry-GM2 to generate pT2-UAS-six3b-pA-γCry-GM2 (further details of constructs will be provided upon request). Transgenic fish were generated using the Sleeping Beauty transposon system by injecting construct DNA (15-20 pg) and synthetic RNA encoding SB10 transposase (200pg) into 1-cell stage embryos, as previously described (Inbal et al., 2006). Founder fish were identified as previously described (Inbal et al., 2006). Other fish lines used in this work were Tg[gsc:Gal4-VP16]vu160 (Inbal et al., 2006), and Tg[hsp70l:Gal4-VP16]vu22 (provided by J.S. and B. Appel).
Heat Shock Treatment
Embryos were placed in pre-warmed 30% Danieau's solution in a water bath at 37°C for 30 minutes. Subsequently, the embryos were allowed to develop at 28.5°C or room temperature until reaching the desired developmental stage, when they were fixed in 4% paraformaldehyde.
Morpholino Oligonucleotide (MO) Information, RNA Synthesis and Injection
MOsix7 targets the six7 gene 5′ untranslated region (UTR), whereas MOsix7-ATG was designed to overlap the six7 gene translation start site. Injection of either MO into six3bvu87/vu87 embryos resulted in reduced eye phenotype, however, MOsix7-ATG was less effective and yielded non-specific defects more often. Therefore, we used MOsix7 in this work, at 1 ng per embryo. MO sequences: MOsix7: 5′ CCAACGGCATTCCAGTGTGAGTAAC 3′; MOsix7-ATG: 5′ TGAACATCGGCAGAGGAAACATGGC 3′. Spaw-MO1 has been described (Long et al., 2003) and was used at 10 ng per embryo.
MO efficacy and specificity: injection of 30 pg synthetic six7 RNA caused severe dorsalization and embryo death during segmentation. Coinjection of MOsix7 and 300 pg synthetic six7 RNA containing the MO binding site did not have any effect on embryos, whereas coinjection of MOsix7 and synthetic six7 RNA lacking the MOsix7 binding site resulted in the same effects as injection of six7 RNA alone.
Synthetic capped RNA was prepared using the mMESSAGE mMACHINE Kit (Ambion) from six3b (Kobayashi et al., 1998) and lft1 (antivin) (Thisse and Thisse, 1999) expression constructs. pCS2-six7: total mRNA was extracted from gastrulation stage embryos using Trizol, and six7 coding sequence with partial 5′ UTR was amplified by RT-PCR (SuperScript, Invitrogen). The amplified fragment was cloned into pCS2+ between BamHI and XbaI sites. To generate pCS2-six7-ATG, which lacks the MOsix7 binding site, only the coding sequence of six7 was amplified by PCR from pCS2-six7, and was cloned into pCS2+ between BamHI and XbaI sites. Sequence was confirmed for both constructs, and synthetic mRNA was prepared by NotI digestion and transcription with SP6 RNA polymerase. MOs and synthetic RNA were injected into 1-4 cell stage embryos.
In Situ Hybridization and Imaging
Single and double whole mount in situ hybridization using riboprobes was performed according to standard protocols. BMPurple (Roche) was used as blue substrate and either Fast Red (Roche) or INT (Iodonitrotetrazolium chloride, Sigma) and BCIP (Roche) mixture were used as red substrates. Images were acquired using Zeiss Axiophot compound or Zeiss dissecting microscope and Axiocam digital camera.
Statistical Analysis
In all cases we used the non-parametric Chi Square analysis to test for goodness of fit. P values were determined according to the Chi Square value and degrees of freedom.
Supplementary Material
Acknowledgments
We thank J. Gamse and S. Wilson for suggesting experiments; M. Carl and S. Wilson for discussions and sharing unpublished data and S. Wilson, C. Wright, J. Gamse, B. Appel, G. Levkowitz, X. Geng and LSK group members for critically reading the manuscript. We thank A. Bradshaw for technical assistance and our entire fish facility staff for excellent fish care. This work is supported by NIH and the Zebrafish Initiative – Vanderbilt University Academic Capital Venture Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aizawa H, Bianco IH, Hamaoka T, Miyashita T, Uemura O, Concha ML, Russell C, Wilson SW, Okamoto H. Laterotopic representation of left-right information onto the dorso-ventral axis of a zebrafish midbrain target nucleus. Curr Biol. 2005;15:238–243. doi: 10.1016/j.cub.2005.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove BW, Essner JJ, Yost HJ. Regulation of midline development by antagonism of lefty and nodal signaling. Development. 1999;126:3253–3262. doi: 10.1242/dev.126.14.3253. [DOI] [PubMed] [Google Scholar]
- Bisgrove BW, Essner JJ, Yost HJ. Multiple pathways in the midline regulate concordant brain, heart and gut left-right asymmetry. Development. 2000;127:3567–3579. doi: 10.1242/dev.127.16.3567. [DOI] [PubMed] [Google Scholar]
- Braun MM, Etheridge A, Bernard A, Robertson CP, Roelink H. Wnt signaling is required at distinct stages of development for the induction of the posterior forebrain. Development. 2003;130:5579–5587. doi: 10.1242/dev.00685. [DOI] [PubMed] [Google Scholar]
- Campione M, Steinbeisser H, Schweickert A, Deissler K, van Bebber F, Lowe LA, Nowotschin S, Viebahn C, Haffter P, Kuehn MR, Blum M. The homeobox gene Pitx2: mediator of asymmetric left-right signaling in vertebrate heart and gut looping. Development. 1999;126:1225–1234. doi: 10.1242/dev.126.6.1225. [DOI] [PubMed] [Google Scholar]
- Carl M, Loosli F, Wittbrodt J. Six3 inactivation reveals its essential role for the formation and patterning of the vertebrate eye. Development. 2002;129:4057–4063. doi: 10.1242/dev.129.17.4057. [DOI] [PubMed] [Google Scholar]
- Cohen MM., Jr Holoprosencephaly: clinical, anatomic, and molecular dimensions. Birth Defects Res A Clin Mol Teratol. 2006;76:658–673. doi: 10.1002/bdra.20295. [DOI] [PubMed] [Google Scholar]
- Concha ML, Burdine RD, Russell C, Schier AF, Wilson SW. A nodal signaling pathway regulates the laterality of neuroanatomical asymmetries in the zebrafish forebrain. Neuron. 2000;28:399–409. doi: 10.1016/s0896-6273(00)00120-3. [DOI] [PubMed] [Google Scholar]
- Concha ML, Russell C, Regan JC, Tawk M, Sidi S, Gilmour DT, Kapsimali M, Sumoy L, Goldstone K, Amaya E, et al. Local tissue interactions across the dorsal midline of the forebrain establish CNS laterality. Neuron. 2003;39:423–438. doi: 10.1016/s0896-6273(03)00437-9. [DOI] [PubMed] [Google Scholar]
- Concha ML, Wilson SW. Asymmetry in the epithalamus of vertebrates. J Anat. 2001;199:63–84. doi: 10.1046/j.1469-7580.2001.19910063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, McCallum CM, Stout JL, Slade AJ, Moens CB. A high-throughput method for identifying N-ethyl-N-nitrosourea (ENU)-induced point mutations in zebrafish. Methods Cell Biol. 2004;77:91–112. doi: 10.1016/s0091-679x(04)77005-3. [DOI] [PubMed] [Google Scholar]
- Dufourcq P, Rastegar S, Strahle U, Blader P. Parapineal specific expression of gfi1 in the zebrafish epithalamus. Gene Expr Patterns. 2004;4:53–57. doi: 10.1016/s1567-133x(03)00148-0. [DOI] [PubMed] [Google Scholar]
- Essner JJ, Branford WW, Zhang J, Yost HJ. Mesendoderm and left-right brain, heart and gut development are differentially regulated by pitx2 isoforms. Development. 2000;127:1081–1093. doi: 10.1242/dev.127.5.1081. [DOI] [PubMed] [Google Scholar]
- Gamse JT, Kuan YS, Macurak M, Brosamle C, Thisse B, Thisse C, Halpern ME. Directional asymmetry of the zebrafish epithalamus guides dorsoventral innervation of the midbrain target. Development. 2005;132:4869–4881. doi: 10.1242/dev.02046. [DOI] [PubMed] [Google Scholar]
- Gamse JT, Shen YC, Thisse C, Thisse B, Raymond PA, Halpern ME, Liang JO. Otx5 regulates genes that show circadian expression in the zebrafish pineal complex. Nat Genet. 2002;30:117–121. doi: 10.1038/ng793. [DOI] [PubMed] [Google Scholar]
- Gamse JT, Thisse C, Thisse B, Halpern ME. The parapineal mediates left-right asymmetry in the zebrafish diencephalon. Development. 2003;130:1059–1068. doi: 10.1242/dev.00270. [DOI] [PubMed] [Google Scholar]
- Gritsman K, Zhang J, Cheng S, Heckscher E, Talbot WS, Schier AF. The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell. 1999;97:121–132. doi: 10.1016/s0092-8674(00)80720-5. [DOI] [PubMed] [Google Scholar]
- Halpern ME, Liang JO, Gamse JT. Leaning to the left: laterality in the zebrafish forebrain. Trends Neurosci. 2003;26:308–313. doi: 10.1016/S0166-2236(03)00129-2. [DOI] [PubMed] [Google Scholar]
- Harris JA, Guglielmotti V, Bentivoglio M. Diencephalic asymmetries. Neurosci Biobehav Rev. 1996;20:637–643. doi: 10.1016/0149-7634(95)00077-1. [DOI] [PubMed] [Google Scholar]
- Inbal A, Topczewski J, Solnica-Krezel L. Targeted gene expression in the zebrafish prechordal plate. Genesis. 2006;44:584–588. doi: 10.1002/dvg.20253. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Toyama R, Takeda H, Dawid IB, Kawakami K. Overexpression of the forebrain-specific homeobox gene six3 induces rostral forebrain enlargement in zebrafish. Development. 1998;125:2973–2982. doi: 10.1242/dev.125.15.2973. [DOI] [PubMed] [Google Scholar]
- Lagutin OV, Zhu CC, Kobayashi D, Topczewski J, Shimamura K, Puelles L, Russell HR, McKinnon PJ, Solnica-Krezel L, Oliver G. Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev. 2003;17:368–379. doi: 10.1101/gad.1059403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JO, Etheridge A, Hantsoo L, Rubinstein AL, Nowak SJ, Izpisua Belmonte JC, Halpern ME. Asymmetric nodal signaling in the zebrafish diencephalon positions the pineal organ. Development. 2000;127:5101–5112. doi: 10.1242/dev.127.23.5101. [DOI] [PubMed] [Google Scholar]
- Long S, Ahmad N, Rebagliati M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development. 2003;130:2303–2316. doi: 10.1242/dev.00436. [DOI] [PubMed] [Google Scholar]
- Masai I, Heisenberg CP, Barth KA, Macdonald R, Adamek S, Wilson SW. floating head and masterblind regulate neuronal patterning in the roof of the forebrain. Neuron. 1997;18:43–57. doi: 10.1016/s0896-6273(01)80045-3. [DOI] [PubMed] [Google Scholar]
- Rebagliati MR, Toyama R, Fricke C, Haffter P, Dawid IB. Zebrafish nodal-related genes are implicated in axial patterning and establishing left-right asymmetry. Dev Biol. 1998a;199:261–272. doi: 10.1006/dbio.1998.8935. [DOI] [PubMed] [Google Scholar]
- Rebagliati MR, Toyama R, Haffter P, Dawid IB. cyclops encodes a nodal-related factor involved in midline signaling. Proc Natl Acad Sci U S A. 1998b;95:9932–9937. doi: 10.1073/pnas.95.17.9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Muenke M. Midline and laterality defects: left and right meet in the middle. Bioessays. 2001;23:888–900. doi: 10.1002/bies.1130. [DOI] [PubMed] [Google Scholar]
- Sampath K, Rubinstein AL, Cheng AM, Liang JO, Fekany K, Solnica-Krezel L, Korzh V, Halpern ME, Wright CV. Induction of the zebrafish ventral brain and floorplate requires cyclops/nodal signalling. Nature. 1998;395:185–189. doi: 10.1038/26020. [DOI] [PubMed] [Google Scholar]
- Seo HC, Drivenes, Ellingsen S, Fjose A. Expression of two zebrafish homologues of the murine Six3 gene demarcates the initial eye primordia. Mech Dev. 1998a;73:45–57. doi: 10.1016/s0925-4773(98)00028-8. [DOI] [PubMed] [Google Scholar]
- Seo HC, Drivenes O, Ellingsen S, Fjose A. Transient expression of a novel Six3-related zebrafish gene during gastrulation and eye formation. Gene. 1998b;216:39–46. doi: 10.1016/s0378-1119(98)00328-x. [DOI] [PubMed] [Google Scholar]
- Staudt N, Houart C. The Prethalamus Is Established during Gastrulation and Influences Diencephalic Regionalization. PLoS Biol. 2007;5:e69. doi: 10.1371/journal.pbio.0050069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot WS, Trevarrow B, Halpern ME, Melby AE, Farr G, Postlethwait JH, Jowett T, Kimmel CB, Kimelman D. A homeobox gene essential for zebrafish notochord development. Nature. 1995;378:150–157. doi: 10.1038/378150a0. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. Antivin, a novel and divergent member of the TGFbeta superfamily, negatively regulates mesoderm induction. Development. 1999;126:229–240. doi: 10.1242/dev.126.2.229. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Plasterk RH. Target-selected gene inactivation in zebrafish. Methods Cell Biol. 2004;77:69–90. doi: 10.1016/s0091-679x(04)77004-1. [DOI] [PubMed] [Google Scholar]
- Wilson SW, Houart C. Early steps in the development of the forebrain. Dev Cell. 2004;6:167–181. doi: 10.1016/s1534-5807(04)00027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.