Abstract
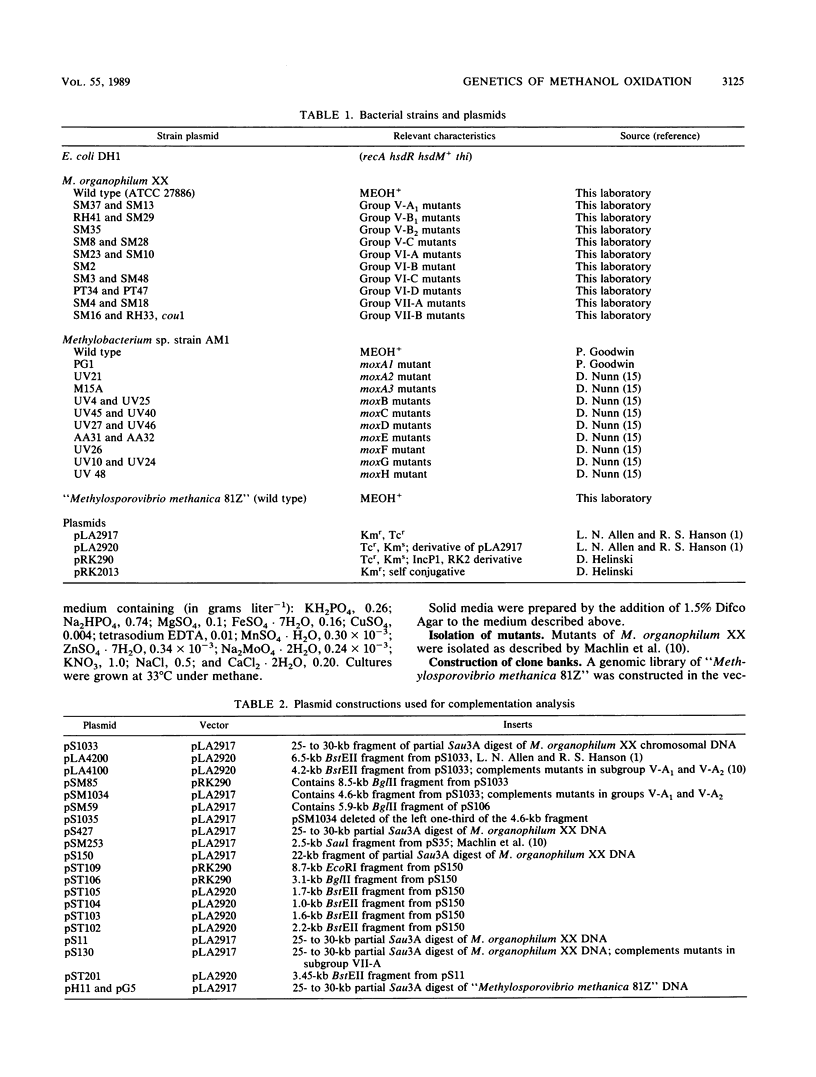
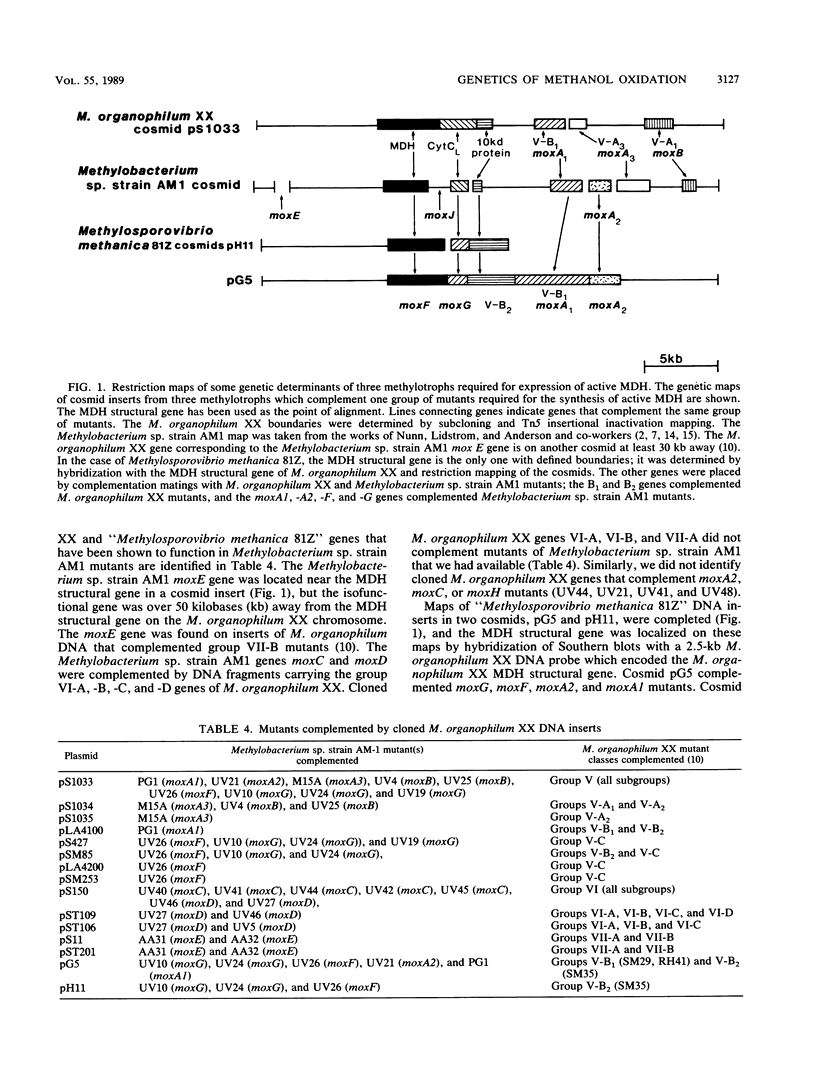
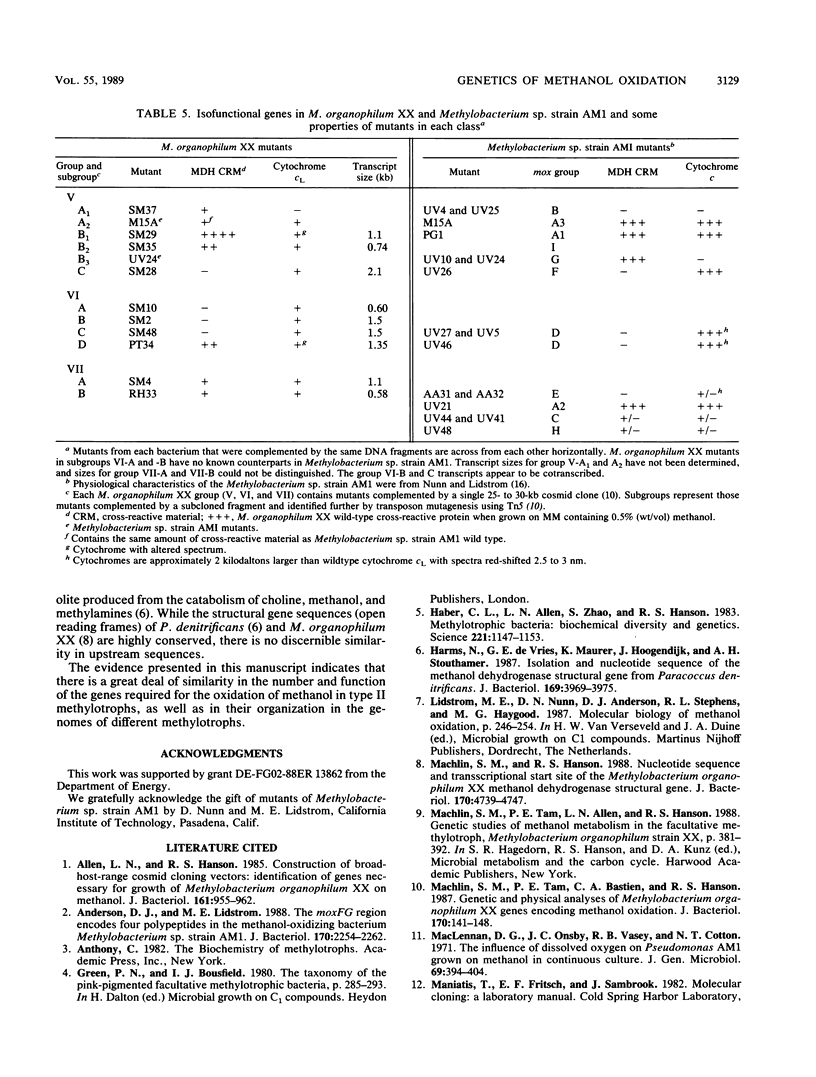
Restriction maps of genes required for the synthesis of active methanol dehydrogenase in Methylobacterium organophilum XX and Methylobacterium sp. strain AM1 have been completed and compared. In these two species of pink-pigmented, type II methylotrophs, 15 genes were identified that were required for the expression of methanol dehydrogenase activity. None of these genes were required for the synthesis of the prosthetic group of methanol dehydrogenase, pyrroloquinoline quinone. The structural gene required for the synthesis of cytochrome cL, an electron acceptor uniquely required for methanol dehydrogenase, and the genes encoding small basic peptides that copurified with methanol dehydrogenases were closely linked to the methanol dehydrogenase structural genes. A cloned 22-kilobase DNA insert from Methylsporovibrio methanica 81Z, an obligate type II methanotroph, complemented mutants that contained lesions in four genes closely linked to the methanol dehydrogenase structural genes. The methanol dehydrogenase and cytochrome cL structural genes were found to be transcribed independently in M. organophilum XX. Only two of the genes required for methanol dehydrogenase synthesis in this bacterium were found to be cotranscribed.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen L. N., Hanson R. S. Construction of broad-host-range cosmid cloning vectors: identification of genes necessary for growth of Methylobacterium organophilum on methanol. J Bacteriol. 1985 Mar;161(3):955–962. doi: 10.1128/jb.161.3.955-962.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. J., Lidstrom M. E. The moxFG region encodes four polypeptides in the methanol-oxidizing bacterium Methylobacterium sp. strain AM1. J Bacteriol. 1988 May;170(5):2254–2262. doi: 10.1128/jb.170.5.2254-2262.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber C. L., Allen L. N., Zhao S., Hanson R. S. Methylotrophic bacteria: biochemical diversity and genetics. Science. 1983 Sep 16;221(4616):1147–1153. doi: 10.1126/science.221.4616.1147. [DOI] [PubMed] [Google Scholar]
- Harms N., de Vries G. E., Maurer K., Hoogendijk J., Stouthamer A. H. Isolation and nucleotide sequence of the methanol dehydrogenase structural gene from Paracoccus denitrificans. J Bacteriol. 1987 Sep;169(9):3969–3975. doi: 10.1128/jb.169.9.3969-3975.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlin S. M., Hanson R. S. Nucleotide sequence and transcriptional start site of the Methylobacterium organophilum XX methanol dehydrogenase structural gene. J Bacteriol. 1988 Oct;170(10):4739–4747. doi: 10.1128/jb.170.10.4739-4747.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlin S. M., Tam P. E., Bastien C. A., Hanson R. S. Genetic and physical analyses of Methylobacterium organophilum XX genes encoding methanol oxidation. J Bacteriol. 1988 Jan;170(1):141–148. doi: 10.1128/jb.170.1.141-148.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclennan D. G., Ousby J. C., Vasey R. B., Cotton N. T. The influence of dissolved oxygen on Pseudomonas AM1 grown on methanol in continuous culture. J Gen Microbiol. 1971 Dec;69(3):395–404. doi: 10.1099/00221287-69-3-395. [DOI] [PubMed] [Google Scholar]
- Nunn D. N., Anthony C. The nucleotide sequence and deduced amino acid sequence of the genes for cytochrome cL and a hypothetical second subunit of the methanol dehydrogenase of Methylobacterium AM1. Nucleic Acids Res. 1988 Aug 11;16(15):7722–7722. doi: 10.1093/nar/16.15.7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D. N., Lidstrom M. E. Isolation and complementation analysis of 10 methanol oxidation mutant classes and identification of the methanol dehydrogenase structural gene of Methylobacterium sp. strain AM1. J Bacteriol. 1986 May;166(2):581–590. doi: 10.1128/jb.166.2.581-590.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D. N., Lidstrom M. E. Phenotypic characterization of 10 methanol oxidation mutant classes in Methylobacterium sp. strain AM1. J Bacteriol. 1986 May;166(2):591–597. doi: 10.1128/jb.166.2.591-597.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M., Wopat A., Hanson R. S. Genetic transformation in Methylobacterium organophilum. J Gen Microbiol. 1977 Jan;98(1):265–272. doi: 10.1099/00221287-98-1-265. [DOI] [PubMed] [Google Scholar]
- O'Keeffe D. T., Anthony C. The two cytochromes c in the facultative methylotroph Pseudomonas am1. Biochem J. 1980 Nov 15;192(2):411–419. doi: 10.1042/bj1920411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEEL D., QUAYLE J. R. Microbial growth on C1 compounds. I. Isolation and characterization of Pseudomonas AM 1. Biochem J. 1961 Dec;81:465–469. doi: 10.1042/bj0810465. [DOI] [PMC free article] [PubMed] [Google Scholar]


