Abstract
TIE2 is a vascular endothelial-specific receptor tyrosine kinase essential for the regulation of vascular network formation and remodeling. Previously, we have shown that the 1.2-kb 5′ flanking region of the TIE2 promoter is capable of directing β-galactosidase reporter gene expression specifically into a subset of endothelial cells (ECs) of transgenic mouse embryos. However, transgene activity was restricted to early embryonic stages and not detectable in adult mice. Herein we describe the identification and characterization of an autonomous endothelial-specific enhancer in the first intron of the mouse TIE2 gene. Furthermore, combination of the TIE2 promoter with an intron fragment containing this enhancer allows it to target reporter gene expression specifically and uniformly to virtually all vascular ECs throughout embryogenesis and adulthood. To our knowledge, this is the first time that an in vivo expression system has been assembled by which heterologous genes can be targeted exclusively to the ECs of the entire vasculature. This should be a valuable tool to address the function of genes during physiological and pathological processes of vascular ECs in vivo. Furthermore, we were able to identify a short region critical for enhancer function in vivo that contains putative binding sites for Ets-like transcription factors. This should, therefore, allow us to determine the molecular mechanisms underlying the vascular-EC-specific expression of the TIE2 gene.
Keywords: transcription, enhancer
Establishment of the circulation is critical for the development of all organ systems of the human body, and endothelial cells (ECs), which line the lumina of all blood vessels, play an essential role in the formation and physiological functions of the circulatory system (for reviews, see refs. 1 and 2). In addition, the importance of the vascular system during pathological conditions such as cancer, atherosclerosis, and wound healing has also been well recognized (for review, see ref. 3). However, little is known about the molecular mechanisms regulating the diverse functions of ECs in vivo.
The recent development of transgenic mouse technology has proven to be a powerful tool for the examination of a number of mammalian developmental processes, and sequences that are able to drive gene expression in ECs of transgenic mice have been described (4–7). However, none of these sequences work uniformly in all ECs of all developmental stages or in the adult animal. Furthermore, some of these expression systems are not strictly EC-specific (5–7). Therefore, we have embarked on the development of an in vivo expression system in which heterologous gene expression can be targeted specifically and uniformly to ECs throughout development and the adult animal.
The TIE2 gene, coding for one of the receptors of the newly identified family of regulators of vascular remodeling, called the angiopoietins (8, 9), appeared to be a promising candidate in the search for EC-specific transcriptional regulatory elements. This is because TIE2 expression becomes detectable as soon as the first ECs arise (late primitive streak stage), remains up-regulated throughout development (i.e., during both vasculogenesis, the formation of blood vessels from in situ-differentiating angioblasts, and angiogenesis, the formation of blood vessels that sprout and split from preexisting vessels), and remains detectable in virtually all tissues of adult specimens (10–14). Furthermore, TIE2 expression becomes superinduced under conditions of increased vessel growth, e.g., during wound healing and tumor angiogenesis (unpublished observation). Therefore, identification of the EC-specific cis-acting sequences of the TIE2 gene should facilitate the development of an EC-specific expression vector for in vivo use. Moreover, characterization of such sequences should provide a starting point for the identification of transcription factors regulating EC-lineage establishment, vasculogenesis, and angiogenesis.
Previously, we have described that TIE2 upstream sequences are active in vivo (4). However, even 7.2 kb of the sequence 5′ to the translational start site were not sufficient to recapitulate the endogenous expression pattern beyond the stages of early vasculogenesis. No reporter gene activity could be detected in areas that are known to be vascularized by predominantly angiogenic processes, such as the limb buds and the neuroectoderm, and in the adult animal. These results suggested that additional cis-acting regulatory elements are present outside the 5′ promoter region and are required for the complete EC-specific expression. Herein, we report the identification and characterization of EC-specific transcriptional enhancer sequences derived from the first intron of the mouse TIE2 gene.
MATERIALS AND METHODS
Reporter Gene Construct.
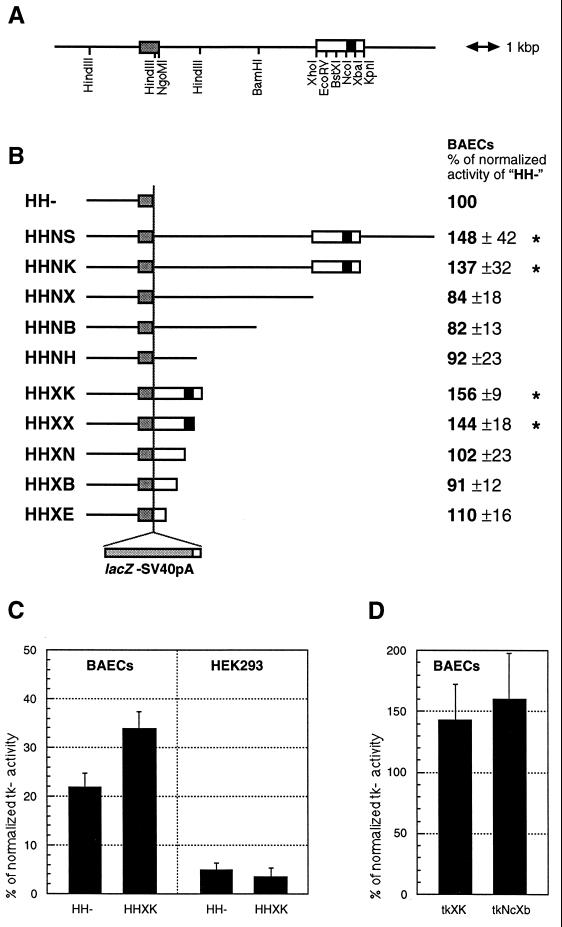
LacZ reporter constructs were based on pBSIIKS(+) or pBSIISK(+) (Stratagene). pHH− consisted of the murine 2.1-kb HindIII TIE2 promoter fragment followed by the LacZ reporter gene and simian virus 40 poly(A) signal sequence from pβ-actinPSDKLacZpA (a gift from Janet Rossant, Mount Sinai Research Institute, Toronto, Canada). In addition to the 2.1-kb promoter, pHHNS contained a mouse TIE2 genomic fragment extending from an NgoMI site at the 3′ end of exon 1 approximately 10 kb into the first intron downstream of the LacZ cassette.
Deletions of pHHNS from the 3′ end of the first intron fragment yielded pHHNH, pHHNB, pHHNX, and pHHNK, respectively (see Fig. 3 for details). pHHXK contained the XhoI–KpnI enhancer fragment downstream of the reporter gene. Systematic 3′ deletions of this XhoI–KpnI fragment yielded pHHXE, pHHXB, pHHXN, and pHHXX, respectively (see Fig. 3 for details). For construction of ptk−, the TIE2 promoter of pHH− was replaced with the herpes simplex virus 1 thymidine kinase gene (tk) minimal promoter (nucleotides −116 to +68) from PMC1POLA (Stratagene). ptkXK and pKXtk contained the XhoI–KpnI enhancer fragment in sense orientation downstream of the reporter gene and in antisense orientation upstream of the tk promoter, respectively. ptkNcXb contained the NcoI–XbaI intron fragment downstream of the tk-promoter driven LacZ. ptkXKΔ was identical to ptkXK, except that it lacked the SacI–XbaI region from the NcoI–XbaI intron fragment. pHHXK-SaXb was identical to ptkXKΔ, except that it contained the 2.1-kb TIE2 promoter instead of tk promoter. Site-directed mutagenesis of the XhoI–KpnI enhancer was performed with the Chameleon kit (Stratagene). The sequence of all constructs was confirmed.
Figure 3.
(A) Structure of the TIE2 locus, including the first exon (shaded box), flanked by upstream and intronic sequences [open box, XhoI–KpnI fragment (XK); solid box, NcoI–XbaI fragment (NcXb)]. (B) Structure of LacZ reporter gene constructs and their respective relative activities (HH− = 100%) in transient transfections of BAECs. Values significantly (P < 0.05; Student’s t test) above 100% have been marked with an asterisk. (C) Activity (in percent of tk core promoter activity) of HH− and HHXK in BAECs and HEK293 cells. (D) Activity of tkXK (tk promoter and 1.7-kb enhancer) and tkNcXb (tk promoter and 303-bp enhancer fragment) in BAECs.
Cell Culture and Transfection Analysis.
Primary bovine aortic EC (BAEC) cultures were kept as described (15). The cells were split 1:5 into 35-mm dishes and transfected 24 h later with 5 μg of DNA [0.5 μg of simian virus 40 enhancer/promoter-driven luciferase plasmid, the respective construct at 0.25 μg/kb, and pBSIIKS(+)], complexed with 10 μl of Lipofectin (GIBCO/BRL). Human embryonic kidney cells (HEK293 cells, American Type Culture Collection catalogue no. 1573) were split 1:3 into 12-well plates and transfected 24 h later with a total of 360 ng of the same DNA mixtures (used for the BAEC transfections) using the MBS transfection kit from Stratagene. Cell lysate preparations and reporter gene activity measurements were performed by using the Galacto Light kit (Tropix, Bedford, MA), the Luciferase assay system (Promega), and a MicroLumat LB96P luminometer (Berthold, Nashua, NH). Background values obtained with lysates from mock-transfections were subtracted, and the β-galactosidase activity of each extract was normalized to the luciferase activity. Each value shown represents the mean from at least six values from at least three experiments.
Transgene Preparation and Transgenic Mice.
Preparation of DNA (lacking any plasmid backbone sequences) for oocyte injection, microinjection, surgical procedures, and genotyping were performed as described (4). However, to genotype transgenic mice carrying tk core-promoter constructs, the upstream forward primer was replaced by 5′-CTCGAGCAAACCCCGCCCAGC.
LacZ-Staining, Immunohistochemistry, and in Situ Hybridization.
Whole-mount LacZ staining of embryos was performed as described (4). For staining of adult organ cryosections, adult mice were first perfused with PBS (10–15 ml), followed by 2% paraformaldehyde/Pipes (30 ml) (4) and finally with PBS. Each organ was dissected, rinsed with PBS for 10 min, and equilibrated with 18% sucrose/PBS at 4°C overnight. Sucrose-treated organs were mounted in OCT (Tissue-Tek), and 10- to 90-μm cryosections were cut and mounted on the slides. Sections were postfixed in 2% paraformaldehyde/Pipes, rinsed twice with PBS (5 min), and stained in 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside. LacZ-stained sections were counterstained with neutral red, mounted in Permount (Fisher Scientific), and photographed with an Axiophot microscope (Zeiss). In situ hybridizations were performed as described with identical probes (4). PECAM-1 immunohistochemistry was performed as described (4, 16). Immunostained sections were photographed (without counterstaining) with an Axiophot microscope (Zeiss) using differential interference contrast optics.
RESULTS
The First Intron of the Murine TIE2 Gene Confers Uniform and EC-Specific Gene Expression in Both Embryo and Adult in Vivo.
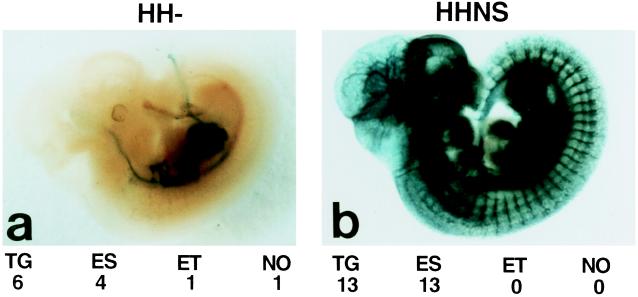
We have reported (4) that the TIE2 promoter alone lacked sequences required for uniform and high-level expression of the LacZ reporter gene in all ECs of transgenic mice, especially in mid to late gestation embryos and postnatal animals. To identify these missing elements, several new reporter gene constructs containing extra sequences from the murine TIE2 gene were tested in transgenic mice. One of these constructs (HHNS) contained an approximately 10-kb fragment from the 5′ half of the first intron in addition to the previously described 2.1-kb TIE2 promoter. When this construct was used to produce transgenic embryos [embryonic day 11.5 (E11.5)], staining revealed strong β-galactosidase activity in virtually all blood vessels (Fig. 1), which was in dramatic contrast to the limited endothelial staining of the mice transgenic with the construct driven by the TIE2 promoter alone (HH−; Fig. 1a and ref. 4). Sectioning of these whole-mount-stained embryos confirmed the EC-specific LacZ expression (data not shown). Furthermore, all 13 independent transgenic embryos exhibited EC-specific LacZ expression, indicating that addition of the intron fragment suppressed the integration site dependency of the LacZ expression. This fragment also protected the TIE2 promoter from being ectopically activated (Fig. 1).
Figure 1.
Whole-mount 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside-stained embryos at E11.5. (a) An embryo transgenic for the construct HH− shows the reduced staining, as described (4). (b) An embryo transgenic for the construct HHNS shows specific staining in virtually all vessels. TG, number of transgenic embryos analyzed; ES, number showing the endothelial-specific staining shown in the respective picture; ET, number showing ectopic staining; NO, number showing no staining at all.
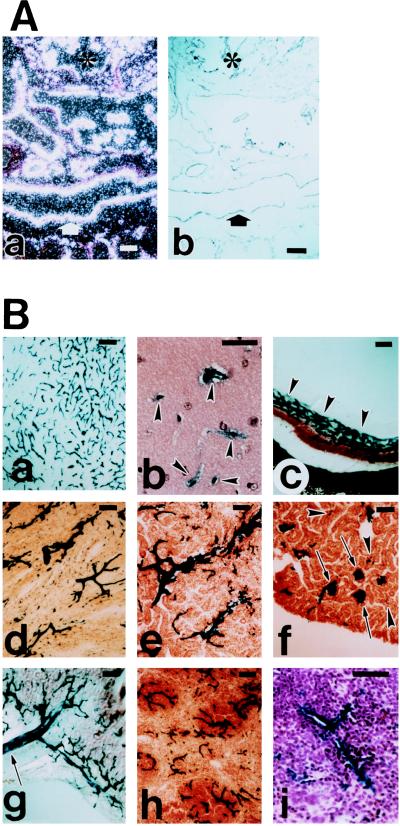
To determine whether this uniform blood-vessel-EC-specific expression of the LacZ persisted in later embryonic stages and adult animals, permanent transgenic mouse lines were established with the HHNS construct. Two transgenic lines derived from founders 182.22 and 182.30 were studied, and uniform and strong LacZ staining was detected in virtually all vessels of embryonic stage E14.5 (data not shown). In situ hybridization with a LacZ probe and immunostaining with an anti-PECAM1 antibody of adjacent sections of E14.5 embryos confirmed the EC-specific, complete, and uniform expression of the LacZ reporter (Fig. 2A).
Figure 2.
LacZ staining of an embryo at later embryonic stage (A) and in adult tissues (B). (Aa) In situ hybridization of embryos at E14.5 with LacZ probe. (b) Adjacent section of a, immunostained with anti-PECAM antibody. Identical staining pattern of a and b confirms specific and uniform endothelial expression of LacZ. ∗, Heart; arrow, dorsal aorta. (B) LacZ staining of adult brain (a, b), eye (c), heart (d), kidney medulla (e), kidney cortex (f), intestine (g), and spleen (h, i). In brain (a), virtually all vessels are stained. In higher magnification of the thinner sections of the brain (b), clearly demonstrates the EC staining (arrowheads). In eye (c), uniform and strong LacZ staining of the capillary plexus is indicated by arrowheads. In heart (d), uniform LacZ staining of the myocardial vasculature is evident. LacZ staining was also observed in endocardium (data not shown). In kidney cortex (f), all of the glomerular microvasculatures (arrows) are strongly LacZ-positive. Other microvasculatures are difficult to see with this low magnification unless they are clustered (arrowheads), but higher magnification confirmed the complete staining of all microvasculatures. In intestine (g), LacZ-positive mesenteric vasculature is indicated (arrow). In spleen, low magnification (h) and higher magnification (i) clearly demonstrates the uniform and EC-specific LacZ staining. (Bar = 100 μm.)
In adult mice, strong and uniform LacZ staining was detected in virtually all blood vessel ECs of many tissues including brain, eye, heart, kidney, intestine, spleen (Fig. 2 Ba–Bh), and other tissues (data not shown).
Identification of Cis-Acting Sequences Within the First Intron That Are Essential for EC-Specific Gene Expression in Vivo.
Nucleotide sequences of the first intron that are essential for EC-specific expression were identified by systematic 3′ truncation of the intron fragment. Analysis of this series of deletion constructs by transient transfection of BAECs in vitro indicated the importance of an internal 1.7-kb XhoI–KpnI fragment (Fig. 3).
This sequence was found to be necessary and sufficient to enhance TIE2 promoter driven LacZ activity by approximately 50% in these cells (Fig. 3B). Transfections with 3′ deletions of the 1.7-kb enhancer revealed an internal 303-bp NcoI–XbaI fragment (NcXb) as an essential region (Fig. 3B). Transcriptional activation by the 1.7-kb XhoI–KpnI fragment appeared to be EC-specific by itself, since it was not active in human embryonic kidney cells (HEK293; see Fig. 3C). In addition, both the 1.7-kb fragment and its 303-bp subfragment were able to up-regulate the heterologous tk promoter by approximately 50% in BAECs (Fig. 3D), showing that these fragments have features of an autonomous EC-specific enhancer.
Sufficiency of these shorter intron fragments for EC-specific transcriptional activation was confirmed by transgenic studies. Transgenic mouse embryos with the construct HHXK were produced and analyzed, and they revealed a LacZ expression pattern indistinguishable from that produced with the HHNS construct at E11.5. However, in adult animals the two independently established HHXK transgenic lines (183.2 and 183.11) showed a less uniform (but still EC-specific) expression pattern (data not shown). Moreover, certain vascular beds such as kidney glomeruli were devoid of LacZ staining (data not shown), suggesting that certain elements lying outside of this 1.7-kb XhoI–KpnI region but within the 10-kb fragment are essential to maximize the enhancer activity in adult tissues.
To investigate whether the 1.7-kb XhoI–KpnI fragment was able to function as a bona fide EC-specific enhancer in vivo, this fragment was placed 3′ and 5′ of the LacZ cassette driven by the heterologous tk promoter, in sense (tkXK) and antisense orientation (KXtk), respectively. Strong and reproducible EC-specific LacZ expression was detected in transgenic embryos (E11.5) derived from both types of constructs (data summarized in Table 1). Furthermore, even the 303-bp NcoI–XbaI fragment (core enhancer) was able to activate the tk promoter (construct tkNcXb) in a EC-specific manner in vivo, although increased integration-site-dependent ectopic expression was observed (Table 1). None of the embryos transgenic for the LacZ reporter gene, driven by the tk promoter alone (construct tk−), showed any EC-specific staining (Table 1).
Table 1.
Summary of the in vivo activity of the enhancer
| Construct | TG | ES | ET | NO |
|---|---|---|---|---|
| HHXK | 14 | 9 | 2 | 5 |
| tkXK | 13 | 6 | 0 | 7 |
| KXtk | 12 | 7 | 2 | 5 |
| tkNcXb | 15 | 9 | 8 | 5 |
| tk- | 9 | 0 | 1 | 8 |
Abbreviations are as in Fig. 1. Data are presented as numbers of embryos.
Importance of the 3′ End of the Core-Enhancer Sequence Including Putative Binding Sites for Ets1, Basic Domain-Leucine Zipper (bZIP), CP2-γ, and PEA3 Transcription Factors.
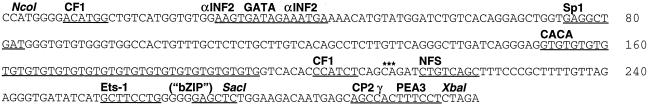
Sequence analysis of the core enhancer (303-bp NcoI–XbaI fragment) revealed the presence of several putative binding sites for general, as well as tissue-specific, transcription factors and other motifs (Fig. 4 and Table 2). The sequence begins with a site for transcription factor Yin Yang 1 (YY1, or CF1). Two αINF2 sites were found to flank a consensus site for GATA transcription factors. Further downstream, there are sites for Sp1 and nuclear factor S (NF-S), a second CF1 site, and a palindromic motif (CAGATCTG; see Discussion). At the 3′ end of the core enhancer, putative sites for transcription factors Ets1, CP2-γ, and PEA3 were identified. In addition, a palindromic motif (GAGCTC) was present that could serve as a binding site for a bZIP type of transcription factor (see Discussion).
Figure 4.
Base sequence of the core enhancer. Sequences matching know transcription factor binding site consensus sequences are underlined: CF1 (consensus, ANATGG), αINF2 (AARKGA), GATA (WGATAR), Sp1 (KRGGCKRRK), NF-S (YGTCAGC), Ets1 (SMGGAWGY), CP2-γ (AGCCACT), and PEA3 (AGGAAR); the CACA microsatellite and a putative bZIP transcription factor binding site (see text) are also underlined. Furthermore, the octameric palindrome (see text) is marked by three asterisks.
Table 2.
Summary of the activity of different mutated constructs in transgenic embryos at E11.5
| Construct (mutated sites) | Wild-type sequence | Mutant sequence | TG | ES |
|---|---|---|---|---|
| tkXK | Wild-type control | No mutations | 13 | 6 |
| tkXK (GATA+αIFN2) | ..AAGTGATAGAAATGA.. | ..AAcgcgtATGA.. | 12 | 7 |
| tkXK (Sp1) | ..GAGGCTGAT.. | ..GAcgcgtAT.. | 12 | 6 |
| tkXK (∗∗∗+NS-F) | ..CAGATCTGTCAGC.. | ..CAGAcgcgtGC.. | 13 | 4* |
| tkXK (Ets1) | ..GCTTCCTG.. | ..GCacgcgTG.. | 9 | 0 |
| tkXK Δ(bZIP, CP2-γ, PEA3) | Deletion of the SacI–XbaI region | 18 | 0 | |
Abbreviations are as in Fig. 1; ∗∗∗, octameric palindrome. Data are presented as numbers of embryos.
Endothelial staining was reduced or enhanced ectopic staining was observed.
Importance of some of these sites for the in vivo activity of the enhancer was challenged by testing constructs in which these sites were altered or deleted. Change of the putative sites for αINF2, GATA, and Sp1 did not significantly affect reporter gene expression in transgenic embryos at E11.5 (data summarized in Table 2). In contrast, the mutation destroying the octameric palindrome described above and the putative NF-S site caused a greater integration-site dependency of the corresponding construct, as fewer transgenic embryos expressed LacZ, expression was weaker and ectopic expression was observed more frequently. The mutation that changed the sequence containing the putative Ets1 site rendered the enhancer completely inactive in vivo, as did the deletion of the SacI–XbaI fragment containing the putative sites for bZIP, CP2-γ, and PEA3. As a control, the latter two mutant forms of the enhancer were also tested in combination with the TIE2 promoter. The expression pattern of these constructs resembled that of HH− (i.e., promoter alone), confirming the effect of these mutations in vivo (data not shown).
DISCUSSION
Our results show that by a combination of upstream and intronic sequences from the murine TIE2 gene, reproducible, strong, uniform, and highly specific expression of genes in virtually all ECs of transgenic embryos and adult mice can be achieved. Although we cannot exclude the possibility that some minor populations of vascular beds did not express the reporter gene, EC targeting with this expression system is by far the most uniform and specific compared with that mediated by other “EC-specific” promoters (5–7).
The apparent discrepancy between the down-regulation of TIE2 expression in adult vessels, as assessed by mRNA in situ hybridization studies (13), and the observed strong LacZ staining may be due to the loss of negative-regulatory elements or to the very low turnover of adult ECs, in which even a small transcriptional activity may be sufficient for the accumulation of significant amounts of a stable protein over time, such as the bacterial β-galactosidase.
Our studies of various intron fragments suggest that several transcriptional regulatory modules are present within the 10-kb intronic fragment. An internal 1.7-kb XhoI–KpnI fragment was identified as an autonomous EC-specific transcriptional enhancer that worked both in vitro and in vivo, in an orientation- and position-independent manner and even when combined with a heterologous promoter. However, the increased integration-site dependency of the HHXK construct (TIE2 promoter plus 1.7-kb enhancer-driven LacZ reporter) compared with the HHNS construct (TIE2 promoter plus 10-kb-fragment-driven LacZ reporter) indicates that additional regulatory sequences exist outside the XhoI–KpnI fragment but within the 10-kb fragment. Sequences outside the 1.7-kb enhancer are also likely to be responsible for the more uniform expression of the HHNS construct in the adult animal.
EPAS1, a recently cloned bHLH/PAS-domain transcription factor highly related to HIF1α, was shown to strongly activate the construct HHNS when both plasmids were cotransfected into HEK293 cells (17). We have reproduced this result, and from our preliminary studies, it seems that intronic sequences outside the 1.7-kb enhancer are necessary for this strong induction. This provides further evidence for the existence of additional intronic transcriptional regulatory sequences.
Furthermore, we have presented evidence that the structure of the 1.7-kb enhancer is complex. An internal 303-bp fragment was identified as sufficient to confer EC-specific activity to the heterologous tk promoter. However, this construct (tkNcXb) was even more integration-site-dependent and more responsive to ectopic activation than the 1.7-kb enhancer-driven construct (tkXK), suggesting that elements necessary for reproducible expression of transgenes, such as a matrix attachment region (18), existed outside the 303-bp region but within the 1.7-kb enhancer.
Several putative transcription factor binding sites, whose presence and functionality have previously been demonstrated in other genes, were identified in the core enhancer sequence: YY1 sites have been implicated in activation (19) and repression (20) of transcription. αINF2-sites have been suggested to play a role in the induction of interferon-β gene expression by viruses (21). GATA sites are known target sites for transcriptional regulators of genes such as von Willebrand factor (22) and endothelial nitric oxide synthase (23), and GATA-2 has been shown to be expressed in ECs (24). An Sp1 site in the endothelial nitric oxide synthase gene is essential for promoter activity (23). A potential site for NF-S (25) in the major histocompatibility complex class II DRA gene promoter becomes occupied upon stimulation with interferon γ (26). The site for CP2-γ (27) in the γ-fibrinogen promoter is a functional CCAAT box (28). The palindromic octamer (CAGATCTG) does not match any described consensus site for transcription factor binding, but the same sequence occurs in the murine TIE1 promoter (7). However, only 7 of the 8 bp are conserved in the homologous region of the human TIE1 promoter. CACA repeats are also part of the human and murine TIE1 promoter (7), and they have been implicated in repression and activation of other genes (29, 30).
Ets1 sites have been shown to be essential for the expression of genes involved in tissue remodeling (31). Furthermore, Ets1 is expressed in migrating and sprouting ECs during embryonic and tumor angiogenesis (31). Close to this Ets1 site, a hexameric palindrome (GAGCTC) has been identified. Interestingly, in the tumor necrosis factor α promoter, the same sequence also lies close to an Ets1 site, where it is part of a putative binding site for the bZIP transcription factor complexes AP-1 or CREB/ATF (32, 33), which cooperate with the Ets1 site in the induction of this promoter by phorbol esters (33). It can therefore be speculated that a bZIP-related transcription factor complex activates the TIE2 intronic enhancer via this hexamer, perhaps in synergy with Ets1.
Head-to-tail clustered PEA3 sites occur in the promoter of TIE1 and TIE2, in regions highly conserved among species (ref. 7 and data not shown). These conservations of the clustered PEA3 sites in both TIE1 and TIE2 gene may imply their importance for the EC-specific gene transcription, although single PEA3 sites, such as that present in the intronic TIE2 enhancer, have also been demonstrated to be functional (34). Besides PEA3, which belongs to the Ets family of transcription factors, Ets1 itself was previously shown to be a candidate for PEA3-site binding (35).
Our mutational analysis of the intronic enhancer showed that the sites for INF2α, GATA, and Sp1 are not critical for in vivo activity at day E11.5. On the other hand, the phenotype of the mutation affecting the NF-S site and the octameric palindrome homologous to the TIE1 promoter suggests that this region contributes to the integration-site independency of the 1.7-kb enhancer. However, the endothelial expression was only slightly weakened by this mutation.
Deletion of the 33-bp SacI–XbaI fragment of the core enhancer, which included the sites for bZIP, CP2γ, and PEA3, and mutation of the Ets1 site just 5′ of the SacI site abolished in vivo activity of the enhancer completely. Preliminary Ets1-cotransfection experiments in endothelial (BAECs) and nonendothelial cells (HEK293) caused only a slight induction of reporter gene expression (tkXK). However, this would be expected if either Ets1 was not a limiting factor or these cell systems lacked putative cofactors.
Identification and characterization of the transcription factors involved in the EC-specific gene expression of the TIE2 gene should lead to a better understanding of the transcriptional mechanisms of the EC-specific expression of TIE2 and other genes expressed in ECs. This type of molecular analysis should also contribute to the understanding of the mechanisms of EC lineage establishment and differentiation and the development of therapies to perturb certain unwanted gene expression in endothelial cells.
Furthermore, by substitution of the LacZ reporter gene in the described expression vectors with other genes such as dominant negative or dominant active mutants, the function of virtually any gene in ECs of transgenic mice can be challenged in vivo. In addition, genes that are usually only expressed under certain conditions such as during an inflammatory response can now be constitutively expressed, and the resulting phenotypes can be analyzed in vivo.
Establishment of an inducible system (36, 37), based on the regulatory sequences described in this report, enabling the switching on and off of transgenes specifically in ECs, could allow investigation into the role of genes with an early lethal knock-out phenotype, such as the vascular endothelial growth factor receptors (38, 39) or TIE2 itself (40, 41) during later stages of development and in adult animals.
Furthermore, the transgenic mouse lines described herein will be a valuable tool for several applications. For instance, the specific sorting of LacZ-positive cells from tissues, which seems feasible from our preliminary experiments, could help to address the differential gene expression of ECs in vivo, which underlies their enormous heterogeneity and plasticity. Also, the endothelial LacZ activity may be a useful marker for the grade of vascularization of a solid tumor of a cancer mouse model, a parameter difficult to assess by other means but important to evaluate the efficacy of antiangiogenesis tumor therapies (for review, see ref. 42). We believe that the transgenic expression vector and transgenic mouse lines described in this report should prove useful in both basic and clinical studies of the cardiovascular system.
Acknowledgments
We thank Dr. Simon Bamforth for comments on the manuscript, and Steve McKnight and David Russell (University of Texas Southwestern Medical Center, Dallas, TX) for communicating their results on EPAS1 and providing us EPAS1 expression vector prior to publication. Initial stages of this project were carried out in the Sato lab at the Roche Institute of Molecular Biology, which was funded by Hoffmann–LaRoche. Work performed in the Sato lab was partly supported by the Department of Medicine, Beth Israel-Deaconess Medical Center.
ABBREVIATIONS
- EC
endothelial cell
- BAEC
bovine aortic EC
- E
embryonic day
- bZIP
basic domain-leucine zipper
- NF-S
nuclear factor S
Note Added in Proof
The TIE2/LacZ Tg mouse (182.30) reported here is now deposited in the Induced Mutant Resource of the Jackson Laboratory (Bar Harbor, ME) for requests.
Footnotes
References
- 1.Risau W, Flamme I. Annu Rev Cell Dev Biol. 1995;11:73–91. doi: 10.1146/annurev.cb.11.110195.000445. [DOI] [PubMed] [Google Scholar]
- 2.Risau W. FASEB J. 1995;9:926–933. [PubMed] [Google Scholar]
- 3.Folkman J. Nat Med. 1995;1:27–31. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- 4.Schlaeger T M, Qin Y, Fujiwara Y, Magram J, Sato T N. Development (Cambridge, UK) 1995;121:1089–1098. doi: 10.1242/dev.121.4.1089. [DOI] [PubMed] [Google Scholar]
- 5.Aird W C, Jahroudi N, Weiler-Guettler H, Rayburn H B, Rosenberg R D. Proc Natl Acad Sci USA. 1995;92:4567–4571. doi: 10.1073/pnas.92.10.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harats D, Kurihara H, Belloni P, Oakley H, Ziober A, Ackley D, Cain G, Kurihara Y, Lawn R, Sigal E. J Clin Invest. 1995;95:1335–1344. doi: 10.1172/JCI117784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korhonen J, Lahtinen I, Halmekyto M, Alhonen L, Janne J, Dumont D, Alitalo K. Blood. 1995;86:1828–1835. [PubMed] [Google Scholar]
- 8.Davis S, Aldrich T H, Jones P F, Acheson A, Bruno J, Radjiewski C,, Compton D, Maisonpierre P C, Yancopoulos G D. Cell. 1996;87:1161–1169. doi: 10.1016/s0092-8674(00)81812-7. [DOI] [PubMed] [Google Scholar]
- 9.Suri C, Jones P F, Patan S, Bartunkova S, Maisonpierre P C, Davis S,, Sato T N, Yancopoulos G D. Cell. 1996;87:1171–1180. doi: 10.1016/s0092-8674(00)81813-9. [DOI] [PubMed] [Google Scholar]
- 10.Dumont D J, Fong G H, Puri M C, Gradwohl G, Alitalo K, Breitman M L. Dev Dyn. 1995;203:80–92. doi: 10.1002/aja.1002030109. [DOI] [PubMed] [Google Scholar]
- 11.Iwama A, Hamaguchi I, Hashiyama M, Murayama Y, Yasunaga K, Suda T. Biochem Biophys Res Comm. 1993;195:301–309. doi: 10.1006/bbrc.1993.2045. [DOI] [PubMed] [Google Scholar]
- 12.Maisonpierre P C, Goldfarb M, Yancopoulos G D, Gao G. Oncology (Basel) 1993;8:1631–1637. [PubMed] [Google Scholar]
- 13.Sato T N, Qin Y, Kozak C A, Audus K L. Proc Natl Acad Sci USA. 1993;90:9355–9358. doi: 10.1073/pnas.90.20.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schnürch H, Risau W. Development (Cambridge, UK) 1993;119:957–968. doi: 10.1242/dev.119.3.957. [DOI] [PubMed] [Google Scholar]
- 15.Rönicke V, Risau W, Breier G. Circ Res. 1996;79:277–285. doi: 10.1161/01.res.79.2.277. [DOI] [PubMed] [Google Scholar]
- 16.Vecchi A, Garlanda C, Lampugnani M G, Resnati M, Matteucci C, Stoppacciaro A, Schnürch H, Risau W, Ruco L, Mantovani A, Dejana E. Eur J Cell Biol. 1994;63:247–254. [PubMed] [Google Scholar]
- 17.Tian H, McKnight S L, Russell D W. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 18.McKnight R A, Shamay A, Sankaran L, Wall R J, Hennighausen L. Proc Natl Acad Sci USA. 1992;89:6943–6947. doi: 10.1073/pnas.89.15.6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riggs K J, Saleque S, Wong K K, Merrell K T, Lee J S, Shi Y, Calame K. Mol Cell Biol. 1993;13:7487–7495. doi: 10.1128/mcb.13.12.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauknecht T, Angel P, Royer H D, zur Hausen H. EMBO J. 1992;11:4607–4617. doi: 10.1002/j.1460-2075.1992.tb05563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujita T, Ohno S, Yasumitsu H, Taniguchi T. Cell. 1985;41:489–496. doi: 10.1016/s0092-8674(85)80022-2. [DOI] [PubMed] [Google Scholar]
- 22.Jahroudi N, Lynch D C. Mol Cell Biol. 1994;14:999–1008. doi: 10.1128/mcb.14.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang R, Min W, Sessa W C. J Biol Chem. 1995;270:15320–15326. doi: 10.1074/jbc.270.25.15320. [DOI] [PubMed] [Google Scholar]
- 24.Dorfman D M, Wilson D B, Bruns G A P, Orkin S H. J Biol Chem. 1991;267:1279–1285. [PubMed] [Google Scholar]
- 25.Kobr M, Reith W, Herrero-Sanchez C, Mach B. Mol Cell Biol. 1990;10:965–971. doi: 10.1128/mcb.10.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright K L, Ting J P. Proc Natl Acad Sci USA. 1992;89:7601–7605. doi: 10.1073/pnas.89.16.7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chodosh L A, Baldwin A S, Carthew R W, Sharp P A. Cell. 1988;53:11–24. doi: 10.1016/0092-8674(88)90483-7. [DOI] [PubMed] [Google Scholar]
- 28.Morgan J G, Courtois G, Fourel G, Chodosh L A, Campbell L, Evans E, Crabtree G R. Mol Cell Biol. 1988;8:2628–2637. doi: 10.1128/mcb.8.6.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berg D T, Walls J D, Reifel-Miller A E, Grinnell B W. Mol Cell Biol. 1989;9:5248–5253. doi: 10.1128/mcb.9.11.5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naylor L H, Clark E M. Nucleic Acids Res. 1990;18:1595–1601. doi: 10.1093/nar/18.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wernert N, Raes M B, Lassalle P, Dehouck M P, Gosselin B, Vandenbunder B, Stehelin D. Am J Pathol. 1992;140:119–127. [PMC free article] [PubMed] [Google Scholar]
- 32.Leitman D C, Ribeiro R C J, Mackow E R, Baxter J D, West B L. J Biol Chem. 1991;266:9343–9346. [PubMed] [Google Scholar]
- 33.Krämer B, Wiegmann K, Kronke M. J Biol Chem. 1995;270:6577–6583. doi: 10.1074/jbc.270.12.6577. [DOI] [PubMed] [Google Scholar]
- 34.Martin M E, Piette J, Yaniv M, Tang W J, Folk W R. Proc Natl Acad Sci USA. 1988;85:5839–5843. doi: 10.1073/pnas.85.16.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fisher R J, Mavrothalassitis G, Kondoh A, Papas T S. Oncology (Basel) 1991;6:2249–2254. [PubMed] [Google Scholar]
- 36.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 37.No D, Yao T P, Evans R M. Proc Natl Acad Sci USA. 1996;93:3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shalaby F, Rossant J, Yamaguchi T P, Gertsenstein M, Wu X-F, Breitman M L, Schuh A C. Nature (London) 1995;376:62–66. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- 39.Fong G-H, Rossant J, Gertsenstein M, Breitman M L. Nature (London) 1995;376:66–70. doi: 10.1038/376066a0. [DOI] [PubMed] [Google Scholar]
- 40.Dumont D J, Gradwohl G, Fong G H, Puri M C, Gertsenstein M, Auerbach A, Breitman M L. Genes Dev. 1994;8:1897–1909. doi: 10.1101/gad.8.16.1897. [DOI] [PubMed] [Google Scholar]
- 41.Sato T N, Tozawa Y, Deutsch U, Wolburg B K, Fujiwara Y, Gendron M M, Gridley T, Wolburg H, Risau W, Qin Y. Nature (London) 1995;376:70–74. doi: 10.1038/376070a0. [DOI] [PubMed] [Google Scholar]
- 42.Folkman J. J Natl Cancer Inst. 1990;82:4–6. doi: 10.1093/jnci/82.1.4. [DOI] [PubMed] [Google Scholar]