Abstract
Development of the vertebrate eye has been found to require the activity of several genes encoding homeodomain proteins (Freund, C., Horsford, D. J. & McInnes, R. R. (1996) Hum. Mol. Genet. 5, 1471–1488). Some of these genes, or portions thereof, are highly conserved across phyla. In this paper, we report the identification of a novel homeobox gene, rax (retina and anterior neural fold homeobox), whose expression pattern suggests an important role in eye development. The predicted amino acid sequence of Rax comprises a protein with a paired-type homeobox, as well as the octapeptide that is found in many paired-type homeobox genes. In addition, in the C terminus of Rax, we found a 15-aa domain that we have named the OAR domain. This domain is also found in several other homeobox genes. In the early mouse embryo, rax is expressed in the anterior neural fold, including areas that will give rise to the ventral forebrain and optic vesicles. Once the optic vesicles form, rax expression is restricted to the ventral diencephalon and the optic vesicles. At later stages, rax expression is found only in the developing retina. After birth, the expression of rax is restricted to the zone of proliferating cells within the retina, and expression gradually decreases as proliferation declines. These findings suggest that rax is one of the molecules that define the eye field during early development and that it has a role in the proliferation and/or differentiation of retinal cells.
During vertebrate development, the central nervous system emerges as a highly complex, patterned structure with an enormous diversity of neuronal and glial cell types. The vertebrate retina is a relatively well described and accessible structure that provides an excellent model system for studies of patterning and cell fate determination within the central nervous system (1). In the early stages of eye development, the optic vesicles evaginate from the ventral forebrain, making contact with the overlying surface ectoderm at embryonic day 9 (E9) in the mouse. Subsequently, the optic vesicle invaginates to form the optic cup, while overlying surface ectoderm gives rise to the lens placode (2, 3). Retinal neurogenesis then proceeds within the inner layer of the optic cup. The mitotic progenitors within the optic cup line the surface apposed to the former lumen of the neural tube, in an area known as the ventricular zone (VZ). Postmitotic progeny of the retina migrate away from the VZ as they differentiate to form the laminar structure of the mature retina. While the different retinal cells are born in a sequence conserved among many species, more than one cell type is born at any one time (4). Lineage analyses in several species have shown that retinal progenitors remain multipotent throughout development (5–8).
The aim of the present study was to identify genes involved in the regulation of retinal development. We have focused on several gene families known to have critical roles in the development of both vertebrate and invertebrate organisms, including those encoding homeodomain proteins. Previous work has indicated that several genes encoding homeodomain proteins are required for proper retinal development (9). pax6 (10, 11), a gene encoding a homeodomain as well as a paired box, has been found to be required for proper development of several ocular tissues in several mammalian species (12–17). In addition, the Drosophila homolog of pax6, eyeless (18), has been found not only to be required for eye development, but to be able to induce ectopic eyes when misexpressed (19). Remarkably, the pax6 gene of mouse also has this activity when misexpressed in Drosophila (19), although it is not known whether it can induce extra eyes when ectopically expressed in vertebrates. Although pax6 is clearly a critical element in eye specification, it cannot be sufficient as it is expressed in multiple locations during mammalian development (10). Presumably, other genes, perhaps upstream of pax6, are required for specification of ocular structures. Multiple genes expressed early in retinal development have been identified, and functional studies have shown that at least one, chx10 (20), another homeobox gene, has a function in retinal proliferation early in development (21). None of the genes identified to date, however, show the specificity of expression that might be expected for specification of the eye field. A gene playing such a role might be expressed fairly early in development, prior to pax6, and might later have its expression restricted to the developing eye.
Here we report the identification of a new paired-type homeobox gene, rax (retina and anterior neural fold homeobox), whose expression profile matches that of a gene that could play a role in specification of the eye field. In situ hybridization has shown that rax RNA is initially expressed in the region of the forebrain where optic vesicles are formed. It is expressed before pax6 and approximately simultaneously with six3 (22), a vertebrate homolog of Drosophila sine oculis, a gene required in Drosophila for visual system development. Later in development, rax is expressed in the VZ of the developing retina, and its expression is limited to the retina throughout development. As both pax6 and six3 are expressed more broadly, specification of the eye field may rely upon rax. Moreover, rax might be involved in the maintenance of retinal fate and/or proliferation of retinal progenitors later during development.
MATERIALS AND METHODS
RT-PCR, Isolation, and Analysis of Mouse rax Genes.
Random-primed cDNA made from rat E18 and P4 retinal mRNA was subjected to 40 cycles of amplification by PCR, at an annealing temperature of 50°C, using fully degenerate primers corresponding the following amino acid sequences: RRSRTTF and QVWF(K/S)NRR. Fourfold degenerate residues were substituted by inosine, and the restriction sites BamHI and EcoRI were included at the 5′ end of each primer. The bands of interest were excised from a 2% agarose gel and subcloned into Bluescript (Stratagene). Of the resulting several hundred clones, several were chosen at random and sequenced. The BamHI-EcoRI inserts of these clones were excised, mixed, and radiolabeled. The mixture of radiolabeled fragments was used as a probe for the clones plated on the original selection plates. Several clones were picked from among those that did not hybridize, and they were sequenced. This procedure was repeated until all of the initial clones showed a hybridization signal. Three novel homeobox genes were obtained.
The rax cDNA fragment so obtained was used to screen a mouse P0-P3 eye cDNA library (23). The longest clone isolated from this library was subcloned into Bluescript and was used to confirm and extend the sequence obtained from the original PCR-amplified cDNA clone of rax.
In Situ Hybridization.
Digoxigenin-labeled riboprobes were synthesized using linearized DNA templates in Bluescript (Stratagene). Transcription reactions were carried out according to the manufacturer’s instructions using either T7 or T3 RNA polymerase in the presence of 11-digoxigenin UTP (Boehringer-Mannheim). The full-length cDNA of rax was used as a probe for in situ hybridization. A mouse otx2 probe, encoding amino acids 84–272 (24), was amplified by reverse transcription–PCR from mouse P0 retinal cDNA using degenerate primers and was cloned into Bluescript. A mouse pax6 probe was obtained from H. Matsunami, as originated by Walther and Gruss (10). Mouse six3 probe was obtained from P. Gruss (22).
Whole mount in situ hybridizations were performed as described by Riddle et al. (25), except mouse embryo powder was used to preabsorb the antibody. In situ hybridization of sections was done according to Yang and Cepko (26), except proteinase K treatment was performed using a concentration of 5 μg/ml proteinase K for 2 min.
Northern Blot Hybridization.
RNA was extracted from adult Swiss-Webster mice using Trizol (GIBCO). Total RNA (10 μg) was electrophoresed in a 1.0% agarose-formaldehyde gel and transferred to a nylon membrane (Zeta-Probe GT, Bio-Rad). A 1.2-kb fragment of rax cDNA (nucleotides 456-1678) was used as a probe both for hybridization of Northern blots and for chromosomal mapping. Hybridization with the rax probe was performed according to the manufacturer’s protocol. Washes with increasing stringency were performed, the last being at 50°C in 0.1× standard saline citrate/0.1% SDS.
Chromosomal Mapping of the Mouse rax Gene.
The rax gene was mapped by analysis of two sets of multilocus crosses: (NFS/N or C58/J × Mus musculus musculus) × M. m. musculus (27) and (NFS/N × Mus spretus) × M. spretus or C58/J (28). DNAs from these crosses have been typed for more than 800 loci, including the chromosome 18 loci Lox (lysyl oxidase), Ii (Ia-associated invariant chain), and Mbp (myelin basic protein). Data from these crosses are stored and analyzed using the program locus developed by C. E. Buckler (National Institutes of Allergy and Infectious Diseases, Bethesda, MD). Percentage recombination and standard errors between specific loci were calculated from the number of recombinants according to Green (29).
RESULTS
Isolation and Characterization of rax cDNA.
Degenerate oligonucleotide primers were used to amplify conserved sequences within specific classes of genes expressed in the rat retina at E18 and P4. The method described by Valenzuela et al. (30) was used with modification to efficiently increase the number of independent clones identified. This method was applied to homeobox genes, using degenerate primers for paired-type homeobox genes. Several known paired-type homeobox genes were isolated, including pax6 (10) and chx10 (20). In addition, three new paired-type or paired-type-related homeobox genes were identified. In this paper, one of these novel homeobox genes, which was named rax, will be described. Other two new homeobox genes will be described elsewhere.
One of the initial clones of rax contained an open reading frame determined to encode a novel paired-type homeodomain. This clone was used to screen a mouse P0-P3 retinal cDNA library (23), and hybridizing clones were sequenced. The largest cDNA insert was 1.7 kb. Sequence analysis showed that this cDNA was from a new gene belonging to the paired-type subclass of homeobox genes. As can be seen in Fig. 1, two putative translation initiation codons are present in the same open reading frame as the homeodomain, with stop codons in all three frames 5′ of them. The second methionine of these putative translation initiation sites shows stronger similarity to the consensus sequence proposed by Kozak (31), including the presence of both the highly conserved purine at position −3 and the guanine at position +4. However, we cannot be certain which of the two initiation codons is the functional translational start site, because a ribosome scanning model would predict that initiation would normally occur at the first initiation codon (32). The stop codon of the predicted Rax protein is also indicated in Fig. 1.
Figure 1.
rax nucleotide and amino acid sequences. Boxed amino acids are the homeodomain sequence. The dashed box shows the octapeptide sequence. Double underline indicates the OAR domain. Possible initiation methionines are in boldface type.
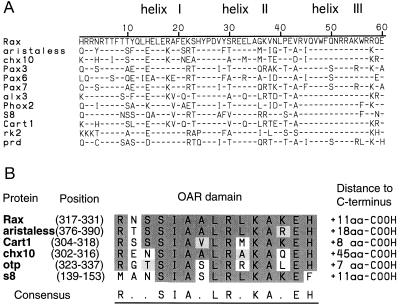
The amino acid sequence of the Rax homeodomain shows a high degree of homology to paired-type homeodomain proteins such as aristaless (33), Chx10 (20), Pax6 (10), and paired (34) (Fig. 2A). In fact, some of the paired-type specific residues are conserved in the homeodomain of Rax protein (43). In addition, Rax has the characteristic octapeptide identified in some of the paired-domain and homeodomain proteins, such as Pax1, 2, 3, 5, 7, 8, and Chx10 (20, 44). However, Rax does not encode the paired-box motif. Interestingly, at the C terminus of the Rax protein, we found a 15-aa stretch that is homologous to the C terminus of the homeodomains of aristaless, cart1 (40), chx10, otp (42), and s8 (39) (Fig. 2B). We have named this conserved region the OAR domain, using the initials of otp, aristaless, and rax. Between the homeodomain and the OAR domain, Rax contains a proline-, serine-, and threonine-rich (PST-rich) domain, which is similar to the homologous region of Pax6 and Oct1 (45). The PST-rich domain of Rax contains 114 amino acids of which 42 (37%) are either proline, serine, or threonine.
Figure 2.
(A) Alignment of homeodomain sequences for the paired-type homeodomain-containing proteins. Only amino acid residues that differ from those of the Rax homeodomain are shown. (B) Sequence alignment of OAR domain sequences. Conserved identical amino acid residues are shown with a dark shadow. Conservative changes of amino acid residues are shown with a light shadow. Sequences of homeodomains and OAR domains are taken from the following references: aristaless (33), chx10 (20), pax3 (35), pax6 (10), pax7 (36), alx3 (37), phox2 (38), s8 (39), cart1 (40), rk2 (41), paired (34), and otp (42).
Chromosomal Localization of rax.
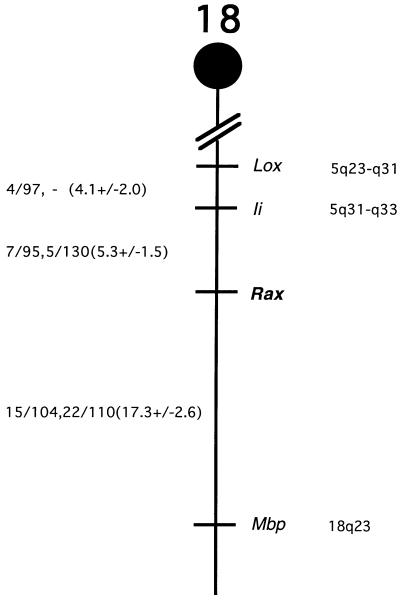
The chromosomal location of rax in the mouse was determined by Southern blot analyses of DNAs from the progeny of two sets of multilocus crosses (see Materials and Methods). EcoRI digestion produced fragments of 9.0 and 3.8 kb in NFS/N and C58/J mice, and 8.3 and 5.7 kb in M. m. musculus. M. spretus produced a ScaI fragment of 11.3 kb, and NFS/N and C58/J produced a fragment of 9.4 kb. Inheritance of the polymorphic fragments in both sets of crosses was compared with inheritance of more than 1,000 markers. The mapping results indicated that rax is located in the distal region of mouse chromosome 18 linked to Lox, Ii, and Mbp (Fig. 3). rax was mapped 5.3 cM distal of Ii.
Figure 3.
Chromosomal location of the rax gene on mouse chromosome 18. (Left) Recombination fractions for adjacent loci: the first fraction represents data from the M. spretus crosses, and the second fraction is from the M. m. musculus crosses. In parentheses are recombinational distances and standard errors calculated according to Green (29). (Right) The positions of loci in human chromosomes, where known.
We have compared our interspecific map of chromosome 18 with a composite mouse linkage map that reports the map location of many uncloned mouse mutations (46). The region encoding rax lacks mouse mutations with a phenotype that might be expected for an alteration in this locus (data not shown). This region of mouse chromosome 18 is homoogous to human chromosome 18p11-q21 or 5q23-q33 (Fig. 3). At present there are no candidate human diseases that map to this region listed in the Online Mendelian Inheritance in Man (The Johns Hopkins University, Baltimore, MD).
Analysis of rax Expression in Mouse Embryos.
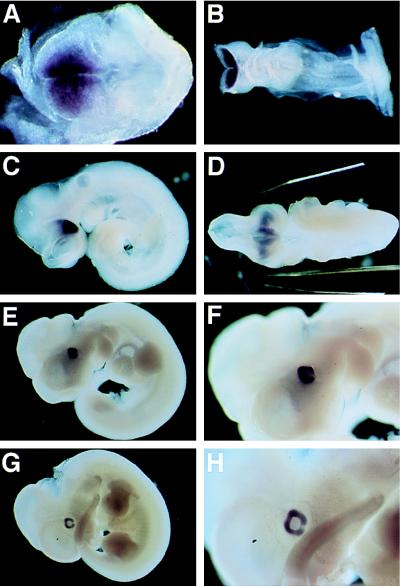
Early mouse embryos were examined for rax expression by whole mount in situ hybridization. No hybridization signal was detected at E6.5 (data not shown). At E7.5, rax expression was observed in the cephalic neural fold (head fold), the area that will later become the forebrain and midbrain (Fig. 4A). At E8.5, rax transcripts were seen in the prospective forebrain region, including the region of the optic placodes (Fig. 4B). The hybridization signal was more intense than on E7.5. On E9.5, rax expression was restricted to the optic vesicles (Fig. 4C), the optic stalk, and the ventral diencephalon (Fig. 4D). On E10.5 and E11.5, the expression of rax was observed only in the retina (Fig. 4 E–H). rax transcripts were not detected in the lens, optic fissure, or elsewhere in the embryo.
Figure 4.
Whole-mount in situ hybridization of E7.5 (A), E8.5 (B), E9.5 (C and D), E10.5 (E and F), and E11.5 mouse embryos (G and H). (Left) Anterior side of the embryos. (A) rax expression is detected in the cephalic head fold. (B) rax expression is at a high level in the prospective forebrain. (C) The embryo has finished turning and the optic vesicles have been formed. The optic vesicles exhibit a high level of expression of rax. (D) A ventral view of the embryo shown in C. rax expression is detected in the optic vesicles, optic stalk, and ventral diencephalon. (E–H) Lateral view of the embryos at E10.5 (E and F) and E11.5 (G and H). rax is expressed in the optic cup. F and H represent a higher magnification of E and G, respectively. Note that the optic fissures are negative for rax expression.
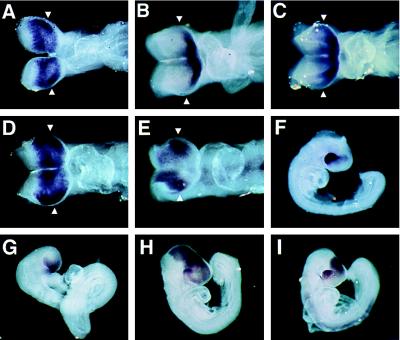
The rax expression pattern was compared with those of the homeobox-containing genes pax6 (10), six3 (22), and otx2 (24). These genes are known to be expressed early in development in a region including the anlagen of the eye. At E8.0, rax, otx2, and six3 were expressed in the anterior central nervous system. pax6 was found to be expressed at a low level at E8.0, barely detectable by whole mount in situ hybridization. At E8.5, all four mRNAs were detected in the anterior head fold (Fig. 5A–E). Six3 RNA was detected at the most anterior region of the forebrain, as previously reported (22). six3 appears not to be highly expressed in the region of the optic evaginations (Fig. 5B). In late E8.5 embryos (i.e., E8.5 embryos that have turned), the domain of six3 expression was seen to extend to the area including the optic evaginations (Fig. 5C). The transcripts of rax, pax6, and otx2 were strongly expressed in the region containing the optic evaginations (Fig. 5 A, D and E). Thus, rax is one of the few known homeobox genes that are expressed in the optic evaginations of the early mouse embryo.
Figure 5.
Comparison of the expression patterns of rax, six3, otx2, and pax6 during optic vesicle formation. (A, B, D, and E) Dorsal views of E8.5 mouse embryos. (C) A dorsal view of an E8.5 embryo after turning. (F–I) Side views of E9.0 embryos. Hybridization probes are as follows: (A and F) rax, (B, C, and G) six3, (D and H) otx2, (E and I) pax6. Arrowheads indicate the level where the optic vesicles begin to evaginate. Note that only rax expression is specific to the optic vesicles and adjacent ventral diencephalon.
At E9.0, rax signal was found in the optic vesicle, optic stalk, and ventral diencephalon, while pax6, six3, and otx2 were found to be expressed in other areas of the embryo (Fig. 5 F–I).
Expression of rax in the Developing Retina.
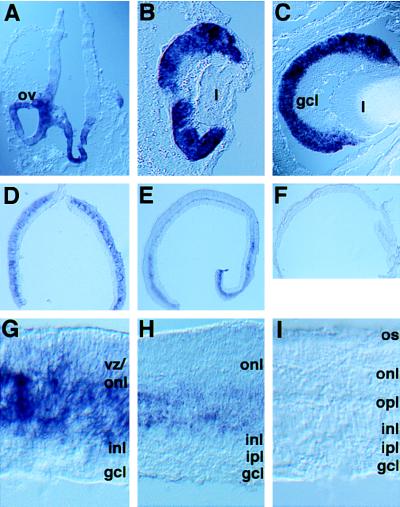
As eye development proceeded, the rax expression pattern shown in Fig. 6 was observed. At E9.5, rax transcripts were found in the optic vesicle and the ventral diencephalon (Fig. 6A). Gradually, rax was found to be down-regulated in all areas except the developing retina, where it was strongly expressed (data not shown). At E11.5 and E18.5, rax transcripts were detected in the neural retina alone (Fig. 6 B and C); unlike pax6, rax was not expressed in the lens or presumptive cornea. After birth, the expression of rax was dramatically reduced (Fig. 6 D–I). At P0, rax expression was observed in the VZ. No hybridization signal was observed in the ganglion cell layer, the layer most advanced in its differentiation. During the postnatal period, the expression of rax appeared to decrease (Fig. 6 G–I). rax expression was relatively stronger near the middle of the developing retinal layers where the most immature cells are located. In this region of the retina at P6, a limited number of cells expressing rax was present (Fig. 6H). rax expression was not detected in the adult retina (Fig. 6I). As the mitotic activity of the rat retina decreases to nearly zero by P8-P10 (47), there is an excellent correlation between the temporal and spatial aspects of mitotic activity and expression of rax.
Figure 6.
rax expression during development of the mouse retina. (A) A coronal section of the forebrain of an E9.5 mouse embryo. The rax hybridization signal was detected in the optic vesicles and the ventral diencephalon. (B) A section through an E11.5 eye. (C) A section through an E18.5 eye. rax expression is not observed in the ganglion cell layer. (D–F) Cross-sections of a P0, P6, and adult mouse retina. Note the stronger signal in the peripheral retina, which lags in development behind that of the central retina. (G–I) Higher magnification of the cross-section of the retina shown in D–F. At P0 and P6, the outer nuclear layer and the inner nuclear layer are developing. Progenitor cells in the VZ are intermixed with these layers. ov, optic vesicle; gcl, ganglion cell layer; l, lens; inl, inner nuclear layer; ipl, inner plexiform layer; onl, outer nuclear layer; opl, outer plexiform layer; os, outer segment; vz, ventricular zone.
Analysis of rax Transcripts in Adult Tissues.
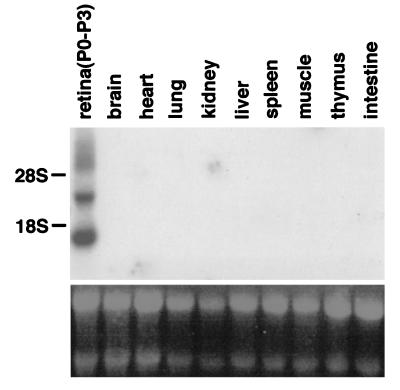
Expression of the rax gene in various adult tissues was examined on Northern blots with a radiolabeled rax cDNA probe. As a control, P0-P3 retinal RNA was used. Two bands of 1.8 kb and 4.0 kb were detected. The 1.8-kb band corresponds to the cDNA characterized here. The larger band has not yet been characterized. The rax probe did not detect a band in the other adult tissues examined, including the retina (Fig. 7), indicating that these tissues do not express rax to a level comparable to that of the developing retina.
Figure 7.
Northern hybridization analysis of rax expression in adult mouse organs. (Upper) Hybridization signals obtained with the mouse rax cDNA. (Lower) Ethidium bromide staining of the RNA. The rax transcripts are ≈4.0 kb and 1.8 kb in the P0-P3 retina, which is used as a control for hybridization.
DISCUSSION
rax Is a Novel Paired-Type Homeobox Gene.
This paper describes the identification and expression of a novel paired-type homeobox gene, rax. Within the amino-terminal portion of the open reading frame, rax encodes the octapeptide motif (44). Furthermore, the C terminus of Rax protein contains a 15-aa domain that we have noticed is conserved among several other genes and that we have named the OAR domain. The octapeptide and/or the OAR domain are observed in other paired-type homeodomain proteins, as well as other homeodomain proteins of other classes. The functions of the octapeptide and the OAR domain have not been reported. However, based on an analysis of deletion mutants of the Otp protein (42), a possible function of the OAR domain might be transactivation. Further analysis of this motif will be required to test this idea.
rax Is a Candidate Molecule for Specification of the Eye Field.
At E7.5, rax was expressed in the cephalic neural folds, the region of the prospective fore- and midbrain. One day later, rax expression was restricted to the region of the forebrain within which all of the cells that contribute to the optic vesicles are likely to originate. By E9.0, rax showed dramatically more restricted expression. It appeared only within the optic vesicle and the ventral diencephalon, from which the optic vesicles protrude. The expression pattern of rax suggests that cells expressing this gene are either competent to become, or are already specified to be, retinal progenitors. Other genes expressed in the anterior neural plate and neural folds during mouse development, as well as later in the retina, have been reported, including pax6 (10), otx2 (24), and six3 (22). Because all of these genes have an expression domain that is larger than that of the presumptive eye, they may have a role in patterning more anterior structures than just the eye, as discussed below.
It has been suggested that pax6, a highly conserved gene expressed in the eyes of all species examined to date, is the master control gene for eye formation. This suggestion comes from work in Drosophila, in which ectopic expression of either mouse pax6 or Drosophila eyeless leads to the formation of extra eyes on legs, wings, and antennae (19). However, pax6 is unlikely to be required for the formation of optic vesicles in mice, because optic vesicles are formed in homozygous sey (“small eye”) mice (48, 49), which are believed to have no functional Pax6 protein. pax6 may be necessary for maintenance of proliferation of retinal progenitors, as growth of optic vesicles is distorted in sey homozygotes (48, 49). pax6 expression starts later than that of rax, suggesting that rax might be directly or indirectly upstream of pax6 in the series of events that lead to optic vesicle formation. otx2 expression is detectable during very early development in mouse embryos, beginning between E5.5 and E5.7, and is observed in the embryonic retina (24). Animals heterozygous for a loss-of-function mutation of otx2 show abnormalities in the anterior portion of the embryo, including dislocation of the retina and the loss of the lens (50). This eye phenotype, however, is likely to be a secondary effect, resulting from a deformity of the entire rostral region, as the homozygous otx2 mutants exhibit loss of both the fore- and midbrain (50–52). six3, a murine homolog of the sine oculis gene in Drosophila, is reported to be specifically expressed in the most anterior ridge region of the forebrain (22), which will give rise to derivatives of the nonneural ectoderm. The expansion of the domain of six3 expression to include the optic pits occurs in relatively later stages, at ≈E8.5. Due to this expression pattern, it is unlikely that six3 functions at the very beginning of optic vesicle formation. rax is the best candidate so far reported to have a role in the specification of the eye before evagination of the optic vesicles. Analysis of the biological function of rax by examination of animals with a loss-of-function or gain-of-function of rax activity is of course required to establish its role(s). In addition, examination of the conservation of rax from vertebrates through invertebrates, as has been done for pax6, six3, and otx2, will be informative.
rax Expression in the Retina Correlates with Proliferation.
The paired-type homeobox genes have been shown to play essential roles in pattern formation and proliferation. At late embryonic stages, rax mRNA is detected exclusively in the retina, and exclusively in proliferating cells within the retina. It is possible, however, that a specific population of cells in some organs express rax and that this was not detected by Northern blot analysis or in situ hybridization. As development proceeds, rax expression in the retina declines; by the adult stage, it is undetectable. rax expression appears to decline as soon as cells leave the VZ and differentiate. It is absent in the ganglion cell layer beginning at approximately E11.5. [3H]thymidine-labeling studies have shown that ganglion cells become postmitotic between E11 and E18 in the mouse (53) and are the first cells to form a layer of differentiating cells. After P0, rax expression gradually declines within the outer retinal layers. The last layer to show rax expression is the last layer to differentiate, the outer nuclear layer, where rod and cone photoreceptor cells reside. rax expression thus appears to be restricted to mitotic progenitors in the retina. Rax protein may regulate the proliferation of retinal progenitors and/or prevent their differentiation. To initiate differentiation, down-regulation of Rax may be necessary.
Interestingly, the expression profile of rax within the developing retina is most similar to that of mouse notch1 (54). notch1 is also expressed in mitotic progenitors in the rodent retina and its expression is reduced during development (48, 55). Because the expression of notch1 starts later than that of rax, rax might be one of the upstream regulators of notch1 in the mouse retina. Another potential target gene of rax is chx10 (20). However, chx10 expression at E9.5 is restricted to the anterior part of the optic vesicles (20). Its regulation would thus require additional factors, because rax is expressed throughout the optic vesicle. Identification of the target genes of Rax, its interactions with other genes active in early eye development, and the roles of these genes in eye specification and morphogenesis should further our understanding of eye development and evolution.
Acknowledgments
We thank Drs. David Feldheim and Xianjie Yang for advice and Julie Zitz and Giuseppe Raviola for technical assistance. We also thank Dr. Peter Gruss for the probes of six3 and pax6. This work was supported by the Howard Hughes Medical Institute.
ABBREVIATION
- VZ
ventricular zone
Footnotes
References
- 1.Cepko C L, Austin C P, Yang X, Alexiades M, Ezzeddine D. Proc Natl Acad Sci USA. 1996;93:589–595. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grant P, Rubin E, Cima C. J Comp Neurol. 1980;189:593–613. doi: 10.1002/cne.901890403. [DOI] [PubMed] [Google Scholar]
- 3.Grainger R M. Trends Genet. 1992;8:349–355. doi: 10.1016/0168-9525(92)90280-h. [DOI] [PubMed] [Google Scholar]
- 4.Altshuler D M, Turner D L, Cepko C L. In: Development of the Visual System. Lam D M-K, Shatz C J, editors. Vol. 3. Cambridge, MA: MIT Press; 1991. pp. 37–58. [Google Scholar]
- 5.Turner D L, Cepko C L. Nature (London) 1987;328:131–136. doi: 10.1038/328131a0. [DOI] [PubMed] [Google Scholar]
- 6.Turner D L, Snyder E Y, Cepko C L. Neuron. 1990;4:833–845. doi: 10.1016/0896-6273(90)90136-4. [DOI] [PubMed] [Google Scholar]
- 7.Holt C E, Bertsch T W, Ellis H M, Harris W A. Neuron. 1988;1:15–26. doi: 10.1016/0896-6273(88)90205-x. [DOI] [PubMed] [Google Scholar]
- 8.Wetts R, Fraser S E. Science. 1988;239:1142–1145. doi: 10.1126/science.2449732. [DOI] [PubMed] [Google Scholar]
- 9.Freund C, Horsford D J, McInnes R R. Hum Mol Genet. 1996;5:1471–1488. doi: 10.1093/hmg/5.supplement_1.1471. [DOI] [PubMed] [Google Scholar]
- 10.Walther C, Gruss P. Development. 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 11.Gruss P, Walther C. Cell. 1992;69:719–722. doi: 10.1016/0092-8674(92)90281-g. [DOI] [PubMed] [Google Scholar]
- 12.Hill R E, Favor J, Hogan B L, Ton C C, Saunders G F, Hanson I M, Prosser J, Jordan T, Hastie N D, van Heyningen V. Nature (London) 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 13.Matsuo T, Osumi-Yamashita N, Noji S, Ohuchi H, Koyama H, Myokai E, Matsuo N, Taniguchi S, Doi H, Iseki S, Ninomiya Y, Fujiwara M, Watanabe T, Eto K. Nat Genet. 1993;3:299–304. doi: 10.1038/ng0493-299. [DOI] [PubMed] [Google Scholar]
- 14.Ton C C, Hirvonen H, Miwa H, Weil M M, Monaghan P, Jordan T, van Heyningen V, Hastie N D, Meijers-Heijboer H, Drechsler M, Royer-Pokora B, Collins F, Swaroop A, Strong L C, Saunders G F. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- 15.Glaser T, Walton D S, Mass R L. Nat Genet. 1992;2:232–238. doi: 10.1038/ng1192-232. [DOI] [PubMed] [Google Scholar]
- 16.Jordan T, Hanson I, Zaletayev D, Hodgson S, Prosser J, Seawright A, Hastie N, van Heyningen V. Nat Genet. 1992;1:328–332. doi: 10.1038/ng0892-328. [DOI] [PubMed] [Google Scholar]
- 17.Hanson I M, Fletcher J M, Jordan T, Brown A, Taylor D, Adams R J, Punnett H, van Heyningen V. Nat Genet. 1994;6:168–173. doi: 10.1038/ng0294-168. [DOI] [PubMed] [Google Scholar]
- 18.Quiring R, Walldorf U, Kloter U, Gehring W J. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 19.Halder G, Callaerts P, Gehring W J. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 20.Liu I S C, Chen J, Ploder L, Vidgen D, van der Kooy D, Kalnins V I, McInnes R R. Neuron. 1994;13:377–393. doi: 10.1016/0896-6273(94)90354-9. [DOI] [PubMed] [Google Scholar]
- 21.Burmeister M, Novak J, Liang M-Y, Basu S, Ploder L, Hawes N L, Vidgen D, Hoover F, Goldman D, Kalnins V I, Roderick T H, Taylor B A, Hankin M H, McInnes R R. Nat Genet. 1996;12:376–383. doi: 10.1038/ng0496-376. [DOI] [PubMed] [Google Scholar]
- 22.Oliver G, Maihos A, Wehr R, Copeland N G, Jenkins N A, Gruss P. Development. 1995;121:4045–4055. doi: 10.1242/dev.121.12.4045. [DOI] [PubMed] [Google Scholar]
- 23.Yang X, Chung D, Cepko C L. J Neurosci. 1993;13:3006–3017. doi: 10.1523/JNEUROSCI.13-07-03006.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simeone A, Acampora D, Mallamaci A, Stornaiuolo A, D’Apice M R, Nigro V, Boncinelli E. EMBO J. 1993;12:2735–2747. doi: 10.1002/j.1460-2075.1993.tb05935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riddle R D, Johnson R L, Laufer E, Tabin C. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- 26.Yang X, Cepko C L. J Neurosci. 1996;16:6089–6099. doi: 10.1523/JNEUROSCI.16-19-06089.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozak C A, Peyser M, Krall M, Mariano T M, Kumar C S, Pestka S, Mock B A. Genomics. 1990;8:519–524. doi: 10.1016/0888-7543(90)90039-w. [DOI] [PubMed] [Google Scholar]
- 28.Adamson M C, Silver J, Kozak C A. Virology. 1991;183:778–781. doi: 10.1016/0042-6822(91)91010-e. [DOI] [PubMed] [Google Scholar]
- 29.Green E L. Genetics and Probability in Animal Breeding Experiments. New York: MacMillian; 1981. pp. 77–113. [Google Scholar]
- 30.Valenzuela D M, Stitt T N, DiStefano P S, Rojas E, Mattsson K, Compton D L, Nuñez L, Park J S, Stark J L, Gies D R, Thomas S, Le Beau M M, Fernald A A, Copeland N G, Jenkins N A, Burden S J, Glass D J, Yancopoulos G D. Neuron. 1995;15:573–584. doi: 10.1016/0896-6273(95)90146-9. [DOI] [PubMed] [Google Scholar]
- 31.Kozak M. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozak M. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneitz K, Spielmann P, Noll M. Genes Dev. 1993;7:114–129. doi: 10.1101/gad.7.1.114. [DOI] [PubMed] [Google Scholar]
- 34.Frigerio G, Burri M, Bopp D, Baumgartner S, Noll M. Cell. 1986;47:735–746. doi: 10.1016/0092-8674(86)90516-7. [DOI] [PubMed] [Google Scholar]
- 35.Goulding M D, Charlepkis G, Deutsch U, Erselius J R, Gruss P. EMBO J. 1991;10:1135–1147. doi: 10.1002/j.1460-2075.1991.tb08054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jostes B, Walther C, Gruss P. Mech Dev. 1991;33:27–38. doi: 10.1016/0925-4773(90)90132-6. [DOI] [PubMed] [Google Scholar]
- 37.Rudnick A, Ling T Y, Odagiri H, Rutter W J, German M S. Proc Natl Acad Sci USA. 1994;91:12203–12207. doi: 10.1073/pnas.91.25.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valalrche I, Tissier-Seta J P, Hirsch M R, Martinez S, Goridis C, Brunet J F. Development. 1993;119:881–896. doi: 10.1242/dev.119.3.881. [DOI] [PubMed] [Google Scholar]
- 39.Opstelten D-J E, Vogels R, Robert B, Kalkhoven E, Zwartkruis F, de Laaf L, Destree O H, Deschamps J, Lawson K A, Meijink F. Mech Dev. 1991;34:29–41. doi: 10.1016/0925-4773(91)90089-o. [DOI] [PubMed] [Google Scholar]
- 40.Zhao G-Q, Zhou X, Eberspaecher H, Solursh M, de Crombrugghe B. Proc Natl Acad Sci USA. 1993;90:8633–8637. doi: 10.1073/pnas.90.18.8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campbell G, Goring H, Lin T, Spana E, Andersson S, Doe C Q, Tomlinson A. Development. 1994;120:2957–2966. doi: 10.1242/dev.120.10.2957. [DOI] [PubMed] [Google Scholar]
- 42.Simeone A, D’Apice M R, Nigro V, Casanova J, Graziani F, Acampora D, Avantaggiato V. Neuron. 1994;13:83–101. doi: 10.1016/0896-6273(94)90461-8. [DOI] [PubMed] [Google Scholar]
- 43.Bopp D, Burri M, Baumgartner S, Frigerio G, Noll M. Cell. 1986;47:1033–1040. doi: 10.1016/0092-8674(86)90818-4. [DOI] [PubMed] [Google Scholar]
- 44.Noll M. Curr Opin Genet Dev. 1993;3:595–605. doi: 10.1016/0959-437x(93)90095-7. [DOI] [PubMed] [Google Scholar]
- 45.Sturm R A, Das G, Herr W. Genes Dev. 1988;2:1582–1599. doi: 10.1101/gad.2.12a.1582. [DOI] [PubMed] [Google Scholar]
- 46.Johnson K R, Davisson M T. Mamm Genome. 1996;6:S300–S308. [PubMed] [Google Scholar]
- 47.Alexiades M, Cepko C. Dev Dyn. 1996;205:293–307. doi: 10.1002/(SICI)1097-0177(199603)205:3<293::AID-AJA9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 48.Hogan B L M, Hirst E M A, Horsburgh G, Hetherington C M. Development. 1988;103:115–119. doi: 10.1242/dev.103.Supplement.115. [DOI] [PubMed] [Google Scholar]
- 49.Grindley J C, Davidson D R, Hill R E. Development. 1995;121:1433–1442. doi: 10.1242/dev.121.5.1433. [DOI] [PubMed] [Google Scholar]
- 50.Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S. Genes Dev. 1995;9:2646–2658. doi: 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- 51.Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, Brulet P. Development. 1995;121:3279–3290. doi: 10.1242/dev.121.10.3279. [DOI] [PubMed] [Google Scholar]
- 52.Ang S L, Jin O, Rhinn M, Daigle N, Stevenson L, Rossant J. Development. 1996;122:243–252. doi: 10.1242/dev.122.1.243. [DOI] [PubMed] [Google Scholar]
- 53.Sidman R L. In: The Structure of the Eye. Smelse G, editor. New York: Academic; 1961. pp. 487–506. [Google Scholar]
- 54.Weinmaster G, Roberts V, Lemke G. Development. 1991;113:199–205. doi: 10.1242/dev.113.1.199. [DOI] [PubMed] [Google Scholar]
- 55.Bao Z-Z, Cepko C L. J Neurosci. 1997;17:1425–1434. doi: 10.1523/JNEUROSCI.17-04-01425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]