Abstract
Mitochondrial function during aging was assessed in isolated rat hepatocytes to avoid the problem of differential lysis when old, fragile mitochondria are isolated. Rhodamine 123, a fluorescent dye that accumulates in mitochondria on the basis of their membrane potential, was used as a probe to determine whether this key function is affected by aging. A marked fluorescent heterogeneity was observed in hepatocytes from old (20–28 months) but not young (3–5 months) rats, suggesting age-associated alterations in mitochondrial membrane potential, the driving force for ATP synthesis. Three distinct cell subpopulations were separated by centrifugal elutriation; each exhibited a unique rhodamine 123 fluorescence pattern, with the largest population from old rats having significantly lower fluorescence than that seen in young rats. This apparent age-associated alteration in mitochondrial membrane potential was confirmed by measurements with radioactive tetraphenylphosphonium bromide. Cells from young rats had a calculated membrane potential of −154 mV, in contrast to that of the three subpopulations from old rats of −70 mV (the largest population), −93 mV, and −154 mV. Production of oxidants was examined using 2′,7′dichlorofluorescin, a dye that forms a fluorescent product upon oxidation. The largest cell subpopulation and a minor one from old animals produced significantly more oxidants than cells from young rats. To investigate the molecular cause(s) for the heterogeneity, we determined the levels of an age-associated mtDNA deletion. No significant differences were seen in the three subpopulations, indicating that the mitochondrial decay is due to other mutations, epigenetic changes, or both.
Keywords: flow cytometry, aging, rhodamine 123, centrifugal cell elutriation, mitochondrial DNA deletions
Mitochondrial dysfunction may be a principal underlying event in aging (1–3). Mitochondria provide energy for basic metabolic processes, and their decay with age impairs cellular metabolism and leads to cellular decline. Oxidative mitochondrial DNA (mtDNA) lesions increase markedly with age (4) and may be responsible for the age-associated increase in mtDNA mutations (3, 5). It is plausible that the accumulation of such mutations leads to decreased gene expression, a decline in oxidative phosphorylation, inefficient electron transport, and increased oxidant flux (6). Increased oxidants may also contribute to alterations in mitochondrial membrane fluidity and phospholipid composition that occur during aging (7–11). These in turn may affect the ability of mitochondria to transport substrates and to generate sufficient proton motive force to meet cellular energy demands.
In contrast to measurements of oxidant-induced damage in mitochondria at the molecular level, some indices of overall mitochondrial function suggest that they adequately meet cellular energy needs with age. Mitochondrial number per cell does not increase with age (12) and the energy charge (3), a measure of the ATP/ADP ratio, does not change. On the other hand, both the mtDNA copy number and the expression of the electron transport chain proteins coded in the nucleus increase with age (13, 14). In addition to mitochondrial turnover, these changes might buffer the impact of mitochondrial oxidative damage and mutation.
Another complexity is whether mitochondrial genetic heteroplasmy, such as occurs in mitochondrial myopathies, also happens during aging (15). A wide variety of altered mtDNA, containing nonlethal mutations, would be expected to occur and accumulate with age. Electron microscopy suggests that morphologically altered mitochondria accumulate in cells, but that these age-associated changes do not affect all cells or all mitochondria within a cell to the same extent; normal and abnormal mitochondria can inhabit the same cell (16, 17). Therefore it is still questionable whether age-related mitochondrial damage is sufficient to cause cellular impairment and organ dysfunction.
To date, studies have generally used isolated mitochondria to determine age-related changes. However, due to their fragility and loss upon isolation, the purified mitochondrial fractions may not accurately represent the mitochondrial diversity found in vivo, leading to misinterpretation of results. In the current study, we examine mitochondrial function in intact rat hepatocytes and show that these cells display a marked degree of age-related variability based on their average mitochondrial membrane potential. We used these differences to separate cells into subpopulations and characterized them according to oxygen consumption, oxidant production, and accumulation of a particular mtDNA deletion.
MATERIALS AND METHODS
Materials.
The following chemicals were used: EGTA [ethylene glycol bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid], digitonin, trypan blue, heparin (sodium salt), Mes (2-[N-morpholino]ethanesulfonic acid) buffer, and rhodamine 123 (R123) (Sigma); silicone oil (density = 1.05 kg/liter) (Aldrich); light white mineral oil (Fisher); [3H]water, [phenyl-3H]tetraphenylphosphonium bromide (TPP) (New England Nuclear), 5,5-dimethyl[2-14C]oxazolidine-2,4-dione (DMO) (Amersham), and [3H]inulin (New England Nuclear); 2′,7′-dichlorofluorescin (DCFH) (Molecular Probes); collagenase (type D) (Boehringer Mannheim); and carbonyl cyanide m-chlorophenylhydrazone (CCCP) (Calbiochem). All other reagents were reagent grade or better. Double-distilled/deionized water was used throughout.
Rats (Fischer 344, male, outbred albino), both young (3–5 months; Simonsen, Gilroy, CA) or old (20–28 months; National Institute on Aging animal colonies), were fed standard Purina rodent chow and water ad libitum.
Cell Isolation.
Liver tissue was dispersed into single cells by collagenase perfusion (18). Cell number was assessed by using a hemocytometer, and viability was determined by trypan blue exclusion. Viability was usually greater than 90% in both age groups.
Cell Elutriation.
Hepatocytes were elutriated using a Beckman J-6M induction-drive centrifuge. Up to 5.0 × 108 cells were loaded into a JE-5 rotor spinning at 1,500 rpm by using a Masterflex peristaltic pump (Cole-Parmer). Krebs–Henseleit medium supplemented with 10% newborn calf serum was continuously passed through this chamber with an initial flow rate of 94 ml/min. Cells were elutriated by decrementally reducing rotor speed and/or increasing the rate of buffer flow. Hepatocytes were separated by using the following conditions: cells designated fraction A were elutriated between 1,500 and 1,470 rpm and a buffer flow rate between 210 and 240 ml/min; cells called fraction B were released between 1,440 and 1,350 rpm and a flow rate of 240 ml/min; cells in fraction C were elutriated between 1,290 and 850 rpm with a flow rate of 240 ml/min. After elutriation, cells were collected by centrifugation at 1,800 rpm for 15 min using a Sorvall A6000A rotor in a Beckman RC-3B centrifuge.
Flow Cytometry.
Hepatocytes (2.0 × 106 cells) were incubated with R123 (0.01 mg/ml) for 30 min at 37°C and then subjected to flow cytometry using an instrument constructed according to the design of Steinkamp et al. (19). Stained cells were passed through a flow chamber at ≈500 cells per sec in a stream that intersected the beam of an argon ion laser tuned to 488 nm. Emitted light, collected at 90° from the laser beam and cell stream, was collected through a band pass filter centered at 550 nm with a spread of 10 nm and a transmittance of 50%. Signals from the photomultiplier were collected in an Oxford multichannel analyzer after analog-to-digital conversion. Nonspecific light scatter was subtracted and all pertinent data were graphed as the number of cells showing a particular fluorescence. Because different numbers of cells were analyzed in separate experiments, all data were normalized to cell number.
Quantitation of Mitochondrial Membrane Potential and Proton Motive Force.
The mitochondrial membrane potential was quantified as described (20, 21). To determine nonspecific binding, cells (1.0 × 106 cells per ml) were boiled for 15 min and incubated with the radioactive indicator molecule, [3H]TPP, as described (20, 21). As a further control, CCCP (20 μM, final concentration), a mitochondrial uncoupler, was added to some cells simultaneously with the radioactive marker. A digitonin concentration of 0.12 mg/ml was used for permeabilizing hepatocytes from young rats, but a lower concentration (0.08 mg/ml) best permeabilized cells from old rats. All samples were assayed in triplicate. The membrane potential (ΔΨ) was quantified using the Nernst equation: ΔΨ = (−RT/F) ln Cr, where Cr is the concentration ratio of TPP in the cytosolic versus the mitochondrial fraction relative to extracellular TPP concentration.
The contribution to the proton motive force from the pH differential across the inner mitochondrial membrane was monitored using 5,5-dimethyl[2-14C]oxazolidine-2,4-dione (0.5 μCi/ml; 1 μCi = 37 kBq) as described (20, 22). Mitochondrial volume was determined (20, 22).
Oxygen Consumption Studies.
Mitochondrial oxygen consumption and respiratory characteristics were analyzed using a Yellow Springs Instruments 5300 oxygen electrode and monitor (23), using glutamate (2.5 mM) and ADP (180 μM) as substrates. In some experiments, cells (1.0 × 107) were permeabilized with digitonin, as described above, prior to measuring O2 consumption.
DCFH Measurement.
Formation of oxidants in cells were determined by using DCFH (24). Duplicate samples were routinely analyzed. Fluorescence was monitored using a Cytofluor 2350 fluorescent measurement system (Millipore) using standard fluorescein filters and cytocalc software. Oxygen consumption was measured and data were expressed as the fluorescence per μM of O2 consumed per min per 106 cells.
mtDNA Deletion Analysis.
Total DNA was isolated from hepatocytes by a standard solvent extraction/proteinase K procedure (25). The polymerase chain reaction (PCR) mixture consisted of 10 mM Tris·HCl (pH 8.3), 50 mM KCl, 0.001% gelatin, 200 μM each dNTP, 2.5 mM MgCl2, 1.25 units of AmpliTaq DNA polymerase (Perkin–Elmer), 10 pmol of each primer, and 500 ng of total DNA in 50 μl; for 1 cycle of 94°C for 2 min, followed by 40 cycles of 94°C for 60 sec, 60°C for 30 sec, and 72°C for 60 sec, and 1 cycle of 72°C for 5 min. Primer 1 (5′-GCTTAGAGCGTTAACCTTTTAAG-3′; nucleotides 7687–7709) and primer 2 (5′-CAGCAGTTTGGATGTGGCG-3′; nucleotides 12962–12981) were used for detection of the 16-nt direct repeat-associated mtDNA deletion at nucleotides 8103 and 12937 (26) of the rat mtDNA sequence. Primer 3 (5′-GGTTCTTACTTCAGGGGCCATC-3′; nucleotides 15768–15789) and primer 4 (5′-GTGGAATTTTCTGAGGGTAGGC-3′; nucleotides 16268–16288) were used for the amplification of wild-type mtDNA. PCR products were cloned into the vector pCRII (Invitrogen), and the insert was sequenced in both directions with the PRISM Ready Reaction DyeDeoxy Terminator Cycle Sequencing Kit (Perkin–Elmer) and a 373 Automated DNA Sequencer (Applied Biosystems). Deletion breakpoints were identified by alignment of PCR product sequences with the rat mtDNA sequence (27). PCR products were quantitated by competitive PCR (28) using a P/ACE 5510 capillary electrophoresis machine with laser-induced fluorescence, a 27-cm-long 100-μm internal diameter eCAP DNA capillary, and dsDNA 1000 buffer that contained 0.04% EN3HANCE (Beckman). The percentage deleted mtDNA was obtained from the ratio of deleted and wild-type mtDNA measured in each sample.
Statistical Analysis.
Statistical significance was determined using the paired Student’s t test or single-factor ANOVA. Results are expressed as the mean ± SEM.
RESULTS
R123 Characteristics of Isolated Rat Hepatocytes in Flow Cytometry.
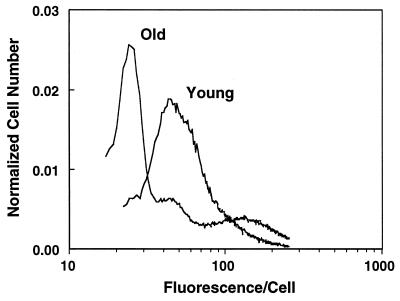
Isolated hepatocytes from old rats (n > 6) exhibited marked differences in R123 staining when compared with cells from young rats. Most cells from old rats showed a decline in average fluorescence, with a small fraction having the same level of fluorescence as young cells (Fig. 1). The heterogeneity in average R123 fluorescence in old rats implies that only a small percentage of hepatocytes have mitochondria with the same membrane potential as that in young rats.
Figure 1.
Hepatocytes isolated from old rats exhibit marked R123 fluorescent heterogeneity. Cells, from either young or old rats, were incubated with R123 30 min before analysis by flow cytometry. Hepatocytes from old rats contained mitochondria that have significantly altered average membrane potential. Shown is a fluorogram typical of that seen in at least six experiments.
Isolation of Hepatocyte Cell Subpopulations by Elutriation and Characterization of Their Average Mitochondrial Membrane Potential.
A centrifugal cell elutriation procedure was developed to separate cells into three subpopulations; the fluorescence levels of these subpopulations correlate with those seen using flow cytometry. Elutriation allows large numbers of cells to be separated into unique subpopulations, providing sufficient quantities of cells to be characterized. In addition, such cells have not been subjected to laser-induced damage that may cause artifactual changes in mitochondrial function.
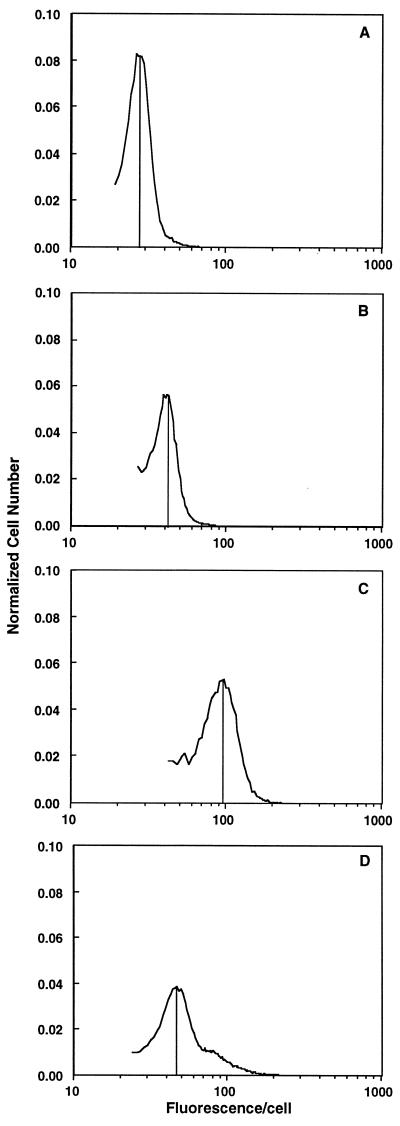
Elutriated hepatocytes from young rats yielded a single cell population (n = 5), consistent with flow cytometry data (Fig. 1 and Fig. 2D). In contrast, two-thirds (65% ± 15%; n = 6) of the elutriated cells from old rats, designated fraction A, had significantly lower fluorescence per cell than that seen in cells from young rats (Fig. 2A). This decrease represents a 40% decline in fluorescence from that seen in cells from young rats. A smaller subset of cells (fraction B; 25% ± 5% of the cells isolated) had only slightly lower fluorescence than cells from young rat hepatocytes (Fig. 2B). A small number cells (fraction C) had equal or greater fluorescence than hepatocytes isolated from young rats (Fig. 2C). Fraction C contained numerous binucleated cells which were very large; it also had the most variability in R123 staining, sometimes appearing as a distinct peak and other times as a broad band of cells with higher fluorescence (Figs. 1 and 2C). Since elutriation separates cells primarily on the basis of hydrodynamic drag, these results imply that the R123 fluorescence parameters being measured are correlated with this property of cells. In general, hydrodynamic drag is a function of cell size, which suggests that mitochondrial changes with age alter cell surface architecture.
Figure 2.
R123 fluorescence profiles of cells from young and old rats after centrifugal cell elutriation. Elutriation of cells from young rats resulted in only one population of cells, while elutriation of cells from old rats yielded one major and two smaller cell subpopulations. (A) The cells that had the lowest R123 fluorescence (fraction A). (B) Cells that had average R123 staining patterns similar to hepatocytes from young rats (fraction B). (C) Cells of this fraction had mitochondria with membrane potential equivalent to or higher than hepatocytes from young rats (fraction C). (D) Cells from young rats. Shown is a typical fluorogram after cell separation. The vertical lines indicate the mean fluorescence value for the majority of cells in the isolated fraction.
Use of R123 provides an assessment of the mitochondrial membrane potential, which is not necessarily a quantitative measure. Interpretation of R123 data assumes that the cellular volume occupied by mitochondria and the mitochondrial number are the same in all fractions. Because it is not known whether these parameters change in hepatocytes with age, we sought to corroborate the changes in membrane potential by using a more quantitative technique, which measures the intracellular compartmentalization of radioactive TPP. This lipophilic ion accumulates preferentially in the negatively charged mitochondrial compartment of the cell, and its distribution can be converted into a membrane potential value (ΔΨ) by using the Nernst equation. Hepatocytes from young rats had a ΔΨ of −154.3 ± 20.4 mV (Table 1), which is in agreement with previously reported values (20, 21). CCCP, a mitochondrial uncoupling agent that disperses the mitochondrial membrane potential, was used to validate whether TPP compartmentalization was based on mitochondrial membrane potential. The addition of 20 μM CCCP lowered the mitochondrial membrane potential to −20.2 ± 6.3 mV (n = 3), which is 87% lower than in the absence of CCCP.
Table 1.
Mitochondrial proton motive force in hepatocytes from young and old rats
| Condition | n | ΔΨ, mV | ΔpH, mV | Δρ (ΔΨ + ΔpH), mV |
|---|---|---|---|---|
| Young | 3 | −154.3 ± 20.4 | −37.1 ± 3.3 | −191.4 ± 23.7 |
| Old | ||||
| Unelutriated | 4 | −101.1 ± 18.4* | −34.0 ± 12.2 | −135.1 ± 30.6* |
| Fraction A | 3 | −70.4 ± 8.9* | −33.8 ± 1.3 | −104.2 ± 10.2* |
| Fraction B | 3 | −92.6 ± 7.0* | −38.0 ± 1.9 | −130.6 ± 8.9* |
| Fraction C | 3 | −154.4 ± 18.3 | −34.0 ± 3.1 | −188.3 ± 21.5 |
ΔΨ denotes the mitochondrial membrane potential. Δρ denotes the mitochondrial proton motive force. ∗, Statistically significant difference from the values shown for hepatocytes from young rats (P ≤ 0.019).
The subpopulations from old rats had average mitochondrial membrane potentials of −70.4 ± 8.9 mV (fraction A), −92.6 ± 7.0 mV (fraction B), and −154.4 ± 18.3 mV (fraction C) (Table 1). These values represent declines of 54.2%, 32%, and 0% relative to cells from young rats, respectively (Table 1). Comparison of these results with R123 staining patterns (Fig. 2) shows that R123 overestimates the relative mitochondrial membrane potential, particularly in fractions B and C. The reasons for this discrepancy are unclear; they may be due to greater nonspecific binding of R123 to various macromolecules with age. However, incubation of cells with CCCP prior to the addition of TPP caused a similar decline in membrane potential (fraction A, −23.1 ± 6.0 mV; fraction B, −19.7 ± 3.2 mV; and fraction C, −24.2 ± 3.3 mV) in the subpopulations from old rats as seen in cells from young animals.
In contrast to the distinct age-related changes in the ΔΨ component of the proton motive force, there was no significant change in the pH differential (ΔpH) across the inner mitochondrial membrane (Table 1). Thus, age-related changes to mitochondria appear to affect only the ΔΨ component of the proton motive force.
Oxygen Consumption Characteristics.
Oxygen consumption in hepatocytes from young rats was 480 ± 60 μM of O2 per min per 107 cells (n = 5). These values are similar to those previously reported for rat liver (29). In contrast, hepatocytes in fraction A had rates of oxygen consumption that were significantly lower (P ≤ 0.047) than that of liver cells from young rats. Overall oxygen consumption declined by 36% to 305 ± 12 μM of O2 per min per 107 cells (n = 3). Fractions B and C did not have significantly different oxygen consumption characteristics compared with young animals. Thus, two-thirds of the hepatocytes from aged rats display significantly lower values of both average mitochondrial membrane potential and overall oxygen consumption.
Determination of state 3/state 4 respiratory control ratios indicates the level of coupling of mitochondrial electron transport to ATP production (23). Measurements made using glutamate and ADP as substrates showed an overall decline in cells from old rats. Fractions A and C were notably uncoupled with respiratory control ratios of 5.4 ± 0.8 (n = 5) and 4.5 ± 0.7 (n = 4), respectively. These results were significantly different (P ≤ 0.028) from the ratios found for fraction B (7.0 ± 0.8; n = 3) or for hepatocytes taken from young rats (8.9 ± 0.9; n = 4).
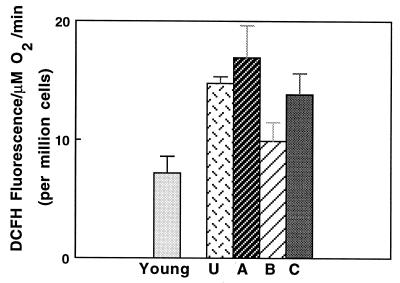
We examined whether the decline in respiratory control ratios resulted in greater oxidant production by incubating cells with DCFH, a dye that is converted to a fluorescent product upon oxidation (24). Oxidant production in fractions A and C was approximately double the rate of cells from young rats or from cells in fraction B (Fig. 3). Thus, electron transport in fractions A and C appears to be inefficiently coupled to ATP synthesis, which suggests an increased rate of superoxide production.
Figure 3.
Hepatocytes from old rats produce significantly higher rates of oxidant production as measured by DCFH. Results show that fractions A and C from old rats had significantly higher rates of oxidant production (P ≤ 0.01) than cells from young rats. U denotes unelutriated cells.
Screening and Quantification of mtDNA Deletions in Hepatocyte Subpopulations.
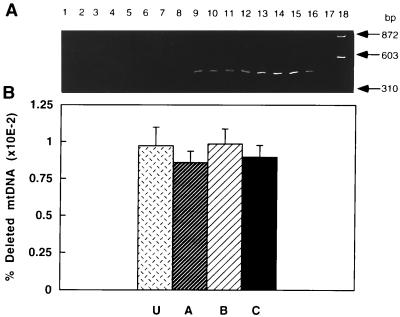
The level of the mtDNA deletion associated with a 16-nt direct repeat was estimated in cells from young and old animals to determine whether there was a relationship between any of the subpopulations and this mutation. This deletion was chosen because it removes several genes that code for subunits involved in complex I formation and its incidence increases with age. The deletion was detected only in old rats (Fig. 4A, lanes 9–16 compared with lanes 1–8). The lack of detection in young rats was not due to PCR inhibition because wild-type mtDNA was successfully amplified (data not shown). No significant difference (P > 0.5) in the mtDNA deletion level was found between any of the rat hepatocyte subpopulations. The average level of this mutation in unelutriated and hepatocyte subpopulations was 0.009% of total mtDNA genomes (Fig. 4B).
Figure 4.
Detection and quantification of the 16-nt direct repeat-associated mtDNA deletion in rat hepatocytes. (A) PCR was performed on total DNA isolated from two young (lanes 1–4 and 5–8) and two old (lanes 9–12 and 13–16) rats. Unelutriated (lanes 1, 5, 9, and 13) or cells from fractions A (2, 6, 10, and 14), B (3, 7, 11, and 15), and C (lanes 4, 8, 12, and 16) were used. Lane 17, negative PCR control; lane 18, molecular weight markers of HaeIII-digested φX174 DNA. (B) Quantification of the mtDNA deletion in unelutriated hepatocytes (U) or hepatocyte subpopulations A–C from old rats (n = 3 or 4).
DISCUSSION
The degree of cellular dysfunction due to mitochondrial damage or how such mitochondrial changes affect organ function is still unclear. This is due in part to difficulties in interpretation because differential lysis of mitochondria occurs when they are isolated from old animals. Thus, any isolated mitochondrial fraction may not be representative of the in vivo situation. That this is a problem is buttressed by studies (16, 17) where a portion of liver tissue from young and old mice was fixed for histological analysis prior to homogenization, followed by mitochondrial isolation from the remaining tissue. A significant proportion of variant mitochondria from old animals were lost, and only mitochondria resembling those from young animals were obtained. Furthermore, mitochondrial isolation from a whole organ cannot assess alterations that occur in specific cells within an organ and therefore would not accurately reflect the extent of age-associated alterations at the cellular level (16, 17). We have avoided these problems by assaying for mitochondrial changes in hepatocytes freshly isolated from young and old rats.
The fluorescent dye R123 was used as a probe for mitochondrial membrane potential because the dye is well characterized, causes no loss of mitochondrial coupling, and is not toxic at low concentrations (30). Also, dye accumulation and fluorescence intensity are stable, allowing accurate measurement of fluorescence characteristics. In the present study using R123 as a probe, a marked fluorescent heterogeneity was observed in hepatocytes from old rats, suggesting that three subpopulations exist that have different mitochondrial membrane properties. The major subpopulation (fraction A) showed decreased R123 fluorescence compared with cells taken from young animals, with only 30% of the hepatocytes from old rats (fractions B and C) showing fluorescence characteristics similar to those seen in young rat cells. Characterization of the subpopulations in terms of oxidant production measured by DCFH showed that fractions A and C released higher levels of oxidants, while fraction B was similar to cells from young hepatocytes.
Previous use of R123 has been with cells from young animals or cells in tissue culture (31, 32, 36). Its potential limitations as a probe to measure age-related changes in mitochondrial membrane potential have not been adequately explored. Furthermore, R123 experiments could be influenced by decreased uptake of the dye into cells from old rats and/or the number of mitochondria per cell could change with age. While unable to gauge this latter parameter directly, [3H]TPP not only quantifies age-associated differences in membrane potential but also determines both the mitochondrial volume per cell and the relative level of radioactivity in the mitochondrial and cytosolic compartments. Hence, the results obtained using R123 were verified by quantitating the proton motive force by using [3H]TPP. There was no significant change in the volume that mitochondria occupy per cell, and the relative cellular uptake of radioactivity did not change with age (data not shown). These studies confirm that R123 staining essentially measures membrane potential differences, with some overestimation of membrane potential, particularly in the smaller cell subpopulations. Therefore, some caution is needed in using R123 as the sole measure of mitochondrial membrane potential in aging studies.
Our results agree with those of Martinez et al. (33, 34), who showed that human fibroblasts in culture developed two distinct cell populations, as judged on the basis of mitochondrial membrane potential after 43 population doublings. Further characterization indicated that these replicative senescent cells had altered numerical densities of mitochondria and a reduced ability for cell replication (33, 34). These in vitro studies, while unable to mimic the diverse age-related changes seen in vivo, nevertheless, point to the same type of mitochondrial heterogeneity as we have noted. In contrast, a study by Maftah et al. (35) showed no age-related decline in mitochondrial membrane potential in isolated human skin epithelial cells. The cause of this difference in results is presently unclear, but may be due to intrinsic properties of the cell types used. Human epithelial cells turn over quite rapidly compared with rat hepatocytes. Cells that turn over rapidly may remove damaged mitochondria before phenotypic changes occur. This theory would predict that postmitotic tissue where mtDNA mutations could accumulate over time may generate substantially more age-related variability than that shown in the present work.
The causes of the heterogeneity in mitochondrial membrane potential observed are unclear and are probably multifactorial, including both genetic and epigenetic changes. While the incidence of the mtDNA deletion examined increases with age, it did not correlate with any particular cell subpopulation. However, only one type of mtDNA deletion was studied, and it is plausible that other types of deletions or point mutations could contribute to the formation of the observed hepatocyte subpopulations. The incidence of mtDNA point mutations also increases with age, and they may exert a physiological effect by creating mitochondria with altered components of the electron transport chain. These mutations may result in varying degrees of inefficient electron transport and a coincident increase in superoxide production. This concept is consistent with our findings of enhanced oxidant production in cells from aged rats in general and the varying degrees of oxidant production in the isolated cell subpopulations in particular.
Such genetic changes that result in enhanced efflux of superoxide could cause epigenetic changes as well. Oxidation of critical thiol groups in key proteins or loss of critical cofactors, such as cardiolipin, may adversely affect transport of substrates necessary for mitochondrial function. These changes may have a direct impact on the ability of mitochondria to maintain their membrane potential and lead to the age-related changes observed. While the compromised mitochondria may continue to perform their basic metabolic function, the consequences for the cell may be increased oxidative insult and a decreased level of energy-linked metabolic reactions.
Acknowledgments
We thank M.-H. Song, J. Wang, E. Silver, and V. Vinarsky for their excellent technical help and M. Mack, P. Walter, K. Beckman, G. Ames, and H. Helbock for their review of the manuscript. This work was supported by a grant from the Sandoz Gerontological Foundation (T.M.H.), by National Cancer Institute Outstanding Investigator Grant CA39910 and National Institute of Environmental Health Sciences Center Grant ES01896 (B.N.A.), and by Office of Energy Research, Office of Health and Environmental Research, Health Effects Research Division of the U.S. Department of Energy under Contract DE-AC03-76SF00098 (J.C.B.).
ABBREVIATIONS
- mtDNA
mitochondrial DNA
- TPP
tetraphenylphosphonium bromide
- R123
rhodamine 123
- DCFH
2′,7′-dichlorofluorescin diacetate
- CCCP
carbonyl cyanide m-chlorophenylhydrazone
References
- 1.Harmon D. Age (Chester PA) 1983;6:86–94. [Google Scholar]
- 2.Miguel J, Fleming J E. Exp Gerontol. 1984;19:31–36. doi: 10.1016/0531-5565(84)90029-9. [DOI] [PubMed] [Google Scholar]
- 3.Shigenaga M K, Hagen T M, Ames B N. Proc Natl Acad Sci USA. 1994;91:10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mecocci P, MacGarvey U, Kaufman A E, Koontz D, Schoffner J M, Wallace D C, Beal M F. Ann Neurol. 1993;34:609–616. doi: 10.1002/ana.410340416. [DOI] [PubMed] [Google Scholar]
- 5.Richter C, Park J W, Ames B N. Proc Natl Acad Sci USA. 1988;85:6465–6467. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallace D C. Science. 1992;256:628–632. doi: 10.1126/science.1533953. [DOI] [PubMed] [Google Scholar]
- 7.Huber L A, Xu Q B, Jurgens G, Bock G, Buhler E, Gey K F, Schonitzer D, Traill K N, Wick G. Eur J Immunol. 1991;21:2761–2765. doi: 10.1002/eji.1830211117. [DOI] [PubMed] [Google Scholar]
- 8.von Zglinicki T, Bimmler M. J Microsc (Oxford) 1987;146:77–85. doi: 10.1111/j.1365-2818.1987.tb01328.x. [DOI] [PubMed] [Google Scholar]
- 9.von Zglinicki T. J Theor Biol. 1987;127:127–132. doi: 10.1016/s0022-5193(87)80123-6. [DOI] [PubMed] [Google Scholar]
- 10.Hoch F L. Biochim Biophys Acta. 1992;1113:71–133. doi: 10.1016/0304-4157(92)90035-9. [DOI] [PubMed] [Google Scholar]
- 11.Guan Z Z, Soderberg M, Sindelar P, Edlund C. Neurochem Intl. 1994;25:295–300. doi: 10.1016/0197-0186(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 12.Tauchi H, Sato T. J Gerontol. 1968;23:454–461. doi: 10.1093/geronj/23.4.454. [DOI] [PubMed] [Google Scholar]
- 13.Corral-Debrinski M, Shoffner J M, Lott M T, Wallace D C. Mutat Res. 1992;275:169–180. doi: 10.1016/0921-8734(92)90021-g. [DOI] [PubMed] [Google Scholar]
- 14.Gadaleta M N, Rainaldi G, Lezza A M, Milella F, Fracasso F, Cantatore P. Mutat Res. 1992;275:181–193. doi: 10.1016/0921-8734(92)90022-h. [DOI] [PubMed] [Google Scholar]
- 15.Wallace D C. Annu Rev Biochem. 1992;61:1175–1212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- 16.Wilson P D, Franks L M. Adv Exp Med Biol. 1975;53:171–183. doi: 10.1007/978-1-4757-0731-1_13. [DOI] [PubMed] [Google Scholar]
- 17.Wilson P D, Franks L M. Biochem Soc Trans. 1975;3:126–128. doi: 10.1042/bst0030126. [DOI] [PubMed] [Google Scholar]
- 18.Moldéus, P., Hogberg, J. & Orrenius, S. (1978) 51, 60–70. [DOI] [PubMed]
- 19.Steinkamp J A, Fulwyler M J, Coulter J R, Hiebert R D, Horney J L, Mullaney P F. Rev Sci Instrum. 1973;44:1301–1310. doi: 10.1063/1.1686375. [DOI] [PubMed] [Google Scholar]
- 20.Andersson B S, Jones D P. Anal Biochem. 1985;146:164–172. doi: 10.1016/0003-2697(85)90411-7. [DOI] [PubMed] [Google Scholar]
- 21.Hoek J B, Nicholls D G, Williamson J R. J Biol Chem. 1980;255:1458–1464. [PubMed] [Google Scholar]
- 22.Waddell W J, Butler T C. J Clin Invest. 1959;38:720–729. doi: 10.1172/JCI103852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Estabrook R W. Methods Enzymol. 1967;10:41–47. [Google Scholar]
- 24.LeBel, C. P., Ishchiropoulos, H. & Bondy, S. C. (1992) 5, 227–231. [DOI] [PubMed]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 9.16–9.19. [Google Scholar]
- 26.Gadaleta M N, Rainaldi G, Lezza A M S, Milella F, Fracasso F, Cantatore P. Mutat Res. 1992;275:181–193. doi: 10.1016/0921-8734(92)90022-h. [DOI] [PubMed] [Google Scholar]
- 27.Gadaleta G, Pepe G, De Candia G, Quagliariello C, Sbisa E, Saccone C. J Mol Evol. 1989;28:497–516. doi: 10.1007/BF02602930. [DOI] [PubMed] [Google Scholar]
- 28.Gilliland G, Perrin S, Blanchard K, Bunn H F. Proc Natl Acad Sci USA. 1990;87:2725–2729. doi: 10.1073/pnas.87.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rickwood D, Wilson M T, Darley-Usmar V M, editors. Mitochondria: A Practical Approach. Oxford: IRL; 1987. pp. 1–16. [Google Scholar]
- 30.Johnson L V, Walsh M L, Chen L B. Proc Natl Acad Sci USA. 1980;77:880–884. [Google Scholar]
- 31.James T W, Bohman R. J Cell Biol. 1980;89:256–260. doi: 10.1083/jcb.89.2.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leprat P, Ratinaud M H, Maftah A, Petit J M, Julien R. Exp Cell Res. 1990;186:130–137. doi: 10.1016/0014-4827(90)90219-z. [DOI] [PubMed] [Google Scholar]
- 33.Martinez A O, Vara C, Castro J. Mech Ageing Dev. 1987;39:1–9. doi: 10.1016/0047-6374(87)90081-9. [DOI] [PubMed] [Google Scholar]
- 34.Martinez A O, Over D, Armstrong L S, Manzano L, Taylor R, Chambers J. Growth Dev Aging. 1991;55:185–191. [PubMed] [Google Scholar]
- 35.Maftah A, Ratinaud M H, Dumas M, Bonte F, Meybeck A, Julien R. Mech Ageing Dev. 1994;77:83–96. doi: 10.1016/0047-6374(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 36.Leprat P, Ratinaud M H, Julien R. Mech Ageing Dev. 1990;52:149–167. doi: 10.1016/0047-6374(90)90121-u. [DOI] [PubMed] [Google Scholar]