Abstract
We have examined differences in diversity at 60 microsatellite loci among human population samples from three major continental groups to evaluate the hypothesis of greater African diversity in this rapidly evolving class of loci. Application of a statistical test that assumes equal mutation rates at all loci fails to demonstrate differences in microsatellite diversity, while a randomization test that does not make this assumption finds that Africans have significantly greater microsatellite diversity (P < 10−8) than do Asians and Europeans. Greater African diversity is most apparent at loci with smaller overall variance in allele size, suggesting that the record of population history has been erased at repeat loci with higher mutation rates. A power analysis shows that only 35–40 microsatellites are needed to establish this difference statistically, demonstrating the considerable evolutionary information contained in these systems. On average, African populations have ≈20% greater microsatellite diversity than do Asian and European populations. A comparison of continental diversity differences in microsatellites and mtDNA sequences suggests earlier demographic expansion of the ancestors of Africans.
Greater genetic diversity in human populations from Africa has been a cornerstone of the argument for an African origin of modern humans, since this greater diversity suggests a greater “age” for African populations. In demographic terms, the greater age hypothesis has been interpreted in at least two ways: first, that human populations outside Africa are descendants of emigrants from a large African population (1); and, second, that all modern humans are derived from expansions from a restricted founding population and that the expansion of African ancestors occurred much earlier than those of populations outside Africa (2, 3).
Many recent empirical studies have demonstrated greater genetic diversity in African populations than in non-African populations. Some of the strongest evidence in support of increased African diversity comes from studies of mtDNA (4), the CD4 microsatellite and a nearby Alu polymorphism (1), and D16S309, a minisatellite system (5). However, each of these systems represents essentially a single locus subject to large stochastic variation. Relethford and Harpending (6) report that a multivariate generalization of variance is greater in craniometric traits from African populations than in populations from Europe and Asia, but the effects of environmental sources of variance were not known. Bowcock et al. (7) showed that heterozygosity in Africans was significantly greater in a sample of 30 dinucleotide microsatellite loci. However, the loci they analyzed were all on chromosomes 13 and 15, violating independence assumptions of their statistical test. Jorde et al. (8) also demonstrated greater African heterozygosity in a sample of 30 tetranucleotide microsatellite loci, but this difference was not statistically significant.
In contrast, classical markers and nuclear restriction fragment length polymorphisms do not show elevated African diversity (9, 10). For these loci, Europeans and occasionally Asians are the most diverse population group, perhaps reflecting ascertainment bias in discovering these systems because they were polymorphic in Europeans (11). This ascertainment bias can be large enough to account for excess heterozygosity in systems in which the average heterozygosity is <0.35 (12). Most classical markers and restriction fragment length polymorphisms fall into this category, making ascertainment bias a possible explanation for the lack of excess African diversity in these systems.
The published genetic data, while mostly supportive of this pattern of elevated African genetic diversity at loci with high mutation rates, are by no means definitive. There is little statistical support for the pattern. To test further the hypothesis of increased African diversity, we have examined 60 unlinked microsatellite loci in a worldwide sample of humans. Using methods appropriate for such systems, we demonstrate a statistically significant elevation of African genetic diversity.
MATERIALS AND METHODS
Subjects.
The study population consists of 72 Africans, 63 Asians, and 120 Europeans. Further details about these subjects are given in a previous publication (8). In addition to the subjects included in the earlier study, 20 Finnish and 10 Polish subjects have been added in the present study. For some analyses, the three continental populations were divided into subpopulations. The African subpopulations consist of San, Sotho/Tswana, Mbuti Pygmy, Biaka Pygmy, Tsonga, and Nguni. The Asian subpopulations consist of Malay, Vietnamese, Cambodian, Chinese, and Japanese. The European subpopulations consist of French, Poles, Finns, and Northern Europeans.
Data.
Thirty microsatellite polymorphisms were reported in an earlier study (8). In the present study, an additional 30 nuclear microsatellite systems have been analyzed. The GenBank database accession numbers for these systems are: D1S1162, D2S1279, D2S272, D3S2304, D3S2322, D4S1531, D5S1347, D6S942, D7S618, D12S297, D22S417, D21S1410, D12S296, D3S1542, D3S1530, D5S1392, D4S1527, D1S404, D5S612, D8S386, D5S1354, D7S1526, D10S521, D13S252, D22S528, D2S1268, D4S2295, D2S274, D9S769, D2S1247, and D13S248 [locus D13S248 replaces locus HRAS1 used in the earlier study (8)]. The systems were chosen to insure that none of them are closely linked, maximizing statistical independence.
Statistical Analysis.
Allele frequencies for each microsatellite system were estimated directly by gene counting. Heterozygosity for each of these systems was estimated as 1 − Σxi2, where xi is the estimated frequency of the ith allele in the system. Standard errors of these estimates were obtained by using equation 8.7 in Nei (13). Heterozygosity was averaged across all loci and estimated separately for Africans, Asians, and Europeans.
Genetic diversity was also estimated using the variance of the allele size distribution of each microsatellite system. Allele size variance provides a valid estimate of genetic diversity if microsatellite systems conform to a stepwise mutation model (14), which they generally do (15–18). Under this process the mean number of repeats in a population is indeterminate, but the variance of the number of repeats is expected to be 2Nμ, where N is the effective population size and μ is the mutation rate per generation. For some microsatellites, there are likely to be selective or mechanical constraints on allele size not accounted for by the stepwise mutation model (19). We calculated the within-population variance Vij for each locus i and each population j and, from these, the ratio Rij of Vij to the mean of Vi1, Vi2, and Vi3. The average of Rij across loci, Rj, measures variance within population j relative to the mean within-population variance. Finally, we calculated the ratio S of the largest Rj to the mean of the other two.
To assess the statistical significance of the excess African diversity, we performed a randomization test (20). In each iteration of the test, we partitioned the data set into three random subdivisions, each equal in size to one of the continental populations in the original data, and then we calculated S as described above. [Programs to do this test were written independently by two of us (A.R.R. and H.C.H.), in two different languages. The results were the same from each version.]
A power analysis was conducted to determine how many loci are needed to reject the null hypothesis of no difference in gene diversity. For each test, 100 subsets of loci from the data set were extracted randomly by sampling with replacement, and a permutation procedure was conducted. Thus, 100 one-locus subsets were generated, 100 two-locus subsets, 100 three-locus subsets, and so on. Type I error (the probability of falsely rejecting the null hypothesis) was fixed at 0.05.
Genetic distances among the 15 populations were estimated using the method of Shriver et al. (21), which assumes a stepwise mutation model. The Mantel matrix comparison technique (22, 23) was used to compare genetic distance matrices.
RESULTS
The first column of Table 1 gives the average heterozygosity values for Africans, Asians, and Europeans. The standard errors provide no indication that the differences between these groups are significant. However, the standard heterozygosity measure does not take variation in microsatellite repeat number (allele size) into account. Theory suggests that the logarithm of allele size variance within populations should approach normality (24). The mean logarithmic variances in Africans, Asians, and Europeans are 1.04, 0.87, and 0.84 respectively. A test of the differences in the mean log variance of these populations by one-way ANOVA gives F(2,177) = 0.56 (P > 0.55). Thus, there is no evidence of population differences in diversity by this statistical test. A test that should be weaker gives a slightly stronger result: at the 60 loci, the largest within-population variances are found in Africans, Asians, and Europeans in 28, 14, and 18 loci respectively, with an associated χ2 significance level between 0.05 and 0.10.
Table 1.
Estimates of heterozygosity (±SE) and variance ratio in repeat number for 60 microsatellite polymorphisms
| Continental group | Heterozygosity | Mean variance ratio, Rj |
|---|---|---|
| Africans | 0.76 ± 0.02 | 1.13 |
| Asians | 0.70 ± 0.02 | 0.92 |
| Europeans | 0.73 ± 0.02 | 0.94 |
The second column of Table 1 gives the allele size variance ratios described in the Materials and Methods section. The African variance ratio is 21% larger than the average of the European and Asian ratios. In 100 million randomizations, the maximum observed ratio was 1.14, far below the observed ratio of 1.21. Thus, the excess African diversity is highly significant by this test (P < 10−8). There are two reasons why the randomization test yielded a significant result while the ANOVA did not. First, randomization tests are often more powerful than tests based on normal theory (20). Second, the ANOVA test compared the variances at each locus and therefore implicitly assumes that all of the microsatellite loci have the same mutation rate. This is unlikely to be the case. The randomization test overcomes this problem by comparing variance ratios, in which mutation rates cancel out.
Indeed, an interesting result emerges if the loci are evaluated separately. African diversity exceeds that of the other groups at loci with small size variance, while at loci with large size variance, there is no apparent pattern. The average size variance at loci with greatest African diversity is 3.4, but it is 5.4 at loci where diversity is greatest in Asia or Europe.
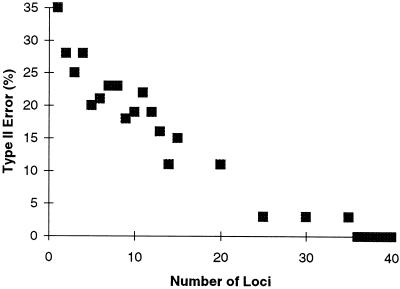
A power analysis (Fig. 1) shows that type II error (the probability of falsely accepting the null hypothesis of no difference in allele size variances among the three populations) decreases rapidly with increasing numbers of loci. It falls below 5% when 25 loci are used, and it declines to zero when 36 or more loci are used. This analysis shows that 60 loci are more than sufficient to test the hypothesis of differences in microsatellite diversity among the three major continental populations.
Figure 1.
A plot of type II error (the probability of falsely accepting the null hypothesis of no increase in genetic diversity) as a function of the number of microsatellite loci. Type I error (the probability of falsely rejecting the null hypothesis) has been fixed at 0.05.
The information content of these systems was also tested by comparing the genetic distance matrix generated from the previously published 30 microsatellites (8) with the 30 loci added in the present study. These matrices were generated for the 15 subpopulations described above. The correlation between the two distance matrices was 0.79, and the Mantel permutation procedure showed that this correlation is highly significant (P < 10−4). This result further supports the consistency and informativeness of the microsatellite systems.
The microsatellite distance matrix (based on 60 polymorphisms) is also highly concordant with a distance matrix based on 30 restriction fragment length polymorphisms (correlation = 0.85, P < 10−4; ref. 8) and with a matrix based on five trinucleotide polymorphisms (0.74, P < 10−4; ref. 25). However, the correlation between the microsatellite distance matrix and a distance matrix based on 600 bp of mtDNA control region sequence data (M.B., W.S.W., S.S., B. R. Bhaskara, and L.B.J., unpublished data) is substantially lower (0.55) but still statistically significant (P < 0.002). The diminished congruence between nuclear and mtDNA genetic distances is largely the result of greater between-population mtDNA diversity in Africa than in Europe and Asia, a pattern that is far less evident in the nuclear restriction fragment length polymorphisms and microsatellites.
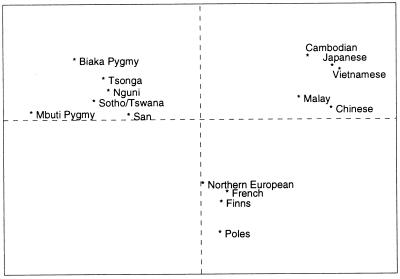
To assess further the pattern of within- and between-continent microsatellite diversity, a principal coordinates analysis of the average allele sizes was performed (Fig. 2). This is the optimal two-dimensional representation of the genetic differences among the subpopulations. The first two coordinates account for 46% of the variance of the data, largely capturing the information about subpopulation relationships. Clustering of subpopulations into continental groups is apparent, even though populations within these three apparently tight clusters are often geographically distant from one another. The consistent grouping of populations according to continent has been observed in other studies of microsatellite polymorphisms (7, 27, 28). Such a pattern would not be expected if the differences among populations reflected just isolation by distance in a widely distributed species with gene exchange among neighboring demes.
Figure 2.
A plot of the first two principal coordinates of the mean allele sizes of each population, showing the genetic relationships of 15 subpopulations.
DISCUSSION
Although most recent comparisons of genetic variation in human populations have shown excess African diversity, few, if any, have demonstrated a statistically significant increase. The mitochondrial data, as well as the CD4 haplotype data, have in fact been criticized for their lack of statistical reliability (29–31). Similar criticisms have been leveled at attempts to estimate coalescence dates from Y chromosome data (32). The present study demonstrates that microsatellite data, when appropriately analyzed, reveal a significant increase in African diversity. It is reassuring that only a modest number of loci (<40) are needed to demonstrate this difference. This implies that microsatellite systems contain a large amount of information about evolutionary history.
There are some caveats, however. More loci are likely to be needed to answer questions on a smaller evolutionary scale (e.g., hypotheses about evolutionary relationships within the major continental populations). Zhivotovsky and Feldman (24) have estimated that as many as several hundred microsatellite systems are needed for fine-scale resolution of population relationships.
A second caution is that some microsatellite loci, because of their high variances, are not likely to be useful for comparisons of major continental populations. This probably reflects differences in mutation rates, which in turn may be influenced by specific repeat motifs and by interruptions of the repeat sequence (19, 33). It is plausible that, at loci with higher mutation rates, allele sizes have reached some selective or mechanical range constraint so that the signature of greater African diversity has been overwritten. At the loci with lower variance this signature (possibly the result of larger African effective size in the past) still remains.
The microsatellite systems reveal only a modest (20%) increase in African diversity relative to Asians and Europeans. Craniometric data reveal a similar degree of excess African diversity (6). In contrast, mtDNA data typically show African populations to be 2–3 times more diverse than others (7, 8). This may reflect the fact that effective population size is four times as large for an autosomal as for a mitochondrial locus. Consequently, the timing of past bottlenecks and expansions will affect mitochondrial and nuclear diversity quite differently. Specifically, if a population expanded from a small size N at T generations in the past, then at a mitochondrial locus the diversity (mean pairwise sequence divergence) is expected to be proportional to N + 2T (34). At a nuclear microsatellite locus the diversity (allele size variance) is expected to be proportional to 2N + T under the stepwise mutation model (14). For example, suppose that the African population and the European population were each of size n = 5,000 before expansion, and the African population expanded earlier than did the European population (T = 3,000 versus 1,000 generations). The ratio of African to European mtDNA diversity would be substantially greater (1.57) than the ratio of African to European nuclear diversity (1.18). Thus, if T is similar in magnitude to N, the ratios of mitochondrial and nuclear diversity can be very different for populations with different expansion times.
Another possible explanation involves natural selection. While the microsatellite systems generally conform well to a neutral mutation/drift model (15), there is evidence for significant deviations from selective neutrality in human mtDNA (M.B., W.S.W., S.S., B. R. Bhaskara, and L.B.J., unpublished data; refs. 26, 35, and 36). A selective sweep of the mitochondrial genome may have occurred in modern humans outside Africa as they adapted to altered climatic conditions. A selective sweep, which would reduce diversity throughout the nonrecombining mitochondrial genome, is roughly equivalent to reducing the initial population size N for mtDNA, but not the nuclear microsatellites. Thus, selection could also account for the observed pattern. More light will be shed on this hypothesis as data accumulate for genetic systems unlikely to experience the same selective forces as mtDNA (e.g., Y chromosome DNA, Alu systems, etc.).
Our findings, while consistent with an African origin of modern Homo sapiens, offer only qualified support for this hypothesis. Increased African diversity could be the result of several different factors, including a larger effective population size in Africa (35, 36). On the other hand, the difference in diversity ratios between mtDNA and the nuclear markers we have described is consistent with an earlier demographic expansion of African ancestors and a much later expansion of the ancestors of the other continental groups.
Acknowledgments
We thank Drs. John Relethford, Andrew Clark, and Sarah Tishkoff for comments and suggestions on the manuscript. Some of the Asian samples were contributed by Drs. Ken and Judy Kidd, and some of the African samples were contributed by Dr. Trefor Jenkins. This research was supported by National Science Foundation Grants DBS-9310105, SBR-9514733, and SBR-9512178; National Institutes of Health (NIH) Grant RR-00064; the Technology Access Center of the Utah Human Genome Project (NIH HG00199); and a supplemental equipment grant from the University of Utah. M.B. was supported by a Clinical Associate Physician fellowship (NIH M01-00064).
References
- 1.Tishkoff S A, Dietzsch E, Speed W, Pakstis A J, Kidd J R, Cheung K, Bonné-Tamir B, Santachiara-Benerecetti A S, Moral P, Krings M, Pääbo S, Watson E, Risch N, Jenkins T, Kidd K K. Science. 1996;271:1380–1387. doi: 10.1126/science.271.5254.1380. [DOI] [PubMed] [Google Scholar]
- 2.Harpending H C, Sherry S T, Rogers A R, Stoneking M. Curr Anthropol. 1993;34:483–496. [Google Scholar]
- 3.Lahr M M, Foley R. Evol Anthropol. 1994;3:48–60. doi: 10.1002/evan.21405. [DOI] [PubMed] [Google Scholar]
- 4.Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson A C. Science. 1991;253:1503–1507. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- 5.Armour J A L, Anttinen T, May C A, Vega E E, Sajantila A, Kidd J R, Kidd K K, Bertranpetit J, Pääbo S, Jeffreys A J. Nat Genet. 1996;13:154–160. doi: 10.1038/ng0696-154. [DOI] [PubMed] [Google Scholar]
- 6.Relethford J H, Harpending H C. Am J Phys Anthropol. 1994;95:249–270. doi: 10.1002/ajpa.1330950302. [DOI] [PubMed] [Google Scholar]
- 7.Bowcock A M, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd J R, Cavalli-Sforza L L. Nature (London) 1994;368:455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- 8.Jorde L B, Bamshad M J, Watkins W S, Zenger R, Fraley A E, Krakowiak P A, Carpenter K D, Soodyall H, Jenkins T, Rogers A R. Am J Hum Genet. 1995;57:523–538. doi: 10.1002/ajmg.1320570340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nei M, Roychoudhury A K. Mol Biol Evol. 1993;10:927–943. doi: 10.1093/oxfordjournals.molbev.a040059. [DOI] [PubMed] [Google Scholar]
- 10.Bowcock A M, Kidd J R, Mountain J L, Hebert J M, Carotenuto L, Kidd K K, Cavalli-Sforza L L. Proc Natl Acad Sci USA. 1991;88:839–843. doi: 10.1073/pnas.88.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mountain J L, Cavalli-Sforza L L. Proc Natl Acad Sci USA. 1994;91:6515–6519. doi: 10.1073/pnas.91.14.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rogers A R, Jorde L B. Am J Hum Genet. 1996;58:1033–1041. [PMC free article] [PubMed] [Google Scholar]
- 13.Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ. Press; 1987. [Google Scholar]
- 14.Slatkin M. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shriver M D, Jin L, Chakraborty R, Boerwinkle E. Genetics. 1993;134:983–993. doi: 10.1093/genetics/134.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valdes A M, Slatkin M, Freimer N B. Genetics. 1993;133:737–749. doi: 10.1093/genetics/133.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber J L, Wong C. Hum Mol Genet. 1993;2:1123–1128. doi: 10.1093/hmg/2.8.1123. [DOI] [PubMed] [Google Scholar]
- 18.Di Rienzo A, Peterson A C, Garza J C, Valdes A M, Slatkin M, Freimer N B. Proc Natl Acad Sci USA. 1994;91:3166–3170. doi: 10.1073/pnas.91.8.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garza J C, Slatkin M, Freimer N B. Mol Biol Evol. 1995;12:594–603. doi: 10.1093/oxfordjournals.molbev.a040239. [DOI] [PubMed] [Google Scholar]
- 20.Efron B, Tibshirani R. An Introduction to the Bootstrap. New York: Chapman & Hall; 1993. [Google Scholar]
- 21.Shriver M D, Jin L, Boerwinkle E, Deka R, Ferrell R E, Chakraborty R. Mol Biol Evol. 1995;12:914–920. doi: 10.1093/oxfordjournals.molbev.a040268. [DOI] [PubMed] [Google Scholar]
- 22.Mantel N. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- 23.Smouse P E, Long J C, Sokal R R. Syst Zool. 1986;35:627–632. [Google Scholar]
- 24.Zhivotovsky L A, Feldman M W. Proc Natl Acad Sci USA. 1995;92:11549–11552. doi: 10.1073/pnas.92.25.11549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watkins W S, Bamshad M J, Jorde L B. Hum Mol Genet. 1995;4:1485–1491. doi: 10.1093/hmg/4.9.1485. [DOI] [PubMed] [Google Scholar]
- 26.Hey, J. (1997) Mol. Biol. Evol., in press.
- 27.Deka R, Jin L, Shriver M D, Yu L M, DeCroo S, Hundrieser J, Bunker C H, Ferrell R E, Chakraborty R. Am J Hum Genet. 1995;56:461–474. [PMC free article] [PubMed] [Google Scholar]
- 28.Nei M, Takezaki N. Mol Biol Evol. 1996;13:170–177. doi: 10.1093/oxfordjournals.molbev.a025553. [DOI] [PubMed] [Google Scholar]
- 29.Templeton A R. Am Anthropol. 1993;95:51–72. [Google Scholar]
- 30.Nei M, Livshits G. Hum Hered. 1989;39:276–281. doi: 10.1159/000153872. [DOI] [PubMed] [Google Scholar]
- 31.Pritchard J K, Feldman M W. Science. 1996;274:1548–1549. [PubMed] [Google Scholar]
- 32.Weiss G, von Haeseler A. Science. 1996;272:1359–1360. doi: 10.1126/science.272.5266.1359. [DOI] [PubMed] [Google Scholar]
- 33.Jin L, Macaubas C, Hallmayer J, Kimura A, Mignot E. Proc Natl Acad Sci USA. 1996;93:15285–15288. doi: 10.1073/pnas.93.26.15285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W-H. Genetics. 1977;85:331–337. doi: 10.1093/genetics/85.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merriwether D A, Clark A G, Ballinger S W, Schurr T G, Soodyall H, Jenkins T, Sherry S T, Wallace D C. J Mol Evol. 1991;33:543–555. doi: 10.1007/BF02102807. [DOI] [PubMed] [Google Scholar]
- 36.Nachman M W, Brown W M, Stoneking M, Aquadro C F. Genetics. 1996;142:953–963. doi: 10.1093/genetics/142.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]