Abstract
Receptor dimerization is the key signaling event for many cytokines, including erythropoietin. A system has been recently developed that permits intracellular protein dimerization to be reversibly activated in response to a lipid-soluble dimeric form of the drug FK506, called FK1012. FK1012 is used as a pharmacological mediator of dimerization to bring together FK506 binding domains, taken from the endogenous protein FKBP12. In experiments reported herein, FK1012-induced dimerization of a fusion protein containing the intracellular portion of the erythropoietin receptor allowed cells normally dependent on interleukin 3 to proliferate in its absence. FK506 competitively reversed the proliferative effect of FK1012 but had no influence on the proliferative effect of interleukin 3. Signaling pathways activated by FK1012 mimicked those activated by erythropoietin, because both JAK2 and STAT5 were phosphorylated in response to FK1012. This approach may provide a means to specifically and reversibly stimulate the proliferation of genetically modified cell populations in vitro or in vivo.
A major obstacle to the success of gene therapy for inherited blood-cell disorders is the extremely low efficiency of stem cell transduction using currently available gene delivery systems (1, 2). A strategy for increasing the frequency of modified stem cells is to specifically direct their preferential expansion through selection. The basis for selection is provided by bicistronic vectors in which the nonselectable therapeutic gene is linked to a second gene encoding a selectable product. Selection is then applied either ex vivo (3, 4) or, if a clinically tolerable strategy for selection were devised, repeated cycles of selection could be applied in vivo (5).
Current strategies for in vivo selection involve the transfer of a gene conferring drug resistance, followed by the subsequent administration of the corresponding cytotoxic drug in vivo (5). This approach is limited by the nonhematological toxicities of the drugs and by the resistance of normal hematopoietic stem cells and progenitors to killing by many cytotoxic agents (6, 7). An alternative approach would be to confer a direct proliferative advantage to the genetically modified cells relative to their nontransduced counterparts. The practical application of this strategy would require that the proliferative stimulus (i) be restricted to the genetically modified population and (ii) be completely reversible.
Cytokines drive hematopoietic cell proliferation; their effects are mediated via ligand-specific surface membrane receptors. Many members of the cytokine receptor superfamily are composed of multiple subunits; however, the receptors for erythropoietin, granulocyte colony-stimulating factor, thrombopoietin, prolactin, and growth hormone consist of single chains that are activated through ligand-mediated homodimerization (8). A system has been recently developed that permits intracellular protein dimerization to be reversibly activated in response to a lipid-soluble dimeric form of the drug FK506, called FK1012 (9–14). FK1012 is used as a pharmacological mediator of dimerization to bring together two FK506 binding domains that are taken from the endogenous protein FKBP12. Thus, fusion proteins consisting of a cytokine receptor signaling domain linked to an FKBP12 domain may be dimerized and thereby activated with FK1012 (9).
Among the best characterized of the cytokine receptors is the receptor for erythropoietin (15). Much of what is known about the function of the erythropoietin receptor (EpoR) has been deduced from studies in transfected interleukin 3 (IL-3)-dependent murine cell lines (16–28). Ectopic expression of a functional EpoR frees these cells from the requirement for IL-3 if erythropoietin is provided. In aggregate, these studies have suggested that mitogenic signaling is mediated through the membrane-proximal portion of the EpoR cytoplasmic domain (17–19), in a region containing docking sites for the Janus family kinase JAK2 (21) and the signal transducer and activator of transcription STAT5 (26, 27).
Studies were performed to determine whether EpoR-mediated proliferative signaling could be stimulated in the IL-3-dependent murine cell line Ba/F3, by using FK1012. Results demonstrate that Ba/F3 cells expressing a membrane-targeted chimeric protein containing the intracellular domain of the EpoR linked to the FK506 binding domain of FKBP12 are capable of FK1012-dependent proliferation in the absence of IL-3. To our knowledge, these results provide the first example of cell proliferation that is dependent on a synthetic drug. In the context of the proper signaling molecule, a similar approach may have applications for gene therapy.
METHODS
Plasmid Construction.
F3, also designated pMF(PK)3E (12) is a modified form of pMF3E (9). FKBP12 has been modified to contain the mutations G89P and I90K. These mutations abrogate the ability of the FK506 complexes of this mutant FKBP to interact with calcineurin (29) and have a reduced propensity to interact with cellular proteins (12). The 708- or 309-bp fragments of the murine EpoR (15) were amplified by PCR using the plasmid pXM(EpoR)-190 (a gift of J. Prchal) as a template and the following primers: 5′-CCCCTCGAGTCCCACCGCCGGACTCTG; 3′ (708 bp), 5′-CCCCTCGAGGGAGCAGGCCACATAGC; 3′(309 bp), 5′-CCCCTCGAGCAACCACTTATCCAATAC. The PCR-amplified fragments were digested with XhoI, gel-purified, and ligated into the phosphatase-treated SalI-digested plasmids F3 or F1 to generate the plasmids F3EpoR236, F3EpoR103, and F1EpoR236. The plasmids were sequenced by using the PRISM system (Applied Biosystems). Plasmids were purified by two rounds of cesium chloride centrifugation prior to transfection.
Electroporation.
Ba/F3 cells (a gift of K. Kaushansky) were maintained in RPMI 1640 medium supplemented with glutamine, pyruvate, penicillin, streptomycin, 10% fetal calf serum, and 10% WEHI conditioned medium. Cells were split 1:2 on the day prior to transfection. Electroporations were performed essentially as described (22), with modifications. Briefly, 1 × 107 cells in 0.8 ml of phosphate-buffered saline (PBS) were mixed with 20 μg of nonlinearized test plasmid and 5 μg of the plasmid pMC1Neo (Stratagene). Electroporation was performed with the Bio-Rad GenePulser apparatus using settings of 350 V and 960 μF. After 48 h, cells were plated at limiting dilution in 96-well plates using RPMI with the above components plus G418 (700 mg/ml of active compound). Clones were expanded and maintained in G418 and WEHI-containing medium until analysis.
Western Blotting Analysis.
Cells were washed twice with PBS then approximately 1 × 106 cells were lysed in 100 μl of 50 mM Tris·HCl, pH 8/150 mM NaCl/1% Nonidet P-40/0.5% deoxycholate/0.1% SDS and placed on ice for 30 min before freezing. Cleared supernatant was assayed for protein content by using a Bradford assay (Bio-Rad) and equal amounts of protein were used for analysis. Lysates were boiled for 5 min in sample buffer containing 50 mM Tris·HCl, pH 6.8/1% 2-mercaptoethanol/1% SDS/10% glycerol immediately prior to loading and size-fractionated by electrophoresis through 7.5% polyacrylamide gels (30). After electrotransfer to nitrocelluose (Schleicher & Schuell, 0.45-μm pore size) blots were incubated with mAb HA.11 (Berkeley Antibody, Richmond, CA) and then with goat anti-mouse IgG horseradish peroxidase (Bio-Rad). Proteins were detected after a 10-sec exposure to x-ray film with ECL reagents (Amersham).
Immunoprecipitation Assay.
Polyclonal rabbit antisera against JAK2 and mAb against phosphotyrosine (4G10) were purchased from Upstate Biotechnology. Polyclonal rabbit antisera specific for STAT5B (31) was a generous gift of Chris Saris (Amgen, Thousand Oaks, CA). Cell lines were incubated at 37°C in serum-free cytokine-free medium for 16 h and then stimulated for 20 min with 100 nM FK1012 or recombinant human erythropoietin at 10 units/ml (a gift of Ortho Pharmaceuticals). The reaction was stopped by adding ice-cold PBS and washing the cells twice at 4°C. Protein lysates were prepared by solubilization in Triton X-100 as described (32). Protein concentration was determined by using the Protein/DC assay (Bio-Rad) to assure equal amounts of protein per lysate (500 μg per immunoprecipitation). Lysates were diluted 1:5 with additional lysis buffer, and then JAK2 or STAT5B antisera were added (3 μl of crude antisera per 500 μg of total protein). After a 2-h incubation at 4°C, immune complexes were collected by adding 20 μl of protein A-Sepharose beads (Santa Cruz Biotechnology) and incubating on a rocker at 4°C for 1 h. Beads were washed three times in lysis buffer and then boiled in sample buffer containing (final concentrations) 62.5 mM Tris·HCl (pH 6.8), 1% 2-mercaptoethanol, 1% SDS, and 10% glycerol. Immunoprecipitated proteins were separated on a 7.5% acrylamide gel. The blot was probed with anti-phosphotyrosine antibody (1:2000 dilution) for 2 h and a goat anti-mouse coupled to horseradish peroxidase (1:5000 dilution) for 1 h. Proteins were detected by ECL as described above.
Cell Proliferation Assay.
Ba/F3 cells were washed twice with PBS and cultured for approximately 14 h in WEHI- and G418- deficient medium. On the next morning, medium was exchanged and live cells were counted. Approximately 1 × 104 cells per well were plated in 96-well plates and medium containing either WEHI or FK1012 was added to a final volume of 100 μl. FK1012, previously designated FK1012A (9), was a gift of S. Schreiber. Plates were incubated at 37°C in 5% CO2/95% air for 40 h, and then 25 μl of a solution of MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) (Sigma) at 5 mg/ml was added to each well. After thorough mixing, cells were incubated for an additional 5 h at 37°C with 5% CO2/95% air, then 100 μl of lysis buffer (20% SDS/40% dimethylformamide/2% glacial acetic acid, pH 4.7) was added, and plates were incubated at 37°C for an additional 2 h prior to assay. OD570–630 value was determined with an ELISA plate reader.
RESULTS
Development of an FK1012-Dependent Cell Line.
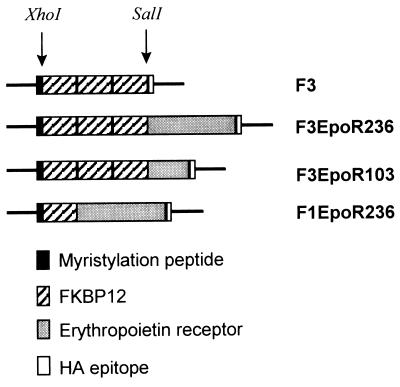
Studies were performed to determine whether genetically modified cells can be induced to proliferate with FK1012. To test the feasibility of this approach, the construct F3EpoR236 was produced by inserting the cytoplasmic domain of the murine EpoR into the SalI site of the plasmid F3, previously designated pMFpk3E (9, 12) (Fig. 1). F3EpoR236 encodes a chimeric protein containing a 14-amino acid myristylation-targeting domain from c-Src (33) to direct localization to the inner surface of the cell membrane, three copies of the 107-amino acid FKBP12 (34) to bind the drug FK506, the entire 236-amino acid intracellular domain of the EpoR (15), and a 9-amino acid influenza hemagglutinin epitope tag (35) to allow detection of the recombinant protein by Western blot analysis.
Figure 1.
Schematic representation of constructs. F3, previously designated pMFPK3E (12), is a modified version of pMF3E (9), where the FKBP12 domain has been modified to contain the mutations G89P and I90K (12). Murine EpoR sequences encode either the full-length 236-amino acid cytoplasmic domain or the membrane-proximal 103-amino acid cytoplasmic domain.
Ba/F3 cells are an IL-3-dependent murine pro-B-cell line (36). Ectopic expression of a functional EpoR releases these cells from the requirement for IL-3 if erythropoietin is provided (16). Experiments were performed to determine whether Ba/F3 cells expressing the F3EpoR236 fusion protein could, after withdrawal of IL-3, be rescued by FK1012.
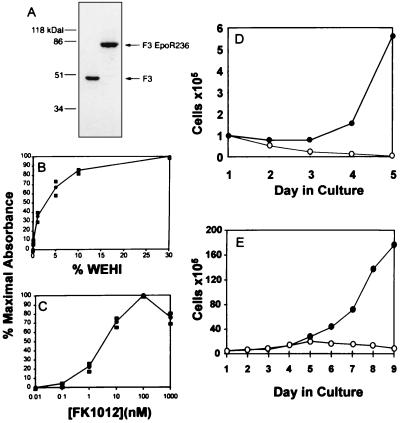
Ba/F3 cells were cotransfected with F3EpoR236 and a plasmid encoding neomycin phosphotransferase. G418-resistant clones were expanded and evaluated for expression of the chimeric protein by Western blot analysis. Twenty-eight clones were analyzed and 1 clone (clone 26) expressed high levels of the chimeric protein (Fig. 2A). The growth characteristics of this clone in response to IL-3 or FK1012 were tested in a colorimetric cell proliferation assay. As seen in Fig. 2B, proliferation did not occur in the absence of IL-3-containing WEHI conditioned medium, demonstrating that expression of the F3EpoR236 construct did not produce factor-independent growth. As expected, WEHI conditioned medium stimulated cell proliferation in a concentration-dependent manner.
Figure 2.
Development of an FK1012-dependent cell line. (A) Western blot of protein extracts from Ba/F3 cell clones expressing the F3 and F3EpoR236 constructs display protein bands of predicted sizes. (B and C) A clonal population of Ba/F3 cells expressing F3EpoR236 was washed and cultured overnight in the absence of IL-3. Cells were then counted and plated at a range of concentrations of either IL-3-containing WEHI-conditioned medium (B) or FK1012 (C). After 40 h, MTT was added. OD570–630 value was determined by using a microplate reader. Results are shown as a percentage of maximal absorbance. (D) Ba/F3 cells expressing F3EpoR236 can be passaged indefinitely in IL-3-deficient FK1012-containing medium. Numbers of cells were counted during culture without IL-3, in the absence (open circles) or in the presence (solid circles) of 100 nM FK1012. (E) FK1012’s proliferative effect is reversible. Cells grown in the presence of FK1012 (from D) were extensively washed and cultured in the absence (open circles) or presence (solid circles) of 100 nM FK1012.
Proliferation assays were performed on the same cells in the absence of IL-3, but with the addition of FK1012 at concentrations ranging from 10−2 to 104 nM. As shown in Fig. 2C, FK1012 exerted a concentration-dependent proliferative effect. Mitogenic responses were detectable at FK1012 concentrations as low as 1 nM and reached a maximum at concentrations of 100 nM. Higher concentrations of FK1012 were associated with lesser proliferative effects, possibly due to excessive occupancy of the FKBP12 sites by FK1012, thus preventing oligomerization/dimerization of the fusion proteins. As shown in Fig. 2D, FK1012 allowed cells to be expanded in culture without IL-3. In contrast, cells grown in the absence of both FK1012 and IL-3 failed to expand and died over a period of several days. The reversible nature of FK1012’s proliferative effect is shown in Fig. 2E. Cells initially grown in FK1012 underwent approximately two additional cycles of division after drug withdrawal. Nevertheless proliferation had ceased by day 5 after discontinuation of FK1012, and thereafter cell numbers slowly fell.
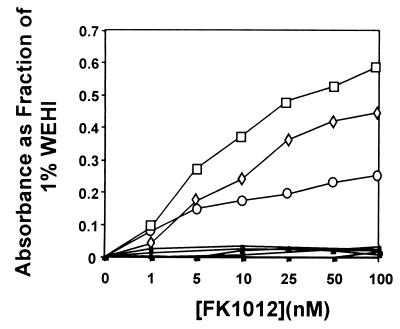
Two additional clones were isolated in an identical manner and produced similar results (Fig. 3). To allow comparisons between the proliferative effects of FK1012 and WEHI conditioned medium, results shown in Fig. 3 indicate absorbance as a fraction of that obtained in parallel cell proliferation assays using 1% WEHI conditioned medium. As controls, Ba/F3 cells were transfected with the construct F3, which does not contain the EpoR (Fig. 1). Among 18 G418-resistant clones tested, 5 were found to express the chimeric protein by Western blot analysis at levels that matched or exceeded the expression of F3EpoR236 Ba/F3 clone 26 (Fig. 2A and data not shown). All F3 clones remained IL-3-dependent; however, none of these clones was capable of proliferation in response to FK1012 (Fig. 3).
Figure 3.
MTT assays for three Ba/F3 clones expressing the F3EpoR236 construct. Each responds to FK1012 in a concentration-dependent manner (open boxes). In contrast five clones expressing the construct F3, which lacks the EpoR, fail to respond to FK1012 (solid boxes). Results are plotted as a fraction of the OD570–630 obtained in 1% WEHI conditioned medium.
FK506 Inhibits FK1012-Dependent Cell Proliferation.
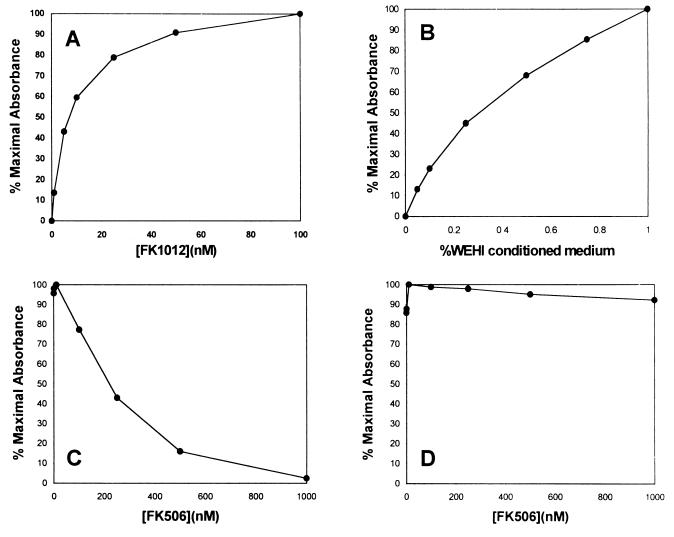
To confirm that FK1012 causes cell proliferation by bringing together individual chains of the chimeric receptor, competition assays were performed with the monomer FK506. FK506 would be expected to inhibit the proliferative effect of FK1012 by competing for binding to FKBP12 sites. In contrast, FK506 would be expected to have no effect on IL-3-mediated proliferative signaling. Results are shown in Fig. 4. Ba/F3 F3EpoR236 clone 26 cells demonstrated a proliferative response to FK1012 that reached a plateau at a drug concentration of 100 nM (Fig. 4A). Proliferation observed at the concentration of 100 nM FK1012 was similar to that obtained with 0.5% WEHI conditioned medium (Fig. 4B). In the presence of 100 nM FK1012, increasing concentrations of FK506 produced a dose-dependent inhibition of cell proliferation (Fig. 4C). At equimolar concentrations of FK506, cell proliferation was reduced by more than 75%, and proliferation was completely abolished in the presence of a 10-fold molar excess of competing monomer. To exclude a nonspecific toxic effect of FK506, the same concentrations were evaluated in clone 26 cells grown in the presence of 0.5% WEHI conditioned medium (Fig. 4D). In sharp contrast to its competitive inhibition of FK1012-dependent cell proliferation, FK506 had no effect on IL-3-mediated cell proliferation. These results confirm that FK1012-mediated aggregation of the chimeric receptor is required for proliferation.
Figure 4.
FK506 competitively inhibits the proliferative effect of FK1012 but has no effect on IL-3-dependent proliferation. MTT assays were performed with a clonal population of Ba/F3 cells expressing F3EpoR236 in the presence of increasing concentrations of FK1012 (A) or of 100 nM FK1012 and increasing concentrations of the competing monomer FK506 (B). IL-3-containing WEHI conditioned medium at the indicated concentrations produced similar levels of proliferation (C). However, in the presence of 0.5% WEHI conditioned medium, FK506 had no inhibitory effect on cell proliferation (D).
A Truncated Form of the Erythropoietin Receptor Is Sufficient for FK1012-Dependent Proliferation.
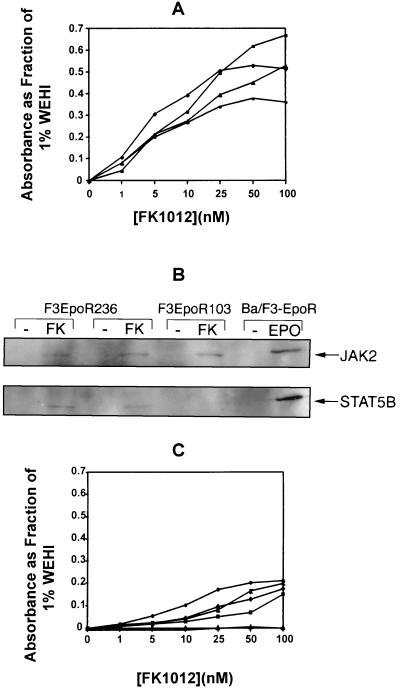
The membrane proximal portion of the EpoR cytoplasmic domain is sufficient for erythropoietin-stimulated proliferation (17–20, 25, 26). This portion of the EpoR retains binding sites for JAK2 (21) and STAT5 (26, 27). Four Ba/F3 clones expressing fusion proteins encoded by the construct F3EpoR103, which contains only 103 amino acids of the membrane-proximal EpoR cytoplasmic domain (Fig. 1) were generated. As shown in Fig. 5A, FK1012 exerted a concentration-dependent proliferative effect in the absence of IL-3. In parallel comparisons with Ba/F3 clones expressing F3EpoR236, which contains the entire EpoR cytoplasmic domain (Fig. 3), no differences in sensitivity to FK1012 were observed.
Figure 5.
(A) The membrane-proximal 103 amino acids of the EpoR intracellular domain are sufficient to mediate proliferative signaling. MTT assays were performed on four Ba/F3 cell clones expressing the construct F3EpoR103. Results are plotted as a fraction of the OD570–630 value measured from the same cells cultured in 1% WEHI conditioned medium. (B) FK1012 activates signaling pathways that are characteristic for a response to erythropoietin. Immunoprecipitations were performed with antibodies directed against JAK2 and STAT5B. After electrophoretic separation, blots were probed with an anti-phosphotyrosine antibody. (C) Receptor dimerization is sufficient to mediate proliferative signaling. MTT assays were performed for five Ba/F3 cell clones expressing the construct F1EpoR236 in the presence of increasing concentrations of FK1012. Results are plotted as a fraction of the OD570–630 value measured from the same cells cultured in 1% WEHI conditioned medium.
FK1012 Stimulates Signaling Pathways That Are Characteristic of Erythropoietin-Mediated Receptor Activation.
Studies were performed to determine whether FK1012 activates erythropoietin-mediated signaling pathways. Binding of erythropoietin to its receptor induces tyrosine phosphorylation of the Janus kinase JAK2 (21) and the transcription factor STAT5 (26, 27). In mice, STAT5 activity is encoded by two highly related chromosomally linked genes, designated STAT5A and STAT5B (37–39), both of which are tyrosine-phosphorylated in response to erythropoietin (40). Immunoprecipitation assays for JAK2 and STAT5B were performed with lysates from Ba/F3 cell clones cultured in the presence or absence of FK1012. Blots were probed with an anti-phosphotyrosine mAb. Two Ba/F3 cell clones expressing F3EpoR236 and a single Ba/F3 cell clone expressing F3EpoR103 were evaluated. Ba/F3 cells expressing the full-length EpoR (a gift of Alan D’Andrea, Harvard University) and exposed to recombinant human erythropoietin provided a positive control. Results are shown in Fig. 5B. As expected, in Ba/F3 cells expressing the full-length EpoR, both JAK2 and STAT5B were tyrosine-phosphorylated in response to erythropoietin. In two Ba/F3 cell clones expressing F3EpoR236, FK1012 activated the same signaling molecules: both JAK2 and STAT5B were tyrosine-phosphorylated. Truncation of the EpoR to 103 amino acids (F3EpoR103) had no effect on tyrosine phosphorylation of JAK2; however, tyrosine phosphorylation of STAT5 was sharply reduced. A potential explanation for this observation is that truncation of the EpoR to 103 amino acids eliminates one of two STAT5 binding sites (27).
Receptor Dimerization Is Sufficient for Proliferative Signaling.
The presence of three FKBP12 domains in the chimeric protein could allow FK1012 to stimulate proliferation either through dimerization of chimeric proteins or through the formation of higher-order oligomers. To distinguish between these possibilities, a construct was produced that contains the 236-amino acid EpoR linked to only a single copy of the FKBP12 domain (F1EpoR236) (Fig. 1). Proliferative signaling by FK1012 in cells expressing this chimeric protein can only be caused by dimerization. As shown in Fig. 5C, four of five clones expressing this construct exhibited FK1012-dependent proliferation, although the magnitude of the proliferative response was less than in Ba/F3 clones containing three copies of the FKBP12 domain (Figs. 3 and 5A). These results confirm that receptor dimerization is sufficient for mitogenic signaling.
DISCUSSION
A vast array of cellular processes are governed by the interactions of proteins. A recently developed method allows interactions between specific intracellular proteins to be reversibly controlled, providing a means for directing intracellular protein trafficking (10–12), stimulating receptor mediated signal transduction (9–12), or activating transcription (12–14). This system provides a powerful new tool for studying intracellular processes (13) and may have novel therapeutic applications.
A theoretical means of compensating for the inefficiency of stem cell transduction is to impart upon the genetically modified cells a proliferative advantage relative to their unmodified counterparts. To be effective, the proliferative stimulus must be specific for the transduced cell population; to be safe, it must be reversible. Experiments presented herein demonstrate that FK1012 can support the proliferation of transfected Ba/F3 cells in the absence of IL-3. The proliferative effect is reversible: removal of FK1012 results in the extinction of mitogenic signaling over a period of several days (Fig. 2E). Furthermore, FK506 competitively inhibits FK1012-mediated proliferation (Fig. 4). In the context of the proper signaling molecule, a similar approach may be envisioned as a method for reversibly immortalizing hematopoietic stem cells.
Although our studies show that FK1012 can mediate proliferation in a factor-dependent cell line, demonstrating a preferential proliferative effect in transduced primary cells in vivo may prove more difficult. In the system presented, withdrawal of IL-3 provides a strong selective pressure that is not reproducible in vivo. Whether FK1012 can provide a proliferative stimulus in transduced primary cells beyond that provided by a physiologic milieu of cytokines remains to be determined. Favorable features of this approach for in vivo applications are that FK1012 lacks the immunosuppressive effects of FK506 (9) and can be administered at biologically effective doses (41).
FK1012 activated signaling pathways that are characteristic of a response to erythropoietin. JAK2 was activated in Ba/F3 cells expressing fusion proteins containing either the full-length or the truncated forms of the EpoR intracellular domain, consistent with retention of the box 1 and box 2 homologous regions within the membrane-proximal 103-amino acid fragment. STAT5 activation was observed in Ba/F3 clones expressing chimeric proteins containing the full-length EpoR cytoplasmic domain; however, STAT5 was activated to a lesser extent in a Ba/F3 clone with a fusion protein containing only the 103-amino acid membrane proximal portion of the EpoR. This truncated version of the EpoR lacks a STAT5 binding site at Tyr-145 but retains a second tyrosine involved in STAT5 binding at position 96 (26, 27). Ba/F3 cells expressing the EpoR have been noted to produce β-globin in response to erythropoietin (22, 23, 28). In three separate experiments, the induction of β-globin mRNA in response to FK1012, as detected by RNase protection assays, was variable (data not shown). These results are consistent with previous studies showing that clones of Ba/F3 cells transfected with the EpoR vary in their capacity for erythropoietin-dependent β-globin synthesis (42) and that β-globin synthesis may require a very narrow range of ligand concentrations (28).
A single copy of the FKBP12 domain was sufficient to transmit a mitogenic signal, confirming results of previous studies that suggested that dimerization is sufficient for proliferative signaling (20, 24). The lower level of proliferation in the clones that we evaluated was not due to differences in level of expression (data not shown); however, it may be due to differences in the probability of dimerization in clones expressing one versus three copies of the FKBP12 domain or may in theory be due to the generation of oligomeric complexes that are more efficient in transmitting a mitogenic signal.
The ability to specifically deliver a mitogenic signal to genetically modified cells has potential therapeutic applications for a large number of tissues. Stem cells and progenitors undergo proliferative expansion and differentiation. The application of pharmacologically induced dimerization to stem-cell gene therapy will require receptors or other proteins that direct the proliferative expansion of stem cells without loss of differentiation potential. Candidate proteins that may fulfill this role include those that are activated upon homodimerization, such as c-kit (43) or flt-3 (44). In addition to attempts to exploit these proteins for therapeutic applications, pharmacological dimerization of these molecules provides a new window for examining their biological roles.
Acknowledgments
We thank S. Schreiber for providing FK1012, J. Prchal for the cDNA encoding the murine EpoR, K. Kaushansky for Ba/F3 cells, Chris Saris for the STAT5B antibody, and Richard Swank for technical advice. This work was supported by National Institutes of Health Grants 5PO1 HL53750 and 5P30 DK47754.
ABBREVIATIONS
- EpoR
erythropoietin receptor
- IL-3
interleukin 3
- MTT
3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide
References
- 1.Brenner M K, Rill D R, Holladay M S, Heslop H E, Moen R C, Buschle M, Krance R A, Santana V M, Anderson W F, Ihle J N. Lancet. 1993;342:1134–1137. doi: 10.1016/0140-6736(93)92122-a. [DOI] [PubMed] [Google Scholar]
- 2.Dunbar C E, Cottler-Fox M, O’Shaughnessy J A, Doren S, Carter C, Berenson R, Brown S, Moen R C, Greenblatt J, Stewart F M, Leitman S F, Wilson W H, Cowan K, Young N S, Nienhuis A W. Blood. 1995;85:3048–3057. [PubMed] [Google Scholar]
- 3.Aran J M, Gottesman M M, Pastan I. Proc Natl Acad Sci USA. 1994;91:3176–3180. doi: 10.1073/pnas.91.8.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Migita M, Medin J A, Qawliuk R, Jacobson S, Nagle J W, Anderson S, Amiri M, Humphries R K, Karlsson S. Proc Natl Acad Sci USA. 1995;92:12075–12079. doi: 10.1073/pnas.92.26.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorrentino B P, Brandt S J, Bodine D, Gottesman M, Pastan I, Cline A, Nienhuis A W. Science. 1992;257:99–103. doi: 10.1126/science.1352414. [DOI] [PubMed] [Google Scholar]
- 6.Blau C A, Neff T, Papayannopoulou Th. Hum Gene Ther. 1996;7:2069–2078. doi: 10.1089/hum.1996.7.17-2069. [DOI] [PubMed] [Google Scholar]
- 7.Blau, C. A., Neff, T. & Papayannopoulou, Th. Blood, in press.
- 8.Ihle J N, Witthuhn B A, Quelle F W, Yamamoto K, Silvennoinen O. Annu Rev Immunol. 1995;13:369–398. doi: 10.1146/annurev.iy.13.040195.002101. [DOI] [PubMed] [Google Scholar]
- 9.Spencer D M, Wandless T J, Schreiber S L, Crabtree G R. Science. 1993;262:1019–1034. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 10.Spencer D M, Graef I, Austin D J, Schreiber S L, Crabtree G R. Proc Natl Acad Sci USA. 1995;92:9805–9809. doi: 10.1073/pnas.92.21.9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holsinger L J, Spencer D M, Austin D J, Schreiber S L, Crabtree G R. Proc Natl Acad Sci USA. 1995;92:9810–9814. doi: 10.1073/pnas.92.21.9810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belshaw P J, Ho S N, Crabtree G R, Schreiber S L. Proc Natl Acad Sci USA. 1996;93:4604–4607. doi: 10.1073/pnas.93.10.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spencer D M. Trends Genet. 1996;12:181–187. doi: 10.1016/0168-9525(96)10013-5. [DOI] [PubMed] [Google Scholar]
- 14.Ho S N, Biggar S, Spencer D M, Schreiber S L, Crabtree G R. Nature (London) 1996;382:822–826. doi: 10.1038/382822a0. [DOI] [PubMed] [Google Scholar]
- 15.D’Andrea A D, Lodish H F, Wong G G. Cell. 1989;57:277–285. doi: 10.1016/0092-8674(89)90965-3. [DOI] [PubMed] [Google Scholar]
- 16.Li J, D’Andrea A D, Lodish H F, Baltimore D. Nature (London) 1990;343:762–764. doi: 10.1038/343762a0. [DOI] [PubMed] [Google Scholar]
- 17.D’ Andrea A D, Yoshimura A, Youssoufian H, Zon L I, Koo J-W, Lodish H F. Mol Cell Biol. 1991;11:1980–1987. doi: 10.1128/mcb.11.4.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miura O, D’Andrea A, Kabat D, Ihle J N. Mol Cell Biol. 1991;11:4895–4902. doi: 10.1128/mcb.11.10.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quelle D E, Wojchowski D M. Proc Natl Acad Sci USA. 1991;88:4801–4805. doi: 10.1073/pnas.88.11.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watowich S S, Yoshimura A, Longmore G D, Hilton D J, Yoshimura Y, Lodish H F. Proc Natl Acad Sci USA. 1992;89:2140–2144. doi: 10.1073/pnas.89.6.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Witthuhn B A, Quelle F W, Silvennoinen O, Yi T, Tang B, Miura O, Ihle J N. Cell. 1993;74:227–236. doi: 10.1016/0092-8674(93)90414-l. [DOI] [PubMed] [Google Scholar]
- 22.Liboi E, Carroll M, D’Andrea A D, Mathey-Prevot B. Proc Natl Acad Sci USA. 1993;90:11351–11355. doi: 10.1073/pnas.90.23.11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiba T, Nagata Y, Kishi A, Sakamaki K, Miyajima A, Yamamoto M, Engel J D, Todokoro K. Proc Natl Acad Sci USA. 1993;90:11593–11597. doi: 10.1073/pnas.90.24.11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watowich S S, Hilton D J, Lodish H F. Mol Cell Biol. 1994;14:3535–3549. doi: 10.1128/mcb.14.6.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maruyama K, Miyata K, Yoshimura A. J Biol Chem. 1994;269:5976–5980. [PubMed] [Google Scholar]
- 26.Damen J E, Wakao H, Miyajima A, Krosl J, Humphries R K, Culter R L, Crystal G. EMBO J. 1995;14:5557–5568. doi: 10.1002/j.1460-2075.1995.tb00243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gobert S, Chretien S, Gouilleux F, Muller O, Pallard C, Dusanter Fourt I, Groner B, Lacombe C, Gisselbrecht S, Mayeux P. EMBO J. 1996;15:69–74. [PMC free article] [PubMed] [Google Scholar]
- 28.Carroll M, Zhu Y, D’Andrea A D. Proc Natl Acad Sci USA. 1995;92:2869–2873. doi: 10.1073/pnas.92.7.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Farmer J J, Lane W S, Friedman J, Weissman I, Schreiber S L. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Mu S X, Xia M, Elliot G, Bogenberger J, Swift S, Bennett L, Lappinga D L, Hecht R, Lee R, Saris C J M. Blood. 1995;86:4532–4543. [PubMed] [Google Scholar]
- 32.Drachman J G, Griffin J D, Kaushansky K. J Biol Chem. 1995;270:4979–4982. doi: 10.1074/jbc.270.10.4979. [DOI] [PubMed] [Google Scholar]
- 33.Cross F R, Garber E A, Pellman D, Hanafusa H. Mol Cell Biol. 1984;4:1834–42. doi: 10.1128/mcb.4.9.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Standaert R F, Galat A, Verdine G L, Schreiber S L. Nature (London) 1990;346:671–674. doi: 10.1038/346671a0. [DOI] [PubMed] [Google Scholar]
- 35.Kolodziej P A, Young R A. Methods Enzymol. 1991;194:508–519. doi: 10.1016/0076-6879(91)94038-e. [DOI] [PubMed] [Google Scholar]
- 36.Palacios R, Steinmetz M. Cell. 1985;41:727–734. doi: 10.1016/s0092-8674(85)80053-2. [DOI] [PubMed] [Google Scholar]
- 37.Azam M H, Erdjument-Bromage H, Kreider B, Xia M, Quelle F W, Basu R, Saris C, Tempst P, Ihle J N, Schindler C. EMBO J. 1995;14:1402–1411. doi: 10.1002/j.1460-2075.1995.tb07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Copeland N G, Gilbert D J, Schindler C, Zhong Z, Wen Z, Darnell J E, Mui A-F, Miyajima A, Quelle F W, Ihle J N, Jenkins N A. Genomics. 1995;29:225–228. doi: 10.1006/geno.1995.1235. [DOI] [PubMed] [Google Scholar]
- 39.Mui A L, Wakao H, Harada N, O’Farrell A M, Miyajima A. J Leukocyte Biol. 1995;57:799–803. doi: 10.1002/jlb.57.5.799. [DOI] [PubMed] [Google Scholar]
- 40.Quelle F W, Wang D, Nosaka T, Thierfelder W E, Stravopodis D, Weinstein Y, Ihle J N. Mol Cell Biol. 1996;16:1622–1631. doi: 10.1128/mcb.16.4.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spencer D M, Belshaw P, Chen L, Ho S N, Randazzo F, Crabtree G R, Schreiber S L. Curr Biol. 1996;6:839–847. doi: 10.1016/s0960-9822(02)00607-3. [DOI] [PubMed] [Google Scholar]
- 42.Yoshimura A. Nature (London) 1994;372:137–138. doi: 10.1038/372137b0. [DOI] [PubMed] [Google Scholar]
- 43.Qiu F H, Ray P, Brown K, Barker P E, Jhanwar S, Ruddle F H, Besmer P. EMBO J. 1988;7:1003–1011. doi: 10.1002/j.1460-2075.1988.tb02907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dosil M, Wang S, Lemischka I R. Mol Cell Biol. 1993;13:6572–6585. doi: 10.1128/mcb.13.10.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]