Abstract
Evolutionary analysis indicates that eubacterial NADP-dependent isocitrate dehydrogenases (EC 1.1.1.42) first evolved from an NAD-dependent precursor about 3.5 billion years ago. Selection in favor of utilizing NADP was probably a result of niche expansion during growth on acetate, where isocitrate dehydrogenase provides 90% of the NADPH necessary for biosynthesis. Amino acids responsible for differing coenzyme specificities were identified from x-ray crystallographic structures of Escherichia coli isocitrate dehydrogenase and the distantly related Thermus thermophilus NAD-dependent isopropylmalate dehydrogenase. Site-directed mutagenesis at sites lining the coenzyme binding pockets has been used to invert the coenzyme specificities of both enzymes. Reconstructed ancestral sequences indicate that these replacements are ancestral. Hence the adaptive history of molecular evolution is amenable to experimental investigation.
Keywords: isopropylmalate dehydrogenase, NAD, NADP, ancient adaptations
Phylogenetic reconstructions using molecular sequences form the basis of many modern evolutionary studies. With use of molecules as markers, many insights have been gained and hypotheses tested. However, the lack of a close association between the sequences and their phenotypic expression compels, with few exceptions (1), the adaptive events of molecular evolution to remain invisible. Yet any claim to an understanding of molecular evolution demands that its adaptive history be investigated. Here, we return form and function to a molecular phylogeny and, in so doing, uncover an adaptive event that occurred between 2 and 3.5 billion years ago (2, 3).
Bacteria growing on energy-rich compounds generate NADPH in various catabolic pathways leading to the Krebs cycle and assorted fermentation pathways (4). Those capable of growth on acetate use isocitrate dehydrogenase (IDH; EC 1.1.1.42) in the Krebs cycle to generate 90% of the NADPH necessary for biosynthesis (5, 6). Indeed, bacteria with NAD-dependent IDHs (NAD-IDHs) are incapable of growth on acetate and lack either a respiratory chain or a complete Krebs cycle (7). There appears to be no other major source of NADPH; the flux through NADP-dependent malic enzyme provides approximately 10% of the NADPH (5, 6), whereas transhydrogenase, a membrane bound enzyme that utilizes energy in the form of the protonmotive force to reduce NADP, has less than 5% of the activity of IDH (8–10). Hence, the NADP dependence of eubacterial IDHs appears to be an adaptation to growth on acetate. Our goal is to identify when this event occurred and the amino acid replacements responsible for it.
MATERIALS AND METHODS
Sequence Alignments.
Initial alignments obtained using clustal w (11) were modified based on a knowledge of the high resolution x-ray crystallographic structures of Escherichia coli NADP-dependent IDH (NADP-IDH) (12) and Thermus thermophilus NAD-dependent isopropylmalate dehydrogenase (NAD-IMDH) (13). Indeed, so divergent are these sequences (between 17% and 24% identical among the four major groups in Fig. 1) that residues known to be critical to substrate binding and catalysis are misaligned for a wide range of gap penalties. Instead, residues critical to substrate binding were identified from high resolution x-ray crystallographic structures of E. coli NADP-IDH with bound isocitrate (14). Residues critical to coenzyme specificity were identified from high resolution x-ray crystallographic structures of E. coli IDH complexed to NADP (15) and T. thermophilus IMDH complexed to NAD (16). Eukaryotic NAD-IDHs and NADP-IDHs were then realigned against this set using residues critical to specificity as key landmarks, and with gaps and insertions introduced into surface loops rather than in the hydrophobic cores of the domains. (The complete alignment of 67 full-length sequences is available from the authors.)
Figure 1.
Unrooted maximum likelihood tree of the decarboxylating dehydrogenases. The numbers adjacent to the arrows are the percentage bootstrap values from 1,000 nearest neighbor joining trees. Roman numerals refer to ancestral nodes, the inferred sequences of which appear in Fig. 3. The square indicates a plausible root for the tree (discussed in the text).
Phylogenetic Reconstructions.
Maximum likelihood trees (17) of the decarboxylating dehydrogenases were reconstructed using a modified point accepted mutation matrix (18) with random orders of taxa input. Consensus trees were calculated based on 1,000 neighbor joining trees (19) or 1,000 parsimony trees (20). Bootstrap values and the evaluation of maximum likelihood trees were used to assess the stability of the phylogenies. Ancestral sequences were reconstructed using a maximum likelihood algorithm (21) that assumes the tree given in Fig. 1 and equal rates of replacement among amino acids.
GenBank/EMBL accession numbers for IDHs are as follows: Anabaena sp. (PCC 7120), A55591A55591; Bacillus subtilis, U05257U05257; Bos taurus, X69432X69432, U07980U07980; Caenorhabditis elegans, C50F4, F59B8, Z46242Z46242; Escherichia coli, J02799J02799; Glycine max, Q06197Q06197, S33612S33612; Homo sapiens, S55282S55282, U52144U52144, X69433X69433, Z68907Z68907; Macaca fascicularis, X74124X74124, X82632X82632, X87172X87172; Medicago sativa, M93672M93672, S28423S28423; Mus musculus, U51167U51167; Nicotiana tabacum, S42892S42892; Rattus norvegicus, L35317L35317, X74125X74125; Saccharomyces cerevisiae, L26312L26312, M57229M57229, M74131M74131, M95203M95203; Solanum tuberosum, X67310X67310, X75638X75638; Sphingomonas yanoikuyae, U37523U37523; Sus scrofa, M86719M86719; Thermus aquaticus, A43934A43934; Thermus aquaticus thermophilus, P33197P33197; Vibrio sp. ABE-1, D14047D14047.
GenBank/EMBL accession numbers for IMDHs are as follows: Acremonium chrysogenum, D50665D50665; Agrobacterium tumefaciens, M38670M38670; Bacillus caldotenax, X04762X04762; Bacillus coagulans, M33099M33099; Bacillus megaterium, X65184X65184; Bacillus subtilis, P05645P05645; Bacteroides fragilis, D45169D45169; Brassica napus, X59970X59970; Buchnera aphidicola, P48572P48572; Candida boidinii, M98832M98832; Candida maltosa, P07139P07139; Clostridium pasteurianum, P31958P31958; Debaryomyces occidentalis, X79823X79823; Escherichia coli, P30125P30125; Haemophilus influenzae, L45625L45625; Kluyveromyces lactis, X55358X55358; Kluyveromyces marxianus, X61490X61490; Lactococcus lactis lactis, M90761M90761; Leptospira interrogans, M59431M59431; Neisseria lactamica, S43888S43888; Neurospora crassa, U01061U01061; Pichia angusta, U00889U00889; Pichia jadinii, M16014M16014; Pichia ohmeri, Z35101Z35101; Pseudomonas aeruginosa, U29655U29655; Saccharomyces cerevisiae, M12909M12909; Salmonella typhimurium, U20795U20795; Schizosaccharomyces pombe, M36910M36910; Spirulina platensis, M75903M75903; Thermus aquaticus, S41223S41223; Thermus aquaticus thermophilus, K01444K01444; Thiobacillus ferrooxidans, JX0286; Yarrowia lipolytica, M37309M37309.
Structural Biology.
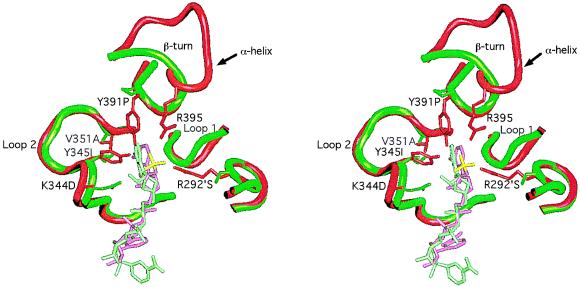
X-ray crystallographic structures were visualized using a Silicon Graphics (Mountain View, CA) 4D 120/GTX running quanta/charmm and grasp software programs. As an aid to aligning the amino acid sequence, the Cα backbone trace of each domain of T. thermophilus IMDH was independently superimposed by least squares on the corresponding domain of E. coli IDH to an rms deviation no greater than 0.42 Å. The nucleotide binding pockets with their respective bound coenzymes were superimposed by least squares to an rms deviation of 0.3 Å using all main chain atoms in loop 2 (Fig. 2, residues Thr-338 through Ala-354) and those of Tyr-391 and Asp-392 common to both IDH and IMDH despite the local difference in secondary structure. Residues in loop 1 (Fig. 2) were not aligned because of obvious and extensive differences in the main chain conformation.
Figure 2.
Stereo view of the E. coli NADP-IDH coenzyme binding pocket (15) (red, with NADP in pink and the 2′-phosphate in yellow) superimposed on that of T. thermophilus NAD-IMDH (16) (green with NAD in light green). Side chains (E. coli IDH numbering) critical to specificity are shown emanating from the backbone worms. The side chain of Lys-344 is disordered in this IDH structure, but it can interact with the 2′-phosphate of NADP.
Coordinates for IDH and IMDH structures can be obtained from the Brookhaven National Laboratory Protein Data Bank. Accession numbers for E. coli IDH are 3icd (apo-IDH), 5icd (IDH with bound Mg2+-isocitrate), 9icd (IDH with NADP bound), and 1ikb (an inactive pseudo-Michaelis complex of IDH with Ca2+-isocitrate and NADP bound). Accession numbers for T. aquaticus thermophilus IMDH are 1ipd (apo-IMDH) and 1hex (IMDH with NAD bound).
RESULTS
Phylogeny of the Decarboxylating Dehydrogenases.
IDHs belong to an ancient and divergent family of decarboxylating dehydrogenases (Fig. 1) that includes NAD-IMDH (22), which catalyzes the penultimate step in leucine biosynthesis (23). An evaluation of maximum likelihood trees and the bootstrap values of nearest neighbor joining and maximum parsimony trees indicate that each of the major groups (eukaryotic NAD-IDHs, eukaryotic NADP-IDHs, eubacterial NADP-IDHs, and the NAD-IMDHs) is monophyletic. Some species within each major group are rearranged by the different algorithms, but the trees remain very similar in overall topology.
Both evaluation of the maximum likelihood trees and the bootstrap value of 67.9% from 1,000 nearest neighbor joining trees indicate that the relative branching order of the major groups is not delimited. The highly divergent eukaryotic NADP-IDHs (<17% identical with eubacterial NADP-IDHs) either are monophyletic with eubacterial NADP-IDHs or branch slightly earlier along the NAD-IMDH limb (as shown). Indeed, the NADP binding pockets of eubacterial NADP-IDHs and eukaryotic NADP-IDHs are sufficiently different that specificity toward NADP may have evolved twice (Fig. 3).
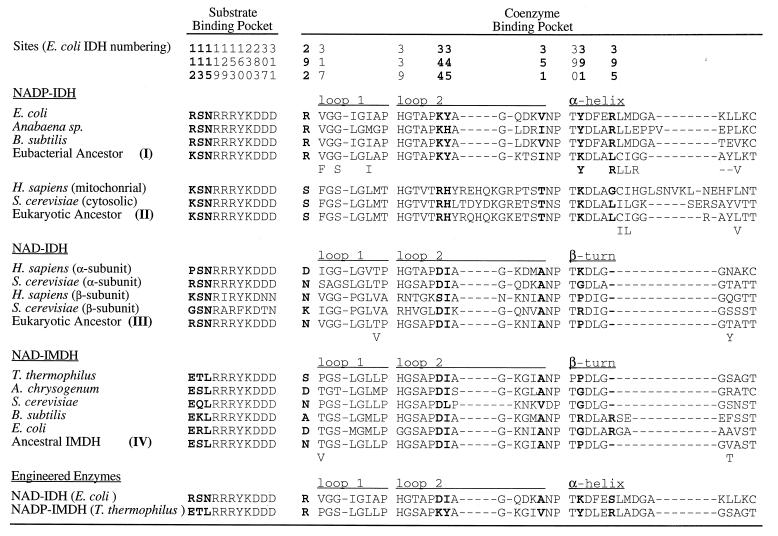
Figure 3.
Amino acids critical to substrate binding and catalysis and sequences surrounding the coenzyme binding pockets in decarboxylating dehydrogenases from several extant species, from two engineered enzymes, and from inferred ancestral sequences (numbers refer to the nodes in Fig. 1) reconstructed using a maximum likelihood algorithm (21) that assumes the tree given in Fig. 1 and equal rates of replacement among amino acids. Residues in boldface type are critical to specificity. Dashes indicate gaps. Below the most likely inferred ancestral sequences are alternative amino acids with likelihoods >10% of the preferred residue.
There is no phylogenetic means to identify the root of the tree in the absence of a defined outgroup or a known ancient ancestral gene duplication. However, the common ancestor of all these taxa was undoubtedly an autotroph, capable of synthesizing both leucine and glutamate, and hence having both IMDH and IDH activities. With this in mind, a root (indicated by the square in Fig. 1) has been tentatively placed along the limb joining the IDHs to the IMDHs. This placement, together with the number and complexity of sequence changes, is consistent with the notion that specificity toward isocitrate may have evolved before specificity toward NADP, and that the latter evolved around the time the eukaryotes first appeared.
Determinants of Coenzyme Specificity.
Residues critical to coenzyme specificity were identified from high resolution x-ray structures of E. coli IDH complexed with NADP (15) and T. thermophilus IMDH complexed with NAD (16). These enzymes share a unique nucleotide binding domain. Instead of the βαβαβ motif characteristic of the Rossmann fold common to most dehydrogenases (24), the binding pocket is constructed from two loops and an α-helix in E. coli IDH, the latter being replaced by a β-turn in T. thermophilus IMDH (Fig. 2).
Specificity in E. coli IDH is conferred by interactions between Arg-395, Lys-344, Tyr-345, Tyr-391, and Arg-292′ (the prime indicates the second subunit of the homodimer) and the 2′-phosphate of bound NADP (15). These residues are conserved in eubacterial NADP-IDHs (Fig. 3). In T. thermophilus IMDH (16), there is no site equivalent to Arg-395 because the α-helix and loop of E. coli NADP-IDH is replaced by a β-turn (Fig. 2). Replacements by Ser-292′, Ile-345, and Pro-391 eliminate all favorable interactions with the 2′-phosphate of NADP. Specificity in IMDH is conferred by the rigidly conserved Asp-344, which forms a double hydrogen bond with the 2′- and 3′-hydroxyls of the adenosine ribose of NAD, shifting its position and changing the ribose pucker from C3′-endo to C2′-endo. The shift in the position of the coenzyme, which is also accommodated by replacements Ile-345 and Ala-351, allows an additional H-bond to form between the N2 of the adenine ring and the main chain amide of residue 352 (data not shown). Not only are these movements incompatible with the strong 2′-phosphate interactions seen in NADP-IDH, but the negative charge on Asp-344 will repel NADP from the pocket in much the same way as phosphorylation of Ser-113, or replacement by Asp or Glu, inactivates IDH through repulsion of the γ-carboxylate of isocitrate (14, 16, 25–30).
Inferred Ancestral Sequences.
Ancestral character states were reconstructed using maximum likelihood (Fig. 3) rather than parsimony (31), which gives less information of the support for any one amino acid over another. The ancestral sequences were inferred with a maximum likelihood algorithm similar to that of Yang et al. (32). Errors in inferred ancestral character states often appear as homoplastic replacements (33). In our case, each inference presented is based on many species, whereas the number of possible character states is large (there are 20 amino acids). This, combined with the slow replacement rate in these proteins, reduces the number of erroneous inferred homoplastic replacements (33).
Ancestral eubacterial NADP-IDHs possess Arg-292, Lys-344, Tyr-345, Lys/Tyr-391, and Arg-395, all of which contact the 2′-phosphate of NADP, and Ile/Val-351, which contacts the adenine ring. All inferred ancestral sequences of NAD-IMDH and NAD-IDH possess Asp-344, which H-bonds to the ribose hydroxyls; Ile-345 and Ala-351, which facilitate H-bond formation by allowing the adenosine to shift in the pocket; and Pro-391, which eliminates an H-bond to the 2′-phosphate of NADP. Arg-395, which forms an H-bond to the 2′-phosphate of NADP bound in E. coli IDH, appears to be present in several NAD-IMDHs (e.g., E. coli and B. subtilis in Fig. 3). However, this site is entirely absent in other NAD-dependent enzymes in which the α-helix is replaced by a β-turn. While reconstructions favor Asn replacing Arg-292, this highly solvated surface residue is frequently replaced by other small polar residues that are too short to contact bound coenzymes.
Ancestral eukaryotic NADP-IDHs possess Arg-344, His-345, and Lys-391, all of which might interact with the 2′-phosphate of NADP. However, replacements Ser-292, Leu-395, and Tyr-351 are unlikely to interact with NADP, and in general, the sequences surrounding the eukaryotic nucleotide binding pocket are so divergent that alternative means to stabilize NADP probably operate. Hence, any conclusions regarding the importance and role of these replacements should be treated with great caution.
DISCUSSION
The Evolution of NADP-IDHs.
Phylogenetic reconstructions reveal that eubacterial NADP-IDHs are monophyletic, include both Gram-positive and Gram-negative species, and diverge near the branch points of the eukaryotic NAD- and NADP-IDHs. This suggests that NADP dependence evolved early in the history of life, on or about the time that the eukaryotes first appeared, between 2 and 3.5 billion years ago (2, 3).
Limitations of Phylogenetic Analyses.
Identifying amino acids responsible for functional differences between similar sequences is trivial. All one needs do is search among those few replacements to identify likely candidates. More sophisticated approaches use maximum parsimony (31) or maximum likelihood (21) to reconstruct ancestral sequences. Nested analyses of variance (34) can also be applied.
However, the problem of identifying the key amino acids becomes intractable by these means when, as in the case of the IDHs, the sequences are highly divergent (35). Under these circumstances, sequence alignments may identify “sites” in one enzyme that have no equivalent in another where local secondary structures differ (e.g., the α-helix and β-turn in Fig. 2). Confusion inevitably arises when sequences sharing little identity, and even encoding different secondary structures, confer the same function (e.g., nucleotide binding pockets of the eubacterial and eukaryotic NADP-IDHs). Even given common secondary structures, critical amino acids need not be conserved because there are frequently various solutions to local steric packing problems (e.g., Ala-351 in the NAD-dependent enzymes is replaced by Val in certain eukaryotic NAD-IMDHs; Fig. 3). Critical amino acids also need not be conserved when particular side chain properties (e.g., a positive charge) are more important than what is actually encoded. Substituting amino acids that are highly conserved in one part of the phylogeny may prove disruptive when they have little or nothing to do with specificity per se, and everything to do with an alternative means to stabilize similar secondary structures (e.g., replacements in loop 1 of IDH in Fig. 2 by residues found in NAD-dependent enzymes disrupt the alignment of the domains that comprise the catalytic site, so reducing catalytic efficiency). Finally, homologous sequences in some regions may be so divergent that alignments based solely on maximizing sequence identities become highly dubious (e.g., alignment of the eukaryotic NADP-IDHs with the other enzymes requires a detailed knowledge of crystallographic structures so that functionally critical residues can be identified and aligned).
Determinants of Coenzyme Specificity.
Three-dimensional structures provide invaluable information in identifying residues critical to specificity (Fig. 2). The high resolution x-ray structure of the binary complex of E. coli IDH with bound NADP (15) reveals that specificity toward NADP is conferred by H-bonds between Arg-395, Lys-344, Tyr-345, Tyr-391, Arg-292′, and the 2′-phosphate of bound coenzyme. These residues are conserved in eubacterial NADP-IDHs (Fig. 3). The high-resolution x-ray structure of the binary complex of T. thermophilus IMDH with bound NAD (16) reveals that all potentially favorable interactions with the 2′-phosphate of NADP are eliminated and that the negatively charged Asp-344, which forms a double hydrogen bond with the 2′- and 3′-hydroxyls of the adenosine ribose of NAD, is in a position to repel the 2′-phosphate of NADP. Although Asp-344 is rigidly conserved in all the NAD-dependent decarboxylating dehydrogenases, Ile-345 is sometimes replaced by Leu, whereas Ala-351, which is critical to H-bonding between Asp-344 by allowing the coenzyme to shift position, is replaced by Val in eukaryotic NAD-IMDHs.
Protein Engineering Confirms Structural Analyses.
Descriptions of the determinants of specificity based on x-ray structures demand experimental verification. Site-directed mutagenesis has been used to invert the coenzyme specificity of E. coli NADP-IDH from a 7,000-fold preference for NADP to a 200-fold preference for NAD (26). Five amino acid replacements (Lys-344 → Asp, Tyr-345 → Ile, Val-351 → Ala, Tyr-391 → Lys, and Arg-395 → Ser) introduced into the nucleotide binding pocket of wild-type E. coli IDH cause a shift in preference from NADP to NAD by a factor exceeding 1 million. Two additional replacements at sites remote to the nucleotide binding pocket improve overall performance to a level comparable with the mitochondrial NAD-IDH from yeast (Table 1).
Table 1.
Kinetic parameters of wild-type and engineered enzymes toward NADP and NAD
| Enzyme | Ref. | NADP
|
NAD
|
Specificity
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Km, μM | kcat, sec−1 | kcat/Km (A), μM−1·sec−1 | Km, μM | kcat, sec−1 | kcat/Km (B), μM−1·sec−1 | (A)/(B) NADP/NAD | (B)/(A) NAD/NADP | ||
| E. coli NADP-IDH | 26 | 17 | 80.5 | 4.7 | 4700 | 3.22 | 0.00069 | 6900 | 0.00015 |
| S. cerevisiae NAD-IDH | 36 | 210 | 40 | 0.190 | |||||
| Engineered NAD-IDH | 37 | 5800 | 4.70 | 0.00081 | 99 | 16.2 | 0.164 | 0.005 | 200 |
| T. thermophilus NAD-IMDH | 27 | 1750 | 0.26 | 0.00015 | 12 | 0.15 | 0.0125 | 0.012 | 80 |
| Engineered NADP-IMDH | 27 | 20 | 0.39 | 0.020 | 25560 | 0.52 | 0.00002 | 1000 | 0.001 |
A = kcatNADP/KmNADP; B = kcatNAD/KmNAD.
Site-directed mutagenesis has also been used to invert the coenzyme specificity of T. thermophilus NAD-IMDH (27). Replacement of the seven residues of the β-turn in IMDH with a 13-residue sequence modeled on the α-helix and loop in E. coli NADP-IDH, together with four additional replacements (Ser-292′ → Arg, Asp-344 → Lys, Ile-345 → Tyr, Ala-351 → Val), causes a shift in preference from NAD to NADP by a factor of 100,000 and a modest improvement in performance (Table 1).
The experimental demonstration that changes at the sites and secondary structures identified by x-ray crystallography are sufficient to generate changes in specificity is crucial. Parallel work on the substrate specificities of these enzymes has yet to yield significant changes while retaining catalytic efficiency (28–30).
That the coenzyme specificities of the decarboxylating dehydrogenases are determined by residues lining the nucleotide binding pocket demonstrates that the many differences outside the nucleotide binding pockets (these enzymes share only 25% sequence identity) contribute little to discrimination between NAD and NADP. Assuming that a similar number of replacements is involved with determining substrate specificity leaves some 240 replacements that play little or no role in determining specificity, toward either substrate or coenzyme. Many may be neutral, but many others are likely to have been subject to selection. For instance, in engineering the NAD-IDH, two replacements remote from the active site improved activity (but not specificity) 16-fold. Such treadmill replacements, selected at one time or another as evolving populations track environmental changes, do not alter the overall function of enzymes in any significant way.
Existing Sequence Differences Reflect Ancestral Adaptive Events.
Ancestral character states were reconstructed using maximum likelihood (Fig. 3). All inferred ancestral sequences of NAD-IMDH and NAD-IDH possess Asp-344, Ile-345, and Ala-351, which were introduced into the engineered E. coli NADP-IDH. Pro-391 and Pro-395 were not introduced because eliminating interactions with the 2′-phosphate of NADP does not require engineering of the α-helix to a β-turn. Arg-292 was retained to prevent a loss of catalytic efficiency that accompanied a further improvement in specificity. Hence, the engineered NAD-IDH contains three inferred ancestral replacements.
Ancestral eubacterial NADP-IDHs possess Arg-292, Lys-344, Tyr-345, Lys-391, and Arg-395, all of which were introduced into the engineered IMDH, and Ile-351, which was replaced by Val in the engineered IMDH and which maximum likelihood suggests Val is also highly plausible. Hence, the engineered NADP-IMDH possesses all the replacements associated with ancient adaptive events which collectively inverted the coenzyme specificity of an ancestral eubacterial IDH from NAD to NADP.
Neither of the engineered enzymes can be considered ancestral. Indeed, inspection of the likelihoods across the 250 or so replacements among the 340 sites (excluding additions and deletions) that characterize the differences between any pair of eubacterial NADP-IDHs and NAD-IMDHs suggests that the probability of accurately reconstructing an ancient ancestral enzyme is vanishingly small.
Activity of the Engineered Enzymes.
The ratio kcat/Km is both a measure of enzyme performance and a measure of the degree to which an enzyme stabilizes the transition state (38). The activity of the engineered NAD-IDH is comparable with that of the mitochondrial NAD-IDH of yeast (Table 1) when fully activated by AMP, yet both are rather less active than the wild-type NADP-dependent enzyme. This is expected. The strong ionic H-bonds formed between Arg-292, Lys-344, and Arg-395 and the 2′-phosphate of NADP of wild-type E. coli IDH must be replaced by weaker H-bonds between Asp-344 and the two ribose hydroxyls. With less energy available to bind NAD, less energy is available to stabilize the transition state and the kcat/Km must drop.
Order of Adaptive Substitutions.
Phylogenetic analyses provide no clue as to the order in which the replacements accumulated during the shift from NAD to NADP utilization. However, during the mutagenesis experiments with T. thermophilus IMDH, the activity toward NADP progressively increased as replacements Arg-292, Asp-344, Ile-345, and Tyr-391 and the Arg-395 of the α-helix were sequentially introduced. This suggests one possible route for the acquisition of specificity toward NADP.
Constraints in Molecular Evolution.
In contrast to the IDHs, all naturally occurring IMDHs are NAD-dependent. Hence, NAD utilization is either favored or constrained. The possibility that fitness is maximized by NAD-IMDH contributing to maintenance of optimal an NAD/NADH ratio seems implausible given that the Leu operon is repressed in the presence of excess Leu (37). A more plausible scenario is that NAD utilization is constrained because mutants displaying intermediate specificities toward NAD and NADP have lower overall (kcat/KmNAD + kcat/KmNADP) activities, as is the case when engineering these enzymes (26, 27). This hypothesis is directly testable; sites crucial to coenzyme specificity have been identified, site-directed mutagenesis can be used to explore possible intermediate states, and chemostat competition experiments can be used to determine Darwinian fitnesses in a model organism such as E. coli (36, 40). Hence, an understanding of the relations between structure and function at the molecular level provides a means to assess the importance of constraints in guiding adaptive evolution.
Conclusion.
The engineered NADP-IMDH possesses all the replacements associated with adaptive events that occurred billions of years ago in an ancestral eubacterial IDH and which permitted growth on acetate as the sole source of carbon and energy. We suggest that much of the adaptive history of molecular evolution is similarly amenable to reconstruction through experimental investigation.
Acknowledgments
We thank Andy Clarke, Dan Hartl, Jack Kirsch, and Phil Bragg for their helpful suggestions. A.M.D. wishes to thank John Gantz for much needed support. This work was financed by Public Health Service Grant GM-48735 from the National Institutes of Health to A.M.D. and by grants from the Natural Sciences and Engineering Research Council (Canada) and Canadian Institute for Advanced Research to G.B.G.
ABBREVIATIONS
- IDH
isocitrate dehydrogenase
- IMDH
isopropylmalate dehydrogenase
- NAD-IMDH
NAD-dependent IMDH
- NADP-IDH
NADP-dependent IDH
References
- 1.Stewart C B, Schilling J W, Wilson A C. Nature (London) 1987;330:401–404. doi: 10.1038/330401a0. [DOI] [PubMed] [Google Scholar]
- 2.Doolittle R F, Feng D-F, Tsang S, Cho G, Little E. Science. 1996;271:470–477. doi: 10.1126/science.271.5248.470. [DOI] [PubMed] [Google Scholar]
- 3.Knoll A H. Science. 1992;256:622–627. doi: 10.1126/science.1585174. [DOI] [PubMed] [Google Scholar]
- 4.Ingraham J L, Maaloe O, Neidhardt F C. Growth of the Bacterial Cell. Sunderland, MA: Sinauer; 1983. [Google Scholar]
- 5.Walsh K, Koshland D E., Jr J Biol Chem. 1984;259:9646–9654. [PubMed] [Google Scholar]
- 6.Walsh K, Koshland D E., Jr J Biol Chem. 1985;260:8430–8437. [PubMed] [Google Scholar]
- 7.Chen R, Gadal P. Plant Physiol Biochem. 1990;28:411–418. [Google Scholar]
- 8.Bragg P D, Davies P L, Hou C. Biochem Biophys Res Commun. 1972;47:1248–1255. doi: 10.1016/0006-291x(72)90969-2. [DOI] [PubMed] [Google Scholar]
- 9.Zahl K J, Rose C, Hanson R L. Arch Biochem Biophys. 1978;190:598–602. doi: 10.1016/0003-9861(78)90315-6. [DOI] [PubMed] [Google Scholar]
- 10.Liang A, Houghton R L. J Bacteriol. 1981;146:997–1002. doi: 10.1128/jb.146.3.997-1002.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurley J H, Thorsness P, Ramalingham V, Helmers N, Koshland D E, Jr, Stroud R M. Proc Natl Acad Sci USA. 1989;86:8635–8639. doi: 10.1073/pnas.86.22.8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imada K, Sato M, Tanaka N, Katsube Y, Matsuura Y, Oshima T. J Mol Biol. 1991;222:725–738. doi: 10.1016/0022-2836(91)90508-4. [DOI] [PubMed] [Google Scholar]
- 14.Hurley, J. H., Dean, A. M., Sohl, J. L., Koshland, D. E., Jr., & Stroud, R. M. Science 249, 1012–1016. [DOI] [PubMed]
- 15.Hurley J H, Dean A M, Koshland D E, Jr, Stroud R M. Biochemistry. 1991;30:8671–8678. doi: 10.1021/bi00099a026. [DOI] [PubMed] [Google Scholar]
- 16.Hurley J H, Dean A M. Structure. 1994;2:1007–1016. doi: 10.1016/s0969-2126(94)00104-9. [DOI] [PubMed] [Google Scholar]
- 17.Adachi J, Hasegawa M. Molphy: Programs for Molecular Phylogenetics; I. Protml: Maximum Likelihood Inference of Protein Phylogeny. Tokyo: Japanese Institute of Statistical Mathematics; 1992. Comput. Sci. Monogr. 27. [Google Scholar]
- 18.Jones D T, Taylor W R, Thornton J M. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 19.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein, J. (1994) phylip (University of Washington, Seattle, WA), Version 3.5.
- 21.Felsenstein J. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 22.Thorsness P E, Koshland D E., Jr J Biol Chem. 1987;264:10422–10425. [PubMed] [Google Scholar]
- 23.Burns R O, Umbarger H E, Gross S R. Biochemistry. 1963;2:1053–1057. doi: 10.1021/bi00905a024. [DOI] [PubMed] [Google Scholar]
- 24.Rossmann M G, Moras D, Olsen K W. Nature (London) 1974;250:194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- 25.Dean A M, Koshland D E., Jr Science. 1990;249:1044–1046. doi: 10.1126/science.2204110. [DOI] [PubMed] [Google Scholar]
- 26.Chen R, Greer A, Dean A M. Proc Natl Acad Sci USA. 1995;92:11666–11670. doi: 10.1073/pnas.92.25.11666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen R, Greer A, Dean A M. Proc Natl Acad Sci USA. 1996;93:12171–12176. doi: 10.1073/pnas.93.22.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dean A M, Dvorak L. Protein Sci. 1995;4:2156–2167. doi: 10.1002/pro.5560041022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen R, Grobler J A, Hurley J H, Dean A M. Protein Sci. 1996;5:287–295. doi: 10.1002/pro.5560050213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dean A M, Shiau A K, Koshland D E., Jr Protein Sci. 1996;5:341–347. doi: 10.1002/pro.5560050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jermann T M, Opitz J G, Stackhouse J, Benner S A. Nature (London) 1995;374:57–59. doi: 10.1038/374057a0. [DOI] [PubMed] [Google Scholar]
- 32.Yang Z, Kumar S, Nei M. Genetics. 1995;141:1641–1650. doi: 10.1093/genetics/141.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frumhoff P C, Reeve H K. Evolution. 1994;48:172–180. doi: 10.1111/j.1558-5646.1994.tb01303.x. [DOI] [PubMed] [Google Scholar]
- 34.Templeton A R, Boerwinkle E, Sing C F. Genetics. 1987;117:343–351. doi: 10.1093/genetics/117.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McClure M A, Vasi T K, Fitch W M. Mol Biol Evol. 1994;11:571–592. doi: 10.1093/oxfordjournals.molbev.a040138. [DOI] [PubMed] [Google Scholar]
- 36.Dean A M. Genetics. 1995;139:19–33. doi: 10.1093/genetics/139.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cupp J R, McAlister-Henn L. Biochemistry. 1993;32:9323–9328. doi: 10.1021/bi00087a010. [DOI] [PubMed] [Google Scholar]
- 38.Hackney D D. Enzymes. 1990;19:1–34. [Google Scholar]
- 39.Umbarger H E. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F, editor. Washington, DC: Am. Soc. Microbiol.; 1987. pp. 352–367. [Google Scholar]
- 40.Dykhuizen D E, Dean A M, Hartl D L. Genetics. 1987;115:25–31. doi: 10.1093/genetics/115.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]