Abstract
Activation of poly(ADP-ribose)polymerases 1 and 2 (PARP-1 and PARP-2) is one of the earliest responses of mammalian cells to DNA damage by numerous genotoxic agents. We have analysed the influence of PARP inhibition, either achieved by over-expression of the DNA binding domain of PARP-1 or by treatment with 3,4-dihydro-5-[4-(1-piperidinyl)butoxyl]-1(2H)-isoquinolinone, on the repair of single-strand breaks (SSB), pyrimidine dimers and oxidative base modifications sensitive to Fpg protein (mostly 8-hydroxyguanine) in mammalian cells at very low, non-cytotoxic levels of DNA damage. The data show that the repair rates of all three types of DNA damage are significantly lower in PARP-inhibited cells. Importantly, the retardation of the repair of base modifications is not associated with accumulation of intermediates such as SSB or abasic sites. Moreover, the influence of the PARP inhibition is not observed in cells deficient in Cockayne syndrome B protein (Csb). The results indicate that PARP activation and Csb are both involved in a novel mechanism that accelerates the global repair of various types of DNA modifications.
INTRODUCTION
Poly(ADP-ribosyl)polymerases (PARPs) are a family of enzymes that catalyse the transfer of ADP-ribose residues from NAD+ to target proteins. The well-characterised original PARP-1 and the more recently identified PARP-2 are nuclear proteins that bind to DNA single-strand breaks (SSB) and possibly other DNA structures (bends, cruciforms, unwound regions) and subsequently attach poly(ADP-ribose) residues to histones and various proteins involved in DNA processing and repair (1–5). Most prominent, however, is PARP-1 automodification, which terminates the catalytic activity of PARP-1 and causes its dissociation from the DNA.
The enzymatic activation by SSB indicates a role of PARP-1 and PARP-2 in the repair of SSB, which is probably independent of other established functions such as the activation of the transcription factor NF-κB (6,7). Indeed, the resealing of SSB generated by alkylating agents was retarded in cells treated with 3-aminobenzamide, which inhibits the enzymatic activity of PARPs by competition with NAD+ (8), in knockout cells either deficient for PARP-1 or PARP-2 (9,10) and in cells in which the PARP activity was inhibited by over-expression of its DNA binding domain (DBD) (11). Similar results were also reported for the repair of SSB induced by ionising radiation (11–13). Interestingly, the kinetic effects on SSB repair were often small or absent, while hypersensitivity to ionising radiation and alkylating agents and genomic instability, which are generally regarded a consequence of the repair defect, appear very pronounced after PARP inhibition and in PARP-1-deficient cells (4,14).
Since SSB are obligatory intermediates in base excision repair (BER) and nucleotide excision repair (NER), an influence of PARP activity on these repair pathways can be expected as well. For BER, this notion is supported by the finding that both PARP-1 and PARP-2 physically interact with other factors involved in BER, e.g. XRCC1, a supposed platform protein, DNA polymerase β and ligase III (10,15,16). The delayed repair of SSB induced by alkylating agents (see above) can be interpreted as an accumulation of repair breaks due to delayed resealing. In vitro experiments with cell-free extracts from PARP-1 knockout cells further support a role of PARP-1 in the late stages of BER. Thus, the polymerisation step following the incision at a single site of base loss (AP site) located in a plasmid was retarded (17). Specifically, the long-patch pathway of BER, which involves strand displacement synthesis prior to ligation and might replace short-patch repair in vivo under certain conditions, was shown to be stimulated by PARP-1 in vitro (17,18). In cells deficient in DNA polymerase β, PARP-1 activity is required for the ligation of repair breaks in a non-transcribed sequence, as was recently shown for transfected plasmids carrying a single 8-hydroxyguanine (8-oxoG) residue (19).
Here, we describe an analysis of the influence of PARP activity on the global repair of DNA base modifications in intact cells under conditions that do not significantly affect replication or transcription. The results indicate that both the repair of pyrimidine dimers (by NER) and that of oxidative base modifications (by BER) are significantly impaired when the PARP activity is inhibited. The retardation is not associated with an accumulation of repair breaks, but is dependent on the presence of Cockayne syndrome B protein (Csb), which originally was shown to participate in the preferential repair of transcribed genes only (20–23). Our findings indicate that PARP activation, independent of its role in SSB resealing during (long-patch) BER, is involved in a novel mechanism that accelerates global BER and NER in intact cells.
MATERIALS AND METHODS
Cell culture and enzymes
COM3 and COR3 cells are stable transfectants of the SV40-transformed hamster cell line CO60 and have been characterised previously (24). COM3 cells contain a construct for the expression of the PARP-1 DBD under control of the glucocorticoid responsive MMTV promoter and a construct for the constitutive over-expression of the human glucocorticoid receptor (Hg0). COR3 are control cells over-expressing the glucocorticoid receptor only. COM3 and COR3 cells were propagated in routine medium [Dulbecco’s modified Eagle medium (DMEM) containing 5% fetal calf serum (FCS), 1 mM l-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin], supplemented with hygromycin B (800 U/ml) in the case of the COR3 cells and hygromycin B (800 U/ml) plus geneticin (800 µg/ml) in the case of the COM3 as described (24).
Spontaneously immortalised mouse embryo fibroblasts (MEFs) of wild-type and csb–/– mice were kindly provided by D. Barnes (Cancer Research UK, London) and A. Klungland (National Hospital, Oslo, Norway). Cells were cultured in DMEM with 15% FCS, 100 U/ml penicillin and 100 µg/ml streptomycin.
Fpg protein (Escherichia coli formamidopyrimidine-DNA glycosylase) was isolated from E.coli strain JM105 harbouring plasmid pFPG230 (25). Endonuclease IV was a kind gift of B. Demple (Boston, USA). T4 endonuclease V was partially purified by the method described by Nakabeppu et al. (26) from an E.coli strain harbouring the denV gene on an inducible expression vector kindly provided by L. Mullenders (Leiden, The Netherlands).
Ro19-8022 ([R]-1-[(10-chloro-4-oxo-3-phenyl-4H-benzo[a] quinolizin-1-yl)-carbonyl]-2-pyrrolidine-methanol) was a kind gift of E. Gocke (Hoffmann-La Roche, Basel, Switzerland).
Inhibition of PARP activity
Trans-dominant PARP inhibition in COM3 cells was achieved by replacing the selection media by routine medium, which after 24 h was supplemented with 0.05 µM dexamethasone (Dex). The cells were incubated in the presence of Dex for 24 h before DNA damage induction (see below), and the glucocorticoid was also present during the repair incubations. The control cells (with or without Dex) were treated analogously.
For the inhibition of the catalytic activity of PARP, 10 µM 3,4-dihydro-5-[4-(1-piperidinyl)butoxyl]-1(2H)-isoquinolinone (DPQ; Alexis Biochemicals, Grünberg, Germany) was added to the medium 30 min before damage induction (27).
Induction and repair of DNA damage
For the predominant induction of oxidative purine modifications sensitive to Fpg protein, cells were exposed to the photosensitiser Ro 19-8022 (0.05 µM) in the presence of visible light from a 1000 W halogen lamp (Philips PF811) at a distance of 38 cm in Ca2+ and Mg2+-free PBS (140 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 1 mM KH2PO4) on ice for 10 min, corresponding to 14 kJ/m2 between 400 and 500 nm. For the induction of pyrimidine dimers by UVB, cells were irradiated with a Philips TL20W/12RS lamp (maximum emission at 306 nm) at 50 cm distance for 10 min corresponding to 2.4 J/m2. To induce SSB, cells were treated with tert-butylhydroperoxide (100 µM) in DMEM without supplements for 15 min at 37°C and 5% CO2.
Cells were washed and incubated at 37°C under culture conditions for the indicated repair times before DNA damage analysis as described below.
Quantification of DNA modifications by alkaline elution
The alkaline elution assay originally described by Kohn et al. (28) with modifications described previously (29,30) was used to quantify SSB, cyclobutane pyrimidine dimers sensitive to T4 endonuclease V, sites of base loss sensitive to endonuclease IV and oxidative purine modifications sensitive to Fpg protein. The sum of DNA modifications sensitive to one of the repair glycosylases and SSB was obtained from experiments, in which the cellular DNA was incubated for 30–60 min at 37°C with Fpg protein (1 µg/ml), endonuclease IV (1 µg/ml) or T4 endonuclease V (0.1 µg/ml) immediately after cell lysis. Under these conditions, the incision by the enzyme at endonuclease-sensitive modifications was shown to be saturated (30). The numbers of modifications incised by the repair glycosylase were obtained by subtraction of the number of SSB observed in experiments without endonuclease treatment. Elution curves obtained with γ-irradiated cells were used for calibration, assuming that 6 Gy generate 1 SSB/106 bp (28). The numbers of DNA modifications observed in untreated control cells (background levels) were subtracted in all cases.
Cleavage activity in cell extracts
Exponentially growing cells were washed with Ca2+- and Mg2+-free PBS and resuspended in 350 µl lysis buffer (20 mM Tris–HCl pH 8.0, 1 mM EDTA, 250 mM NaCl, 0.8 µg/ml antipain, 0.8 µg/ml leupeptin and 0.8 µg/ml aprotinin). The cell suspension was sonicated and after centrifugation (85 000 g, 45 min, 4°C), the protein concentrations of the recovered supernatants were determined according to Bradford.
To determine the cleavage activities of these cell extracts at either 8-oxoG or at a site of base loss, aliquots containing 5 µg total protein were incubated for 30 min at 37°C with a 34mer 5′-32P-labelled double-stranded oligodeoxyribonucleotide, which contained either an 8-oxoG or a tetrahydrofuranyl residue in the labelled strand at position 16 opposite a cytosine. The extent of DNA cleavage was determined following PAGE as described previously (31).
RESULTS
The global repair of various types of DNA modifications is retarded by PARP inhibition
The very sensitive alkaline elution technique was used to determine the repair kinetics of various types of DNA modifications in COM3 Chinese hamster cells, which over-express the PARP-1 DBD under the control of the glucocorticoid receptor and therefore lose PARP activity when treated with Dex (trans-dominant inhibition) (24). Oxidative DNA base modifications were induced by exposure to the photosensitiser Ro19-8022 plus light, which generates predominantly 8-oxoG and only relatively few SSB, sites of base loss and oxidative pyrimidine modifications (32). The number of oxidative purine modifications (including 8-oxoG) was quantified by alkaline elution in combination with the repair glycosylase Fpg as a probe. Fpg is the bacterial functional analogue of Ogg1, the most relevant mammalian repair glycosylase that initiates the BER of 8-oxoG and other oxidative guanine modifications. It should be emphasised that the low level of damage induced in the repair assays (<0.7 lesions/106 bp) was not associated with significant cytotoxicity, i.e. the proliferation rate of the cells was not reduced (data not shown).
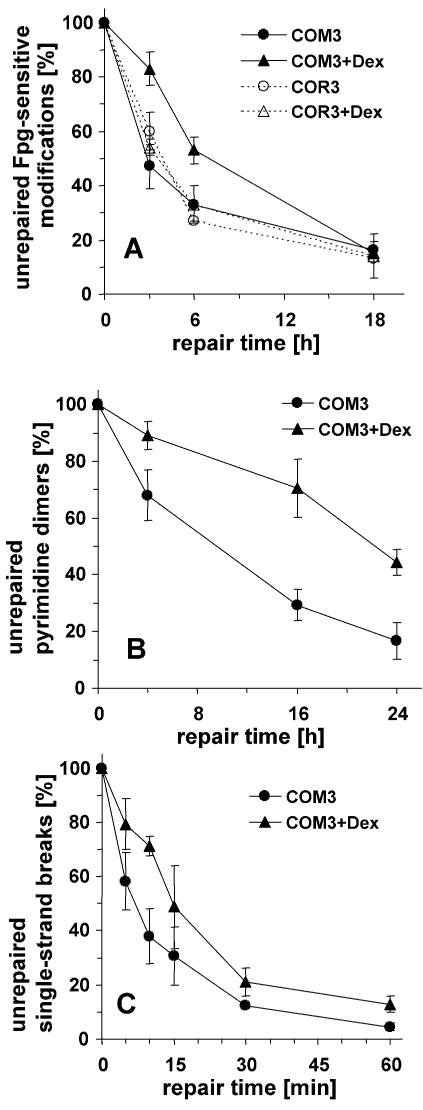
The results shown in Figure 1A indicate that the removal of the Fpg-sensitive modifications is retarded in COM3 cells when the DBD of PARP-1 is expressed by treatment with Dex. No retardation is observed in COR3 control cells, which express the glucocorticoid receptor only, with or without Dex. As shown in Figure 1B, a clear retardation by the trans-dominant inhibition of PARP activity is also observed for the repair of cyclobutane pyrimidine dimers induced by a low dose of UVB (2.4 J/m2), quantified by using T4 endonuclease V as a probe. The repair of SSB induced by treatment with tert-butylhydroperoxide, which overall is very rapid, is retarded to an extent similar to that observed for the DNA base modifications (Fig. 1C).
Figure 1.
Repair kinetics of various oxidative DNA modifications in cells undergoing trans-dominant PARP inhibition (COM3 in the presence of Dex) and control cells (COM3 without Dex, COR3 in the presence or absence of Dex). (A) Repair of Fpg-sensitive oxidative modifications induced by Ro 19-8022 (0.05 µM) plus visible light. (B) Repair of cyclobutane pyrimidine dimers sensitive to T4 endonuclease V induced by UVB (2.4 J/m2). (C) Repair of SSB induced by tert-butylhydroperoxide (100 µM). Data represent means of three to six independent experiments (± SD).
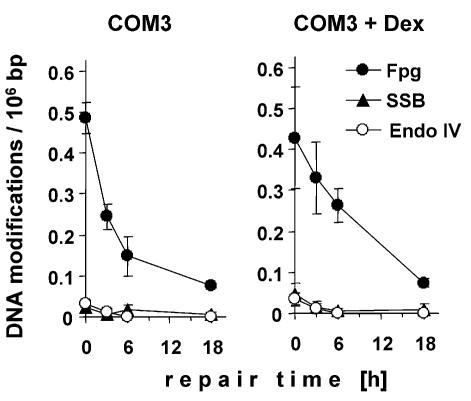
An impaired processing of AP sites and SSB generated during the repair of base modifications is expected to result in an accumulation of these intermediates. As shown in Figure 2, neither the number of SSB nor that of AP sites (quantified by using endonuclease IV as a probe) increased significantly during the repair of oxidative base modifications either with or without trans-dominant inhibition of PARP activity. The low level of these repair intermediates excludes that the incision at AP sites or the resealing of repair breaks has become rate limiting for the overall repair process of the base modifications under conditions of PARP inhibition. The same result was observed for the repair of pyrimidine dimers (data not shown). Therefore, the removal rates of the repair glycosylase-sensitive modifications shown in Figure 1A and B not only apply to the first step of the repair process, but also reflect the overall repair rate.
Figure 2.
Number of SSB and sites of base loss (sensitive to endonuclease IV) during the repair of Fpg-sensitive base modifications induced by Ro 19-8022 (0.05 µM) plus visible light in COM3 cells without (left panel) or with (right panel) trans-dominant inhibition of PARP.
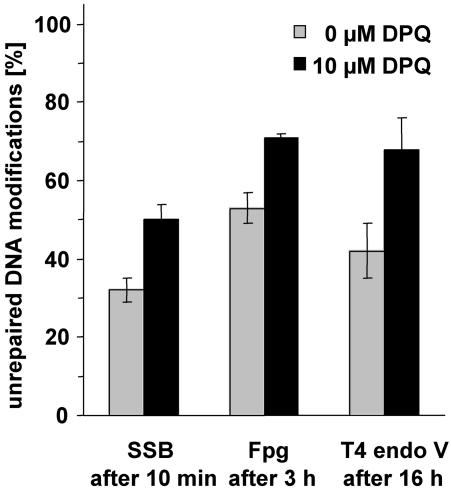
Besides, by over-expression of its DBD, the catalytic activity of PARP-1 can be inhibited by compounds that compete with NAD+ for the binding site at the catalytic domain. We therefore analysed the effects of DPQ, a recently developed efficient PARP inhibitor (27) on the repair of oxidative purine modifications induced by Ro19-8022 plus light, pyrimidine dimers induced by UVB and SSB induced by tert-butylhydroperoxide in COM3 cells (in the absence of Dex) at selected time points. The results shown in Figure 3 confirm those obtained by trans-dominant PARP inhibition, i.e. the repair of all these types of DNA modifications is retarded in the absence of PARP activity.
Figure 3.
Influence of PARP inhibition by DPQ (10 µM) on the repair of SSB, Fpg-sensitive DNA modifications and pyrimidine dimers at selected time points in COM3 cells. Data represent means of three to four independent experiments (± SD).
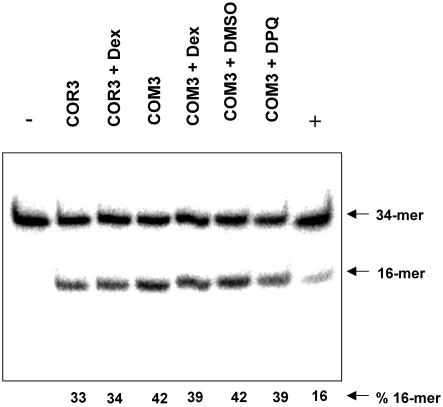
An in vitro assay was carried out to analyse the influence of PARP inhibition on the incision capacity of cell extracts at single 8-oxoG residues located in an oligonucleotide. As shown in Figure 4, total protein extracts from COR3 control cells and from COM3 cells with and without PARP inhibition by either expression of the DBD or by treatment with DPQ all exhibit similar cleavage activities at 8-oxoG. Similar results were obtained with an oligonucleotide carrying a single AP site (data not shown). Therefore, the effects of PARP inhibition on the repair rates shown in Figures 1 and 3 cannot be explained by reduced glycosylase/AP endonuclease activity as detectable in cell-free extracts.
Figure 4.
Cleavage of a 34mer duplex DNA containing a single 8-oxoG residue at position 16 by crude protein extracts of COR3 and COM3 cells, with or without PARP inhibition by pre-treatment of the cells with Dex (0.05 µM) or DPQ (10 µM). The 34mer oligodeoxyribonucleotide was 5′-32P-labelled in the 8-oxoG-containing strand and incubated with 5 µg of the crude cell-free extracts for 30 min at 37°C. The cleavage products were analysed by 20% denaturing PAGE. Controls are [8-oxoG:C] duplex incubated with buffer alone (–) or with 10 ng purified human Ogg1 protein (+).
The acceleration of repair by PARP is dependent on Csb
We have demonstrated recently that Csb protein, which is mutated in Cockayne syndrome B patients, accelerates the global repair of Fpg-sensitive oxidative base modifications and is required for an Ogg1-independent back-up repair mechanism both in vivo and in cultured cells (33). In that study, the absence of Csb had no influence on the incision capacity at 8-oxoG residues in cell-free extracts, an observation similar to that described above for the effects of PARP activity. We therefore analysed the influence of PARP inhibition on the repair in cells deficient in Csb.
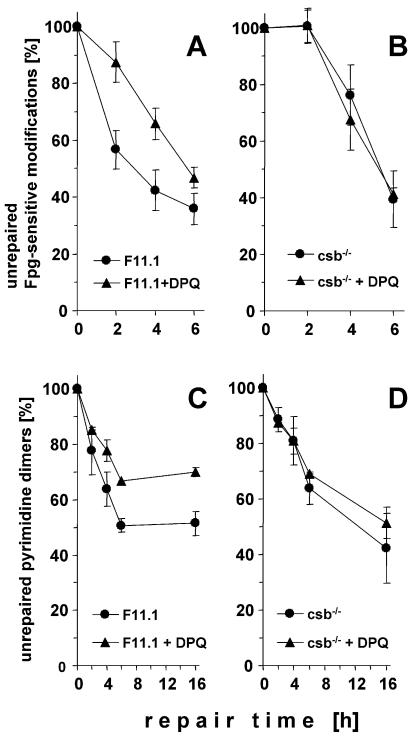
In immortalised mouse embryonic fibroblasts (MEFs) the inhibition of PARP activity by DPQ caused a similar retardation of the repair of Fpg-sensitive oxidative base modifications induced by Ro19-8022 as observed in the COM3 cells (Fig. 5A). In MEFs from csb–/– knockout mice, the repair without DPQ was significantly slower than in the wild-type cells, as described previously (33). Strikingly, inhibition of PARP activity by DPQ did not further reduce the repair rates (Fig. 5B). Therefore, the roles of PARP activity and Csb protein in the stimulation of the global repair of the oxidative base modifications appear to depend on each other.
Figure 5.
Repair of Fpg-sensitive oxidative base modifications induced by Ro 19-8022 plus light (A and B) and of cyclobutane pyrimidine dimers (C and D) in wild-type (F11.1; A and C) and Csb-deficient (B and D) MEFs with and without PARP inhibition by pre-treatment with DPQ (10 µM). Data represent the means of three to four independent experiments (± SD).
The effect of PARP inhibition by DPQ on the repair of cyclobutane pyrimidine dimers in wild-type MEFs was much smaller than in the COM3 cells, but still significant (Fig. 5C). Similarly, the global repair of the pyrimidine dimers in the Csb-deficient fibroblasts was only slightly slower than in wild-type cells. As in the case of the oxidative base modifications induced by photosensitisation, inhibition of PARP by DPQ in the Csb-deficient fibroblasts did not cause further retardation of the repair (Fig. 5D).
Only small effects of DPQ on the rejoining of SSB induced by tert-butylhydroperoxide were observed in both wild-type and Csb-deficient fibroblasts, and the repair rates in the two types of fibroblasts did not differ significantly (data not shown).
DISCUSSION
The results presented demonstrate that PARP activity has a distinct influence on the global repair of DNA base modifications. Both the removal of oxidative purine modifications including 8-oxoG, which in mammalian cells are predominantly subject to BER, and that of cyclobutane pyrimidine dimers, which are repaired by NER only, is significantly slower when PARP activity is inhibited either by over-expression of the PARP-1 DBD or by a low-molecular weight inhibitor of the catalytic domain. This effect cannot be explained by the established role of PARP-1 in SSB resealing during (long-patch) BER, since the level of repair breaks represents only a small fraction of the total number of lesions even under conditions of PARP inhibition (Fig. 2), indicating that the resealing of the repair breaks is not the rate limiting step of the overall repair process when PARP is inhibited. In contrast, the previously reported accumulation of SSB in PARP-inhibited or PARP-deficient cells exposed to alkylating agents probably reflects the impaired resealing of SSB, which in this case may be generated both enzymatically (repair breaks) and non-enzymatically (following depurination).
Although PARP-1 has been shown to act as a regulator of transcription factors such as p53 and NF-κB (6,34–36), the acceleration of BER cannot be explained by up-regulated expression or increased activity of the repair enzymes directly involved in the recognition and excision of the base modifications, as the cleavage activity observable in an in vitro repair assay was not affected by PARP inhibition (Fig. 4). An involvement of p53 also appears unlikely because the effect was also observed in COM3 cells (Figs 1 and 3), which are SV40-transformed and therefore lack functional p53. It should be noted in this context that the global repair of cyclobutane pyrimidine dimers was found to be strongly stimulated by a functionally active p53, which acts as a transactivator of p48, a specific recognition factor for this type of modification (37,38). The relatively efficient and apparently p53- independent repair observed in this study might be explained by the only very low level of DNA damage that was generated.
An interesting clue regarding the role of PARP activity in BER and NER comes from the observation that Csb-deficient cells display a similar repair retardation as PARP-inhibited cells and that the repair in csb–/– cells is not further impaired by PARP inhibition (Fig. 5B and D). Csb protein has long been known for its involvement in the transcription-coupled repair of various lesions including pyrimidine dimers and 8-oxoG (21–23), but its role in the global repair of 8-oxoG has been demonstrated only recently (33,39,40). The underlying mechanism still remains to be established, but in our recent study (33) up-regulation of the cleavage activity at 8-oxoG detectable in cell extracts was not involved, as in the case of the PARP inhibition (Fig. 4). Instead, the recently demonstrated implication of CSB in ATP-dependent chromatin remodelling (41) suggests that CSB might accelerate repair by increasing the accessibility of packaged DNA (42). Direct poly(ADP-ribosyl)ation of histones or PARP-1 automodification leading to non-covalent interaction of histones with poly(ADP-ribose) and thus to chromatin decondensation (43) appear to be involved in the same pathway, since the effects of Csb deficiency and PARP inhibition were not additive (Fig. 5). Conceivably, the two proteins cooperate in the opening of the chromatin that is triggered by a stalled transcription machinery and allows a more rapid repair of DNA modifications situated far away from the transcription site. In accordance with this assumption, the acceleration of global repair mediated by Csb is already observable at early time points (Fig. 5), but requires active transcription, as deduced from its inhibition by α-amanitin (33). The recent suggestion that poly(ADP-ribose) might serve as a supply of ATP (44) is interesting in view of the DNA-dependent ATPase activity of Csb.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Ina Schulz for excellent technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft (Ep11/4), the PROCOPE program and the Association pour la Recherche sur le Cancer (No. 4688 to J.P.R.).
REFERENCES
- 1.Shall S. and de Murcia,G. (2000) Poly(ADP-ribose)polymerase-1: what have we learned from the deficient mouse model? Mutat. Res., 460, 1–15. [DOI] [PubMed] [Google Scholar]
- 2.Bürkle A. (2001) Poly(APD-ribosyl)ation, a DNA damage-driven protein modification and regulator of genomic instability. Cancer Lett., 163, 1–5. [DOI] [PubMed] [Google Scholar]
- 3.Bürkle A. (2001) Physiology and pathophysiology of poly(ADP-ribosyl)ation. Bioessays, 23, 795–806. [DOI] [PubMed] [Google Scholar]
- 4.Herceg Z. and Wang,Z.Q. (2001) Functions of poly(ADP-ribose) polymerase (PARP) in DNA repair, genomic integrity and cell death. Mutat. Res., 477, 97–110. [DOI] [PubMed] [Google Scholar]
- 5.Ziegler M. and Oei,S.L. (2001) A cellular survival switch: poly(ADP-ribosyl)ation stimulates DNA repair and silences transcription. Bioessays, 23, 543–548. [DOI] [PubMed] [Google Scholar]
- 6.Oliver F.J., Menissier-de Murcia,J., Nacci,C., Decker,P., Andriantsitohaina,R., Muller,S., de la Rubia,G., Stoclet,J.C. and de Murcia,G. (1999) Resistance to endotoxic shock as a consequence of defective NF-kappaB activation in poly (ADP-ribose)polymerase-1 deficient mice. EMBO J., 18, 4446–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hassa P.O. and Hottinger,O. (1999) A role of poly (ADP-ribose) polymerase in NF-kappaB transcriptional activation. Biol. Chem., 380, 953–959. [DOI] [PubMed] [Google Scholar]
- 8.Durkacz B.W., Omidiji,O., Gray,D.A. and Shall,S. (1980) (ADP-ribose)n participates in DNA excision repair. Nature, 283, 593–596. [DOI] [PubMed] [Google Scholar]
- 9.Trucco C., Oliver,F.J., de Murcia,G. and Menissier-de Murcia,J. (1998) DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res., 26, 2644–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schreiber V., Ame,J.C., Dolle,P., Schultz,I., Rinaldi,B., Fraulob,V., Menissier-de Murcia,J. and de Murcia,G. (2002) Poly(ADP-ribose)polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem., 277, 23028–23036. [DOI] [PubMed] [Google Scholar]
- 11.Beneke R., Geisen,C., Zevnik,B., Bauch,T., Müller,W.U., Küpper,J.H. and Moroy,T. (2000) DNA excision repair and DNA damage-induced apoptosis are linked to poly(ADP-ribosyl)ation but have different requirements for p53. Mol. Cell. Biol., 20, 6695–6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durkacz B.W., Shall,S. and Irwin,J. (1981) The effect of inhibition of (ADP-ribose)n biosynthesis on DNA repair assayed by the nucleoid technique. Eur. J. Biochem., 121, 65–69. [DOI] [PubMed] [Google Scholar]
- 13.James M.R. and Lehmann,A.R. (1982) Role of poly(adenosine diphosphate ribose) in deoxyribonucleic acid repair in human fibroblasts. Biochemistry, 21, 4007–4013. [DOI] [PubMed] [Google Scholar]
- 14.Vodenicharov M.D., Sallmann,F.R., Satoh,M.S. and Poirier,G.G. (2000) Base excision repair is efficient in cells lacking poly(ADP-ribose)polymerase 1. Nucleic Acids Res., 28, 3887–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caldecott K.W., Aoufouchi,S., Johnson,P. and Shall,S. (1996) XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly(ADP-ribose)polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res., 24, 4387–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masson M., Niedergang,C., Schreiber,V., Muller,S., Menissier-de Murcia,J. and de Murcia,G. (1998) XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol., 18, 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dantzer F., de La Rubia,G., Menissier-De Murcia,J., Hostomsky,Z., de Murcia,G. and Schreiber,V. (2000) Base excision repair is impaired in mammalian cells lacking poly(ADP-ribose)polymerase-1. Biochemistry, 39, 7559–7569. [DOI] [PubMed] [Google Scholar]
- 18.Prasad R., Lavrik,O.I., Kim,S.J., Kedar,P., Yang,X.P., Vande Berg,B.J. and Wilson,S.H. (2001) DNA polymerase beta-mediated long patch base excision repair. Poly(ADP-ribose)polymerase-1 stimulates strand displacement DNA synthesis. J. Biol. Chem., 276, 32411–32414. [DOI] [PubMed] [Google Scholar]
- 19.Le Page F., Schreiber,V., Dherin,C., De Murcia,G. and Boiteux,S. (2003) Poly(ADP-ribose)polymerase-1 (PARP-1) is required in murine cell lines for base excision repair of oxidative DNA damage in the absence of DNA polymerase beta. J. Biol. Chem., 278, 18471–18477. [DOI] [PubMed] [Google Scholar]
- 20.Venema J., Mullenders,L.H., Natarajan,A.T., van Zeeland,A.A. and Mayne,L.V. (1990) The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc. Natl Acad. Sci. USA, 87, 4707–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedberg E.C. (1996) Relationships between DNA repair and transcription. Annu. Rev. Biochem., 65, 15–42. [DOI] [PubMed] [Google Scholar]
- 22.Tornaletti S. and Hanawalt,P.C. (1999) Effect of DNA lesions on transcription elongation. Biochimie, 81, 139–146. [DOI] [PubMed] [Google Scholar]
- 23.Le Page F., Kwoh,E.E., Avrutskaya,A., Gentil,A., Leadon,S.A., Sarasin,A. and Cooper,P.K. (2000) Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell, 101, 159–171. [DOI] [PubMed] [Google Scholar]
- 24.Küpper J.H., Müller,M., Jacobson,M.K., Tatsumi-Miyajima,J., Coyle,D.L., Jacobson,E.L. and Bürkle,A. (1995) Trans-dominant inhibition of poly(ADP-ribosyl)ation sensitizes cells against γ-irradiation and N-methyl-N′-nitro-N-nitrosoguanidine but does not limit DNA replication of a polyomavirus replicon. Mol. Cell. Biol., 15, 3154–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boiteux S., O’Connor,T.R., Lederer,F., Gouyette,A. and Laval,J. (1990) Homogeneous Escherichia coli FPG protein. A DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidinic sites. J. Biol. Chem., 265, 3916–3922. [PubMed] [Google Scholar]
- 26.Nakabeppu Y., Yamashita,K. und Sekiguchi,M. (1982) Purification and characterization of normal and mutant forms of T4 endonuclease V.J. Biol. Chem., 257, 2556–2562. [PubMed] [Google Scholar]
- 27.Suto M.J., Turner,W.R., Arundel-Suto,C.M., Werbel,L.M. and Sebolt-Leopold,J.S. (1991) Dihydroisoquinolinones: the design and synthesis of a new series of potent inhibitors of poly(ADP-ribose) polymerase. Anticancer Drug Des., 6, 107–117. [PubMed] [Google Scholar]
- 28.Kohn K.W., Erickson,L.C., Ewig,R.A.G. and Friedman,C.A. (1976) Fractionation of DNA from mammalian cells by alkaline elution. Biochemistry, 15, 4629–4637. [DOI] [PubMed] [Google Scholar]
- 29.Epe B. and Hegler,J. (1994) Oxidative DNA damage: endonuclease fingerprinting. Methods Enzymol., 234, 122–131. [DOI] [PubMed] [Google Scholar]
- 30.Pflaum M., Will,O. and Epe,B. (1997) Determination of steady-state levels of oxidative DNA base modifications in mammalian cells by means of repair endonucleases. Carcinogenesis, 18, 2225–2231. [DOI] [PubMed] [Google Scholar]
- 31.Vidal A.E., Hickson,I.D., Boiteux,S. and Radicella,J.P. (2001) Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Res., 29, 1285–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulz I., Mahler,H.C., Boiteux,S. and Epe,B. (2000) Oxidative DNA base damage induced by singlet oxygen and photosensitization: recognition by repair endonucleases and mutagenicity. Mutat. Res., 461, 145–156. [DOI] [PubMed] [Google Scholar]
- 33.Osterod M., Larsen,E., Le Page,F., Hengstler,J.G., Van Der Horst,G.T., Boiteux,S., Klungland,A. and Epe,B. (2002) A global DNA repair mechanism involving the Cockayne syndrome B (CSB) gene product can prevent the in vivo accumulation of endogenous oxidative DNA base damage. Oncogene, 21, 8232–8239. [DOI] [PubMed] [Google Scholar]
- 34.Agarwal M.L., Agarwal,A., Taylor,W.R., Wang,Z.Q., Wagner,E.F. and Stark,G.R. (1997) Defective induction but normal activation and function of p53 in mouse cells lacking poly-ADP-ribose polymerase. Oncogene, 15, 1035–1041. [DOI] [PubMed] [Google Scholar]
- 35.Simbulan-Rosenthal C.M., Ly,D.H., Rosenthal,D.S., Konopka,G., Luo,R., Wang,Z.Q., Schultz,P.G. and Smulson,M.E. (2000) Misregulation of gene expression in primary fibroblasts lacking poly(ADP-ribose)polymerase. Proc. Natl Acad. Sci. USA, 97, 11274–11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wieler S., Gagne,J.P., Vaziri,H., Poirier,G.G. and Benchimol,S.J. (2003) Poly(ADP-ribose)polymerase-1 is a positive regulator of the p53-mediated G1 arrest response following ionizing radiation. J. Biol. Chem., 278, 18914–18921. [DOI] [PubMed] [Google Scholar]
- 37.Ford J.M. and Hanawalt,P.C. (1997) Expression of wild-type p53 is required for efficient global genomic nucleotide excision repair in UV-irradiated human fibroblasts. J. Biol. Chem., 272, 28073–28080. [DOI] [PubMed] [Google Scholar]
- 38.Hwang B.J., Ford,J.M., Hanawalt,P.C. and Chu,G. (1999) Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl Acad. Sci. USA, 96, 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tuo J., Muftuoglu,M., Chen,C., Jaruga,P., Selzer,R.R., Brosh,R.M.,Jr, Rodriguez,H., Dizdaroglu,M. and Bohr,V.A. (2001) The Cockayne Syndrome group B gene product is involved in general genome base excision repair of 8-hydroxyguanine in DNA. J. Biol. Chem., 276, 45772–45779. [DOI] [PubMed] [Google Scholar]
- 40.Sunesen M., Stevnsner,T., Brosh,R.M.,Jr, Dianov,G.L. and Bohr,V.A. (2002) Global genome repair of 8-oxoG in hamster cells requires a functional CSB gene product. Oncogene, 21, 3571–3578. [DOI] [PubMed] [Google Scholar]
- 41.Citterio E., Van Den Boom,V., Schnitzler,G., Kanaar,R., Bonte,E., Kingston,R.E., Hoeijmakers,J.H. and Vermeulen,W. (2000) ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell. Biol., 20, 7643–7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ura K. and Hayes,J.J. (2002) Nucleotide excision repair and chromatin remodeling. Eur. J. Biochem., 269, 2288–2293. [DOI] [PubMed] [Google Scholar]
- 43.Realini C.A. and Althaus,F.R. (1992) Histone shuttling by poly(ADP-ribosylation). J. Biol. Chem., 267, 18858–18865. [PubMed] [Google Scholar]
- 44.Oei S.L. and Ziegler,M. (2000) ATP for the DNA ligation step in base excision repair is generated from poly(ADP-ribose). J. Biol. Chem., 275, 23234–23239. [DOI] [PubMed] [Google Scholar]