Abstract
Establishment of the vertebrate body plan requires a variety of signaling molecules. In a search for tyrosine kinases expressed in early zebrafish embryos, a model system for the study of vertebrate development, we discovered Jak1 kinase to be maternally encoded and the mRNA evenly distributed among the cells of blastula-stage embryos. Injection of RNA-encoding dominant-negative Jak1 kinases reduces a specific cell migration, epiboly, and results in the reduction of goosecoid expression and of anterior structures. This work establishes that, in addition to its role in signal transduction of cytokines in adult tissues, Jak1 kinase has a role in early vertebrate development.
One of the earliest events of vertebrate development is the specification of the primordial germ layers, ectoderm, endoderm, and mesoderm, and the extensive cell migrations that immediately follow. These processes are likely to require a variety of signal transduction cascades, and several peptide growth factors have been implicated in these processes (1–3). In this study, we show that Jak1 kinase also is required for these processes.
In a model vertebrate, the zebrafish, developmental events are easily visualized and studied. In a search for signal transducers that might regulate early development, we used a degenerate PCR approach to identify tyrosine kinases expressed during zebrafish gastrulation. Other than the FGF receptor, no other tyrosine kinase has been implicated in the earliest events of vertebrate development. Using degenerate primers to conserved residues in the catalytic domain of tyrosine kinases, we have isolated a homologue of the Jak1 kinase from early gastrula embryos. This is surprising, as Jak1 kinase has been implicated thus far only in signaling for adult tissues in vertebrates.
Jak1 is one of four members of a tyrosine kinase family that possesses several features in common (4, 5). The Jak family members, Jak1, Jak2, Jak3, and Tyk2, lack SH2 and SH3 domains, are cytoplasmic, and possess two kinase domains. The N-terminal kinase domain has only limited homology to the phosphotransferase domains of other tyrosine kinases and the isolated domain, expressed as a glutathione S-transferase (GST) fusion, does not possess kinase activity (6). The C-terminal domain, however, possesses all the motifs found in tyrosine kinase domains and this domain, fused to GST, has phosphotransferase activity (6).
Jak1 kinase is ubiquitously expressed in adult mouse tissues (6) and has been shown to transduce signals from a variety of cytokines (4, 5). Its developmental expression, however, has never been analyzed. Jak1 has been thought to function exclusively in adults, primarily in the modulation of immune function. We show that during early development, Jak1 kinase is exclusively of maternal origin, and through the use of dominant negative Jak1 kinases, that this kinase is required for cell migrations, goosecoid expression, and anterior shield formation. In agreement with our findings, recent studies have shown that Jak kinase plays a role in early Drosophila development (7). We now show that this kinase also functions during early vertebrate development.
MATERIALS AND METHODS
Isolation of Zebrafish Jak1 Kinase, Reverse Transcription–PCR (RT–PCR), and in Situ Hybridization.
Jak1 kinase was isolated using mRNA from 6-h embryos using degenerate PCR, as described (8). Total RNA, isolated from staged embryos or adults, served as template for RT–PCR as described (9). The minus-RT control contained adult RNA, however, reverse transcriptase was omitted from the reaction. Oligonucleotide primers complementary to EF1 alpha have been described (9). Jak1 primers were 5′-ATTGGAGACTTCGGCCTGAC-3′ and 5′-GGGTGTTGCTTCCCAGCATC-3′. Blots were probed with an oligonucleotide complementary to a region between the PCR primers of either Jak1 or EF1 alpha. The oligonucleotide probe for Jak1 was 5′-GTGACCATGTATGAGCTCCT-3′. Filters were subjected to autoradiography and bands quantitated using a Fuji bio-image analyzer. For in situ hybridization, staged embryos were probed with either sense or antisense digoxigenin-labeled RNA probes to Jak1 kinase as described (9).
Site-Directed Mutagenesis.
JakKE was constructed by site-directed mutagenesis using sequential PCR as described (10), using 5′-AGACGCTGACTCAAGGAGTG-3′ and the mutagenic oligonucleotide: 5′-CTGGCTTCAGAGACTCCACAGCCACTAGCTC-3′ in one PCR reaction. The second PCR reaction was performed using the mutagenic oligonucleotide: 5′-AGTGGCTGTGGAGTCTCTGAA-3′ and an oligonucleotide complementary to a sequence in the T7 promoter of pBluescript: 5′-TAATACGACTCACTATAGGG-3′ located 3′ to the Jak1 insert. JakΔKin was constructed by deleting downstream sequences from the XmnI site at position 2810 of the nucleotide sequence.
Baculovirus Expression.
Baculovirus plasmids were constructed in the vector pVL1393, containing maltose-binding protein upstream from the polylinker sequence. Cloning, infection, and cell lysis were performed as described (11). Sf9 cell lysates were purified over amylose resin (New England Biolabs) as described by the manufacturer. Western blot analysis with the antiphosphotyrosine antibody RC20 was performed as described by the manufacturer (Transduction Laboratories, Lexington, KY).
RNA Injections.
Genes encoding JakWt, JakKE, or JakΔKin were cloned into the expression vector pSP64T (12). Capped RNA was synthesized in vitro from these constructs, and 100 pg were injected into one-cell embryos. Embryos then were fixed and analyzed for the expression of ntl, axial, or goosecoid. Ntl expression was assessed using an antibody to ntl protein as described (13). Axial and goosecoid were analyzed by in situ hybridization (14, 15).
RESULTS
Isolation of Jak1 Kinase and Its Temporal Expression During Embryogenesis.
Using degenerate oligonucleotides complementary to the catalytic domain of tyrosine kinases, we performed PCR using cDNA from gastrula embryos as template. The PCR products were cloned, and using one of these clones we isolated the corresponding cDNA from a gastrula cDNA library. Comparison of this sequence with the GenBank database revealed that the kinase we isolated is a Jak1 kinase. Zebrafish Jak1 is 62% identical to human Jak1 at the amino acid level and is most similar to carp Jak1 (86% identity).
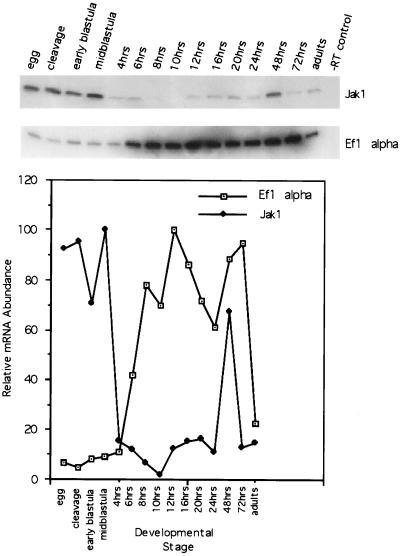
We assayed the levels of Jak1 mRNA in developmental stages of zebrafish using RT–PCR (Fig. 1). The maternal transcript for Jak1 mRNA is stored in unfertilized eggs and is the exclusive source of Jak1 mRNA during early zebrafish development. Maternal transcripts remain roughly constant through the midblastula stage (31/3 h of development). Less than 1 h later, at 4 h of development, the level of Jak1 mRNA drops precipitously, coincident with the midblastula transition (MBT) and the induction of zygotic transcription.
Figure 1.
Temporal expression of Jak1 kinase during zebrafish development. RT–PCR analysis was used to compare the levels of Jak1 mRNA to the housekeeping gene EF1 alpha. The mRNA abundance for Jak1 and EF1 alpha is shown as expression relative to the maximally observed value for that mRNA species from the developmental stages examined. Jak1 mRNA is of maternal origin and rapidly decreases at the beginning of MBT, whereas EF1 alpha mRNA levels increase at the beginning of zygotic gene expression. Hours of development indicate times postfertilization at 28°C.
We compared the levels of Jak1 mRNA to the levels of EF1 alpha mRNA, a housekeeping gene required for translation. Maternally encoded transcripts for EF1 alpha, remain constant through early stages, however, rather than decreasing, this mRNA species increases at the beginning of gastrulation (6 h of development), slightly behind the MBT. These results, and the developmental profile of other genes, argue against the generalized degradation of mRNA at MBT, and suggest that there is a regulated and specific degradation of maternal Jak1 mRNA. The level of Jak1 mRNA continues to decrease until this RNA is undetectable at 10 h of development. At 12 h of development, Jak1 mRNA levels again increase. At 48 h of development the mRNA level dramatically increases. In contrast, the level of EF1 alpha remains constant from 8 h to 72 h of development. In agreement with studies on mice, Jak1 mRNA is found in adults.
Spatial Expression of Jak1 Kinase During Early Embryogenesis.
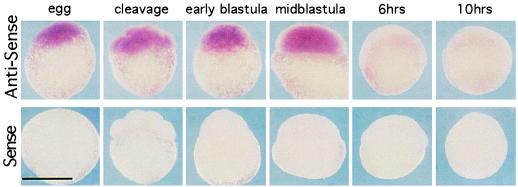
To determine the spatial localization of Jak1 mRNA, we performed whole-mount in situ hybridization with an antisense RNA probe. Fig. 2 shows that Jak1 mRNA appears in unfertilized eggs and is present through blastula stage embryos. The distribution of this RNA is uniform among all blastomeres. In agreement with the RT–PCR analysis, Jak1 mRNA dramatically decreases after the blastula stage and is undetectable until 48 h of development, where this RNA is concentrated in the region of the gill arches (data not shown).
Figure 2.
Spatial expression of Jak1 kinase during early embryogenesis. The developmental profile of Jak1 RNA was revealed by in situ hybridization, using digoxigenin-labeled antisense RNA probes. In agreement with results from RT–PCR quantitation, Jak1 kinase is present through the blastula stage of development and then rapidly disappears. The message is uniformly distributed among all blastomeres. The scale bar equals 0.5 mm.
Construction of Defective Jak1 Kinases.
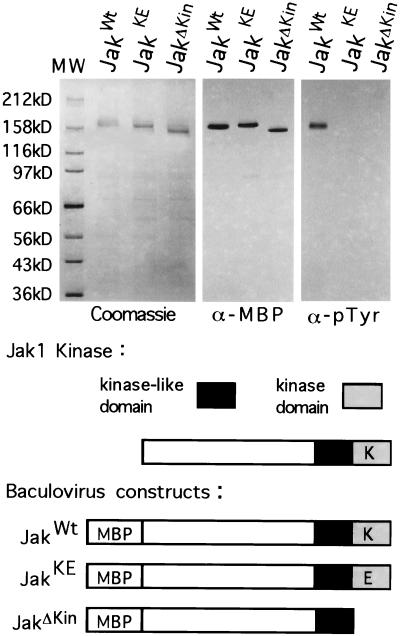
Jak1 signaling occurs by transphosphorylation, thus a dominant–negative approach is possible. Previous studies have used inactive Jak kinases as dominant–negatives to inhibit Jak signaling pathways (16, 17). To assess the role of the Jak1 kinase in early development we constructed two kinase inactive mutants. One construct, JakKE contains a single base change at position 2915, changing a lysine residue to a glutamic acid. All protein kinases possess an invariant lysine in the ATP binding domain that is directly involved in the phosphotransferase reaction. Amino acid substitutions have been made for this lysine in a number of kinases, including Jak2, another member of the Jak kinase family, and in all cases this abolishes kinase activity (18). A second mutant, JakΔKin, was constructed by deleting the C-terminal kinase domain.
To insure that these constructs are kinase deficient, we produced wild-type Jak1 (JakWt), JakKE, and JakΔKin in a baculovirus expression system (Fig. 3). We purified Jak1 proteins from infected Sf9 cells and probed them with an antiphosphotyrosine antibody. Only JakWt possesses phosphotyrosine. Jak proteins are known to autophosphorylate on tyrosine, and this phosphorylation is thought to activate the phosphorylation of exogenous substrates. Normally, autophosphorylation requires ligand binding by Jak1-associated receptors; however, in the baculovirus expression system, the high concentration of Jak1 protein leads to autophosphorylation. Autophosphorylation has been observed by others for Jak2 expressed by baculovirus, similarly a Jak2 lysine to glutamic acid mutant expressed by baculovirus fails to autophosphorylate (18). We detect no phosphorylation for mutant JakKE and also no phosphotyrosine in JakΔKin, which lacks the kinase domain.
Figure 3.
Analysis of the kinase activity of wild-type and mutant Jak1 kinases. Wild-type and mutant Jak1 kinase genes were cloned into a baculovirus expression vector and expressed as a fusion protein with maltose binding protein (MBP), to aid in purification. The JakKE mutant possesses a point mutation at position 2915, changing a lysine (K) to a glutamic acid (E). The JakΔKin mutant was constructed by deleting the C-terminal kinase domain. The fusion proteins were purified to near homogeneity on amylose resin from infected Sf9 cells, and showed little degradation as revealed by Coomassie blue staining and Western analysis with anti-MBP antisera. Purified fusion proteins were analyzed for the presence of phosphotyrosine resulting from autophosphorylation. Only the wild-type kinase possesses phosphotyrosine.
Defective Jak1 Kinases Inhibit Cell Movements of Epiboly and Reduce Anterior Structures.
We injected one-cell embryos with 100 pg of in vitro-transcribed RNA from constructs encoding the JakWt, JakKE, or JakΔKin genes. Until 4 h of development, embryos injected with either of these RNAs appear identical to uninjected embryos. However, at 4 h of development, the first effects upon development appear. At this time cell migrations begin, which are impaired by dominant–negative Jak1 kinases.
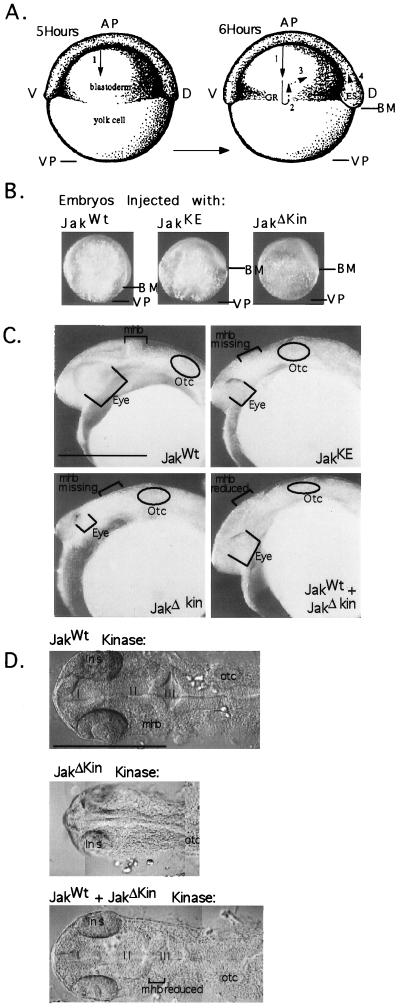
During gastrulation, four cell movements occur. The first of these movements is epiboly (Fig. 4A), blastomeres move down the surface of the centrally located yolk cell from the animal pole toward the vegetal pole, eventually covering the entire surface of the yolk cell, a process that takes 10 h to complete (arrow 1). Shortly after 6 h of development, three additional cell movements begin. During involution, cells at the blastoderm margin curl underneath and migrate between the yolk cell and overlying cells, producing a localized thickening of cells around the equator of the embryo called the germ ring (arrow 2). As involution and epiboly proceed, both involuted and noninvoluted cells move toward the future dorsal side of the embryo in a process known as convergence (arrow 3), producing a thickening of the germ ring known as the embryonic shield. Cells in the embryonic shield undergo a fourth cell movement, known as extension (arrow 4), causing an elongation of the embryonic shield. The four cell movements of gastrulation (epiboly, involution, convergence, and extension) transform a ball of cells into an elongated structure with the major body axes.
Figure 4.
Defective Jak1 kinases inhibit cell movements of epiboly and reduce anterior structures. (A) During early zebrafish development four cell movements occur. Cells of the blastoderm move down the surface of the large yolk cell in a migration called epiboly (arrow 1). Once half of the yolk surface is covered, cells begin three additional cell movements. Involution (arrow 2) results in the formation of a localized thickening of cells known as the germ ring. Convergence of cells (arrow 3) toward the future dorsal side of the embryo creates the embryonic shield and extension (arrow 4) elongates the body axis. (B) One-cell embryos were injected with 100 pg of in vitro-synthesized RNA encoding JakWt, JakKE, and JakΔKin kinases. Until 4 h of development all embryos develop normally, however, at 4 h of development, when the cell migration of epiboly begins, embryos injected with defective forms of Jak1 kinase show impaired cell migration. At 8 h of development (shown above), embryos injected with JakWT have 80% of the yolk surface covered by blastomeres; in contrast, embryos injected with defective forms of Jak1 kinase have 50% of the yolk covered. The blastoderm margin, and hence the extent of cell migration, is indicated by the arrow marked BM. The vegetal pole (VP) is indicated. (C) By 24 h of development (lateral view), embryos injected with defective forms of Jak1 kinase exhibit reduced anterior structures; scale bar equals 0.5 mm. (D) In a dorsal view with Nomarski optics, these embryos possess small eyes, and neural structures anterior to the otocyst develop as a fused neural tube lacking ventricles and a midhind brain boundary. Injection of JakWT and JakΔKin gives partial rescue of anterior structures; scale bar equals 0.25 mm. AP, animal pole; VP, vegetal pole; V, ventral; D, dorsal; GR, germ ring; ES, embryonic shield; BM, blastoderm margin; Otc, otocyst; lns, lens; I, II, III, ventricles I, II, III; mhb, midhind brain boundary; scale bar equals 0.5 mm.
When epiboly commences, embryos injected with RNA from mutant Jak1 constructs show slower cell movements (Fig. 4B); however, the yolk is ultimately covered, epiboly taking 16 h to complete rather than the usual 10 h (Table 1). By 24 h of development, embryos injected with JakWt RNA appear identical to uninjected embryos; however, embryos injected with mutant forms of Jak1 kinase possess anterior defects (Table 1). These embryos have small eyes, and neural structures anterior to the otocyst are reduced, developing as a fused neural tube, lacking ventricles and a midhind brain boundary (Fig. 4 C and D). Despite neural defects, the otocysts develop normally, including more posterior structures, such as the spinal cord and somites. Injection of JakWt and JakΔKin in a 3:1 ratio yields partial rescue of anterior structures; however, the midhind brain boundary is still reduced (Table 1).
Table 1.
Effects of wild-type and defective Jak1 kinases on zebrafish development
| Jak1 kinase | Epiboly defect*
|
Anterior defect†
|
||||
|---|---|---|---|---|---|---|
| No. | n | % | No. | n | % | |
| JakWt | 9 | 234 | 4 | 0 | 35 | 0 |
| JakKE | 90 | 116 | 76 | 23 | 32 | 72 |
| JakΔkin | 150 | 205 | 73 | 37 | 53 | 70 |
| JakWt + JakΔkin | 40 | 148 | 27 | 10 | 36 | 28 |
Embryos that did not complete epiboly by 12 h.
Embryos allowed to develop for 24 h that possess reduced eye and head structures.
Defective Jak1 Kinases Reduce Goosecoid Expression and Anterior Shield Formation.
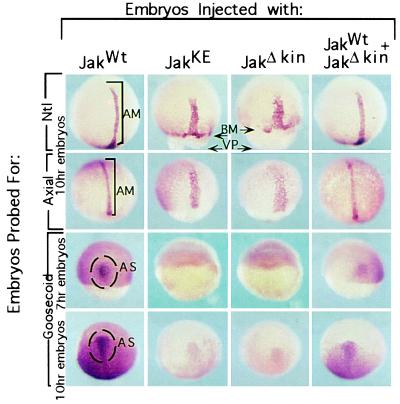
Mesendodermal cells are the first cells to undergo migrations. The cell migration defect seen in embryos injected with Jak1 mutants might be a secondary consequence of improper mesodermal induction. To assess mesoderm formation, we probed embryos injected with RNA from Jak1 constructs for expression of the mesodermal markers: ntl, axial, and goosecoid (Fig. 5).
Figure 5.
Analysis of mesodermal markers in injected embryos. Embryos injected with JakWt RNA possess a band of axial mesoderm (AM), which expresses ntl and axial, extending anteriorly from the vegetal pole (VP). Embryos injected with mutant forms of Jak1 kinase possess broader expression domains for these genes, which are twice as wide and extend half as far anteriorly. Reduced epibolic movements are evident in embryos injected with mutant forms of Jak kinase by the gap between the ntl expressing cells of the blastoderm margin (BM) and the vegetal pole. In a rescue experiment, JakWt and JakΔKin RNAs were injected together in a 3:1 molar ratio. This yields partial rescue, with narrower expression domains that extend more anteriorly. Embryos injected with JakWt RNA show goosecoid staining marking the anterior shield (AS). Embryos injected with mutant forms of Jak1 kinase show reduced goosecoid expression.
These three genes are transcription factors and have unique expression domains. Ntl, the zebrafish homologue of brachyury, is expressed in the mesendodermal cells of the germ ring and later in a narrow band of axial mesoderm extending to the mesencephalon anlage, marking precursors of the future notocord (13). The axial expression domain includes cells expressing ntl, but extends to more anterior axial mesoderm, as far as the diencephalon (14). Goosecoid is expressed in dorsal mesoderm, which populates the most anterior axial mesoderm, eventually giving rise to the precordal plate and pharyngeal endoderm (15). Analysis of these three genes allows us to assess posterior to anterior axial mesoderm formation.
As shown in Fig. 5, 10-h embryos injected with JakWt RNA at the one-cell stage possess a strip of axial mesoderm expressing ntl approximately 5 cells wide extending anteriorly from the vegetal pole. Embryos injected with mutant forms of Jak1 kinase, and showing reduced epibolic movements, possess a broader expression domain, which is twice as wide and extends only half the anterior distance, compared with embryos injected with RNA from the wild-type construct. The broader strip of axial mesoderm is explained by reduced extension of the embryonic shield. An analysis of axial expression gives similar findings. Again, the expression domain is broader and does not extend as far anteriorly. The broader and shortened expression domains for ntl and axial always are accompanied by reduced epibolic movements. These findings indicate that posterior axial mesoderm forms; however, its placement is perturbed by reduced cell movements.
The observation that developmental defects are seen only when mutant forms of Jak1 kinase are injected, and not wild-type kinase, indicates that the defects observed are due to an inactive Jak1 kinase and not a microinjection artifact. A rescue experiment further confirms the specific nature of these defects. We injected JakWt RNA with JakΔKin RNA in a 3:1 molar ratio and of these embryos, only 27% (n = 148) display reduced epibolic movements. The remainder complete epiboly by 12 h and possess narrower expression domains for ntl and axial compared with embryos injected with JakΔKin alone.
Analysis of goosecoid expression allows us to examine anterior axial mesoderm. A domain of goosecoid expression marking the anterior embryonic shield is apparent in 7-h embryos injected with JakWt RNA. Embryos injected with RNA-encoding mutant forms of Jak1 kinase fail to show a region of goosecoid expression, and reduced goosecoid expression always is accompanied by reduced epibolic movements. Injection of JakWt and JakΔKin RNA in a 3:1 molar ratio rescues goosecoid expression, though the level of expression is still less than embryos injected with JakWt RNA alone. The possibility that goosecoid expression is simply delayed in embryos injected with Jak1 mutants is countered by examination of older embryos. At 10 h of development the anterior shield has moved further and takes on a triangular appearance in JakWt-injected embryos. Even at this later time, embryos injected with mutant forms of Jak1 kinase have reduced goosecoid expression; only a small anterior shield of goosecoid expressing cells is seen. Given numerous studies that have shown that anterior mesoderm can induce anterior neural structures (19–22), the neural defects observed later in development can be attributed to reduced anterior-axial mesoderm.
DISCUSSION
Injection of a dominant-negative Jak1 kinase results in three developmental defects: reduced epibolic movements and extension of the embryonic shield, reduced goosecoid expression, and reduced anterior structures. These three developmental defects always are seen together, and it is possible that any one of these defects leads to the appearance of the other two. The primary effect of a dominant–negative Jak1 kinase may be to reduce cell movements, which because of improper cell placement, affects goosecoid expression and anterior shield formation.
Alternatively, the primary defect may be in reducing goosecoid expression, resulting in reduced anterior shield formation. Ectopic expression of goosecoid in Xenopus leads to axis duplication (23–25); however, loss-of-function studies have not been reported in Xenopus, so it is unclear if reduced or absent goosecoid expression would directly lead to a reduction or loss of anterior axial mesoderm. Goosecoid-expressing cells of the dorso-anterior mesoderm are the first cells to involute, are at the leading edge of axial mesoderm migration, and may “set the pace” for extension movements. In Xenopus embryos, goosecoid has been show to control cell migrations during gastrulation (26). Ectopic expression of goosecoid leads to cell movement toward the anterior of the embryo, not only in cells expressing ectopic goosecoid, but also in neighboring nonexpressing cells, which are recruited to follow.
Jak1 kinase is one of several transduction molecules that modulate goosecoid expression. Studies in zebrafish and Xenopus have shown that activin, Wint8, and nodal can induce ectopic goosecoid expression (24, 27–31). Jak kinases transduce signals by the phosphorylation of STAT (signal transducers and activators of transcription) proteins. STAT proteins, of which six mammalian forms are known to date, reside in the cytoplasm, until they are tyrosine phosphorylated. Phosphorylation leads to hetero or homo dimerization, which triggers nuclear translocation. Once located in the nucleus, these proteins bind target sequences and activate transcription (4, 5).
Interestingly, the promoter region of the goosecoid gene, from the three species (zebrafish, Xenopus, and mouse) from which have it has been sequenced (32, 33), all possess STAT DNA-binding sites. Jak1 kinase may synergize with signals from other transduction pathways on common STATs to yield maximal goosecoid expression. In this regard, during interferon signaling, the Jak1 kinase and MAP kinase pathways converge on a single STAT, and phosphorylation of this STAT by both pathways is required for maximal cytokine response (34). Elucidation of the upstream effectors and downstream targets of Jak1 kinase (ligands, receptors, and STATs) will clarify its mode of action in early development.
A defective Jak1 signaling pathway may reduce anterior axial mesoderm by affecting genes other than goosecoid. This would result in the appearance of reduced goosecoid levels. As mentioned above, cells of the anterior shield are the first cells to undergo cell movements, and a reduced number of these cells might reduce the pace of cell migrations. In any case, reduced anterior axial mesoderm would account for the reduced anterior structures observed later in development.
Will the role of Jak1 kinase in early zebrafish embryos be conserved among other species? A homologue of Jak kinase exists in Drosophila, called hopscotch (7). This gene is also maternally expressed, and mutations in this gene lead to early patterning defects. Hopscotch signals through a STAT protein called marelle, and mutations in marelle lead to a phenotype identical to the hopscotch mutation (35, 36). It is difficult, however, to make comparisons between the role of hopscotch in Drosophila and Jak1 in zebrafish, given the very different developmental programs in these two species. Though different species of vertebrates show a diversity of gastrulation processes, the basic mechanisms, and molecular players, TGFβs, FGF, and Wnts, appear conserved. Given the conservation of these transduction pathways, it is likely that Jak1 kinase plays a role in the early development of other vertebrates.
Acknowledgments
We thank members of the Gilbert and Roberts laboratories for helpful discussions. Thanks to Doug Melton and members of his laboratory for helpful advice and use of equipment. G.C. was supported by an American Cancer Society postdoctoral fellowship, and he was a Medical Foundation/Charles A. King Trust Research Fellow. This work was supported in part by grants from the National Institutes of Health to W.G. and R.J.R.
ABBREVIATIONS
- MBT
midblastula transition
- STAT
signal transducers and activators of transcription
- RT–PCR
reverse transcription–PCR.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U8290).
References
- 1.Smith J C. EMBO J. 1993;12:4463–4470. doi: 10.1002/j.1460-2075.1993.tb06135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kessler D S, Melton D A. Science. 1994;266:596–604. doi: 10.1126/science.7939714. [DOI] [PubMed] [Google Scholar]
- 3.Slack J M W. Curr Biol. 1994;4:116–126. doi: 10.1016/s0960-9822(94)00027-8. [DOI] [PubMed] [Google Scholar]
- 4.Schindler C, Darnell J E., Jr Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 5.Ihle J N. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 6.Wilks A F, Harpur A G, Kurban R R, Ralph S J, Zurcher G, Zeimiecki A. Mol Cell Biol. 1991;11:2057–2065. doi: 10.1128/mcb.11.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binari R, Perrimon N. Genes Dev. 1994;8:300–312. doi: 10.1101/gad.8.3.300. [DOI] [PubMed] [Google Scholar]
- 8.Wilks A F. Proc Natl Acad Sci USA. 1989;86:1603–1607. doi: 10.1073/pnas.86.5.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conway G. Mech Dev. 1995;52:383–391. doi: 10.1016/0925-4773(95)00416-x. [DOI] [PubMed] [Google Scholar]
- 10.Cormack B. In: Current Protocols in Molecular Biology. Ausubel E M, Brent R, Kinston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. New York: Wiley; 1991. Suppl. 15, pp. 8.5.7.–8.5.9. [Google Scholar]
- 11.Gruenwald S, Heitz J. Baculovirus Expression Vector System: Procedures and Methods Manual. San Diego: PharMingen; 1993. [Google Scholar]
- 12.Krieg P A, Melton D A. Nucleic Acids Res. 1984;12:7057–7069. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulte-Merker S, Ho R K, Herrman B G, Nusslein-Volhard C. Development (Cambridge, UK) 1992;116:1021–1032. doi: 10.1242/dev.116.4.1021. [DOI] [PubMed] [Google Scholar]
- 14.Strahle U, Blader P, Henrique D, Ingham P W. Genes Dev. 1993;7:1436–1446. doi: 10.1101/gad.7.7b.1436. [DOI] [PubMed] [Google Scholar]
- 15.Stachel S E, Grunwald D J, Myers P Z. Development (Cambridge, UK) 1993;117:1261–1274. doi: 10.1242/dev.117.4.1261. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Patel S V, He T, Sonsteby S K, Niu Z, Wojchowdki D M. J Biol Chem. 1994;269:21411–21414. [PubMed] [Google Scholar]
- 17.Guschin D, Rogers N, Briscoe J, Witthuhn B, Watling D, Horn F, Pellegrini S, Yasukawa K, Heinrich P, Stark G R, Ihle J N, Kerr I M. EMBO J. 1995;14:1421–1429. doi: 10.1002/j.1460-2075.1995.tb07128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quelle F W, Thierfelder W, Witthuhn B A, Tang B, Cohen S, Ihle J N. J Biol Chem. 1995;270:20775–20780. doi: 10.1074/jbc.270.35.20775. [DOI] [PubMed] [Google Scholar]
- 19.Sharpe C R, Gurdon J B. Development (Cambridge, UK) 1990;109:765–774. doi: 10.1242/dev.109.4.765. [DOI] [PubMed] [Google Scholar]
- 20.Hemmati-Brivanlou, Stewart R M, Harland R M. Science. 1990;250:800–802. doi: 10.1126/science.1978411. [DOI] [PubMed] [Google Scholar]
- 21.Saha M S, Grainger R M. Neuron. 1992;8:1003–1014. doi: 10.1016/0896-6273(92)90123-u. [DOI] [PubMed] [Google Scholar]
- 22.Ang S L, Rossant J. Development (Cambridge, UK) 1993;118:139–149. doi: 10.1242/dev.118.1.139. [DOI] [PubMed] [Google Scholar]
- 23.Cho K W, Blumberg B, Steinbeisser H, DeRobertis E M. Cell. 1991;67:1111–1120. doi: 10.1016/0092-8674(91)90288-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinbeisser H, DeRobertis E M, Ku M, Kessler D S, Melton D A. Development (Cambridge, UK) 1993;118:499–507. doi: 10.1242/dev.118.2.499. [DOI] [PubMed] [Google Scholar]
- 25.Niehrs C, Steinbeisser H, DeRobertis E M. Science. 1994;263:817–820. doi: 10.1126/science.7905664. [DOI] [PubMed] [Google Scholar]
- 26.Niehrs C, Keller R, Cho K W, DeRobertis E M. Cell. 1993;72:491–503. doi: 10.1016/0092-8674(93)90069-3. [DOI] [PubMed] [Google Scholar]
- 27.Tadano T, Otani H, Taira M, Dawid I B. Dev Genet. 1993;14:204–211. doi: 10.1002/dvg.1020140307. [DOI] [PubMed] [Google Scholar]
- 28.Green J B, Smith J C, Gerhart J C. Development (Cambridge, UK) 1994;120:2271–2278. doi: 10.1242/dev.120.8.2271. [DOI] [PubMed] [Google Scholar]
- 29.Symes K, Yordin C, Mercola M. Development (Cambridge, UK) 1994;120:2339–2346. doi: 10.1242/dev.120.8.2339. [DOI] [PubMed] [Google Scholar]
- 30.Kelly G M, Greenstein P, Erezyilmas D F, Moon R T. Development (Cambridge, UK) 1995;121:1787–1799. doi: 10.1242/dev.121.6.1787. [DOI] [PubMed] [Google Scholar]
- 31.Toyama R, O’Connel M L, Wright C V E, Kuehn M, Dawid I B. Development (Cambridge, UK) 1995;121:383–391. doi: 10.1242/dev.121.2.383. [DOI] [PubMed] [Google Scholar]
- 32.Watabe T, Kim S, Candia A, Rothbacher U, Hashimoto C, Inoue K, Cho K W Y. Genes Dev. 1995;9:3038–3050. doi: 10.1101/gad.9.24.3038. [DOI] [PubMed] [Google Scholar]
- 33.Joore J, Fasciana C, Speksnijder J E, Kruijer W, Destree O H J, van den Eijnden-vanRaaij A J M, deLaat S W, Zivkovic D. Mech Dev. 1996;55:3–18. doi: 10.1016/0925-4773(95)00481-5. [DOI] [PubMed] [Google Scholar]
- 34.David M, Petricoin D, Benjamin C, Pine R, Weber M J, Larner A C. Science. 1995;269:1721–1723. doi: 10.1126/science.7569900. [DOI] [PubMed] [Google Scholar]
- 35.Hou X S, Melnick M B, Perrimon N. Cell. 1996;84:411–419. doi: 10.1016/s0092-8674(00)81286-6. [DOI] [PubMed] [Google Scholar]
- 36.Yan R, Small S, Desplan C, Dearolf C R, Darnell J E. Cell. 1996;84:421–430. doi: 10.1016/s0092-8674(00)81287-8. [DOI] [PubMed] [Google Scholar]