Abstract
The bric à brac (bab) locus is composed of two paralogous genes, bab1 and bab2, in Drosophila melanogaster. Bab1 and Bab2 are nuclear proteins that contain a broad complex, tramtrack, bric à brac/poxviruses and zinc-finger (BTB/POZ) domain. Many BTB/POZ proteins are transcriptional regulators of which the majority contain C2H2 zinc-finger motifs. There is no detectable zinc-finger motif in either Bab protein. However, they share the Bab conserved domain (BabCD) that is highly conserved between Bab1 and Bab2, and the Bab proteins of several other species, e.g. Anopheles gambiae, Apis mellifera and Drosophila virilis. Here we show that Bab2 binds to several discrete sites on polytene chromosomes including the bab locus, and that the BabCD of both Bab1 and Bab2 binds in vitro to the cis-regulatory regions of bab1 and bab2. Our results indicate that the BabCD binds to A/T-rich regions and that its optimum binding sites contain TA or TAA repeats. The BabCD is a composite DNA binding domain with a psq motif and an AT-Hook motif; both motifs are required for DNA binding activity. Structural similarities suggest that the BabCD may bind to DNA in a similar manner as some prokaryotic recombinases.
INTRODUCTION
During development, many genes are expressed in a temporal and/or tissue-specific manner. Regulated transcription of such genes is achieved by transcription factors that bind specific regulatory sequences in the genome and recruit co-activators or co-repressors to the promoters of target genes. Transcription factors function modularly and can be classified according to the structure of their DNA binding domain.
Many broad complex, tramtrack, bric à brac/poxviruses and zinc-finger (BTB/POZ) proteins are transcriptional regulators. The BTB/POZ domain is an evolutionarily conserved protein–protein interaction domain that is involved in homophilic and heterophilic interactions (1,2). Analysis of promyelocytic leukemia zinc-finger (PLZF) dimers shows that a charged pocket is formed at the BTB/POZ dimer interface (3). Melnick et al. (4) showed that the charged pockets of PLZF and Bcl-6 are required for recruitment of corepressor-HDAC complexes by directly interacting with N-CoR and SMRT. The majority of BTB/POZ transcriptional regulators also contain C2H2 zinc-finger motifs, one of the most common DNA binding domains (5–7).
The functionally related bric à brac 1 (Bab1) and bric à brac 2 (Bab2) proteins are nuclear BTB/POZ proteins that are required during the development of adult ovaries (8–10), legs (8,10,11), antennae (11,12) and abdomen (10,13) in Drosophila melanogaster. Bab1 and Bab2 can homo- and hetero-dimerize in vitro (14). The BTB/POZ domain of Bab proteins binds to BIP-2/TAF3, which is a component of TFIID, suggesting that the Bab proteins can interact directly with the basal transcriptional machinery (14,15). There is no detectable zinc-finger motif in either Bab protein. However, both Bab proteins share a strongly conserved domain that we call the Bab conserved domain (BabCD) (10).
The BabCD (10) contains a pipsqueak (psq) motif (16) and an AT-Hook motif (17). The psq motif was first identified in the Psq protein which contains four tandem repeats of this motif (16). It has been shown that three of these repeats are required for DNA binding (18). A systematical search for the psq motif in protein databases has shown that it is evolutionarily conserved and present in several Drosophila proteins that also have BTB domains (19).
The AT-Hook motif is a positively charged stretch of 10 amino acids containing the invariant core peptide sequence RGRP and is usually flanked by basic residues (17,20). It binds to DNA through the minor groove with the optimal DNA binding site centered at the sequence AA(T/A)T (20,21). The AT-Hook was first identified in the high mobility group of non-histone chromosomal proteins, called HMGA (17). Since its discovery, the AT-Hook motif has been found in either single or multiple copies in a large number of DNA binding proteins, many of which are transcription factors or components of chromatin remodeling complexes from a wide range of organisms (20,22).
In this paper, we show that the BabCD of both Bab1 and Bab2 is able to bind DNA through a composite DNA binding domain. They bind in vitro to several DNA fragments from the cis-regulatory region of bab1 and bab2 genes. Our data indicates that the BabCD binds specifically to A/T-rich regions and that its optimum binding site contains TA or TAA repeats. The psq motif and AT-Hook motif within the BabCD are both required for its DNA binding activity.
MATERIALS AND METHODS
Immunostaining of Drosophila salivary gland polytene chromosomes
Salivary glands were dissected from wild-type and homozygous babAR07 mutant larvae at the third instar. Preparation of polytene chromosomes and immunostaining were performed as described previously (23). The polyclonal rat anti-Bab2-R10 primary antibody (10) was diluted 1/1000. The specificity of this antibody has been demonstrated previously (10). The Vectastain Kit (Vector Laboratories) was used for signal detection. The chromosomes were counterstained with Giemsa. Chromosome in situ hybridizations, using a digoxigenin-labeled bab2 cDNA as a probe, were executed as described in Godt et al. (12).
Expression and purification of GST–BabCD recombinant proteins
To generate fusion proteins between the BabCD and the glutathione-S-transferase (GST), DNA sequences that encode the entire BabCD1 and BabCD2 or parts of the BabCD1 were amplified by PCR, using the bab1 cDNA and bab2 cDNA as templates and the Expand Long Template PCR system (Roche), and were subcloned into a pGEX (Amersham Pharmacia Biotech) expression vector using the appropriate restriction sites. The following Bab peptides were expressed as GST fusion proteins: BabCD1, amino acids 490–672; BabCD1151, amino acids 522–672; BabCD1123, amino acids 550–672; BabCD1119, amino acids 490–608; BabCD193, amino acids 580–672; BabCD173, amino acids 550–622; BabCD159, amino acids 550–608; BabCD2, amino acids 560–742. All constructs were verified by automated DNA sequencing prior to transformation into Escherichia coli BL21.
Production of GST–BabCD fusion proteins in E.coli and purification of recombinant proteins were performed as described in Current Protocols in Molecular Biology (24) with slight modifications. BL21 transformant colonies were inoculated into 100 ml of LB/ampicillin medium and incubated for 3 h at 30°C. Fusion protein expression was induced by adding IPTG to a final concentration of 0.1 mM and further incubating for 3 h. Pellets were resuspended in 10 ml of ice-cold solubilization buffer (50 mM Tris–HCl pH 7.4, 1 mM EDTA, 100 mM NaCl, 10% glycerol, 1% NP-40, 1 mM DTT, 1 nM PMSF, 10 µg/ml aprotinin, 2 µg/ml leupeptin, 2 µg/ml pepstatin, 0.5 mg/ml lysozyme). After sonication, supernatants were incubated for 30 min with 1 ml of 50% glutathione–agarose beads, washed three times in 1 M NaCl, three times in PBS and resuspended in 1 ml of PBS. For DNase I footprinting and electrophoretic mobility shift assay (EMSA) experiments, GST fusion proteins were eluted from beads by incubating for 10 min in 10 mM glutathione/50mM Tris–HCl, pH 9.
In vitro pull-out experiments
bab1 or bab2 genomic DNA fragments, cloned into the Bluescript vector, were digested with HaeIII. The resulting fragments were mixed with GST–BabCD proteins on glutathione–agarose beads in the binding buffer [10 mM HEPES, 50 mM KCl, 1 mM DTT, 2.5 mM MgCl2, 20 µg/ml poly(dG–dC), 7.5% glycerol pH 7.9] and incubated at room temperature for 2 h. Beads were pelleted and washed twice in the binding buffer containing 300 mM NaCl to remove all fragments that were not tightly bound. DNA that remained bound to the beads was extracted by phenol/chloroform, precipitated and resuspended in TE before being analyzed on a 5% non-denaturing polyacrylamide gel.
In vitro DNase I footprinting on genomic bab DNA
DNase I footprinting assays were done according to standard protocols (24). Approximately 100 ng of eluted GST–BabCD1123 were incubated with either the 450 bp bab1 DNA fragment or 222 bp bab2 DNA fragment end-labeled with the T4 polynucleotide kinase (Life Technologies) and [γ-32P]ATP at 3000 Ci/mmol. Both the concentration of DNase I and the time of digestion were empirically determined to obtain optimal results. Products of the Maxam–Gilbert chemical cleavage reaction of either the 450 bp bab1 DNA fragment or the 222 bp bab2 DNA fragment served as reference standards.
Cyclic amplification of selected targets (CAST)
CAST experiments (24) were done using oligonucleotides with an internal variable region (20 random nucleotides). Oligonucleotides retained by the BabCD1 fusion protein were isolated either by in vitro pull-out (IVPO) or by gel-shift. Ten cycles of selection were done and the selected oligos were cloned into pGEMT-T. After sequencing, internal variable regions were analyzed by MEME version 3.0 (25).
Electrophoretic mobility shift assays
Oligonucleotide 5′-3′ sequences of the plus strands are as follows: 1TAA, GTCGACGCCTAAGCCCTGCAG; 2TAA, GTCGACGCTAATAAGCTGCAG; 3TAA, GTCGACTAATAATAACTGCAG; 4TAA, GTCGTAATAATAATAAGCAG; 5TA, GTCGACTATATATATACTGCA; 3TAA*TTA, GTCGACTAATTATAACTGCAG; 3TAA*AAA, GTCGACTAAAAATAACTGCAG; 3TAA*TCA, GTCGACTAATCATAACTGCAG; 3TAA*TAC, GTCGACTAATACTAACT GCAG; 5GA, GTCGACGAGAGAGAGACTGCA; 4GAA, GTCGGAAGAAGAAGAATGCAG; EcRE, AGACAAGGGTTCAATGCACTTGTCCAA. Double-stranded oligonucleotides were end-labeled with T4 polynucleotide kinase (Life Technologies) and [γ-32P]ATP at 3000 Ci/mmol.
Binding reactions were carried out at room temperature in a mixture containing 10 mM HEPES pH 7.9, 1 mM DTT, 100 mM KCl, 2.5 mM MgCl2, 10% glycerol, 0.05 µg/µl poly(dG–dC) (Sigma) in a reaction volume of 20 µl. After incubation for 20 min, the binding mixtures were loaded onto pre-electrophoresed 5% polyacrylamide gels in 0.5× TBE, run at 4°C and 200 V, and autoradiographed.
Generation of mutated BabCD
Amino acid substitutions in the AT-Hook or in the psq domain of BabCD1 were generated using the QuikChange (Stratagene) system according to the manufacturer’s instructions. The forward primers used to generate the BabCDAT-Hook1, changing RGR to RGD and the Bab CDAT-Hook2, changing RGR to DGD, had the sequence CGGCCCAAGGGGCGTGGCGACCCGCAGCGAATC and CGGCCCAAGGGGGATGGCGACCCGCAGCGAATC, respectively. The forward primers used to generate the BabCD1AI576GP and the BabCD1A590P mutants had the sequences TCGCTATCTCAGCCCGCCCGCAAGTACGAC and ACCATGGCGGAGGGCCCTTTCAGTGTGCTA, res pectively. The substituted bases are underlined. The sequence changes and the integrity of the surrounding sequence were verified by DNA sequencing.
RESULTS
The Bab2 protein binds to specific sites on polytene chromosomes
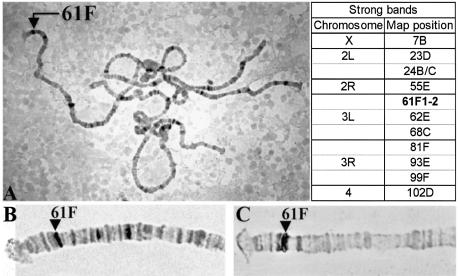
We have generated a Bab2 antibody that detects a 145 kDa protein on western blots from prepupal leg and ovary imaginal discs. This protein is undetectable in babAR07/babAR07 mutants and is truncated in two bab ethylmethane sulfonate alleles (10). This antibody detects a nuclear protein in the terminal filament cells in ovaries and in the tarsus of leg imaginal discs from wild-type larvae but not from babE1 mutants (10). Immunostainings of squashed polytene chromosomes using this Bab2 antibody shows that Bab2 binds to many sites on the chromosome arms but not to the chromocenters (Fig. 1A). This staining pattern was not seen on chromosomes from bab mutant larvae that lack Bab2 (data not shown), which served as a control, demonstrating that the staining pattern indeed represents the binding pattern of the Bab protein. Approximately 66 sites are stained, and the position of the 12 most intensely stained bands are listed in Figure 1. One of the strongest sites corresponds to the bab locus, which maps to 61F1-2 at the tip of the left arm of the third chromosome (Fig. 1B and C). This observation indicates that the Bab proteins are chromosomal proteins and are able to bind directly or indirectly to DNA. It also suggests that the chromosomal binding sites identify loci whose expression is regulated by the Bab proteins and that the Bab proteins regulate the expression of the bab genes.
Figure 1.
Bab2 proteins bind to several loci on polytene chromosomes. (A) The Bab2 protein was detected on squashed salivary gland polytene chromosomes and the strongest binding sites were cytologically mapped. The arrow on the left arm of the third chromosome points to the bab locus at 61F1-2. Indicated on the right are the map positions of the 12 strongest binding sites that include a double band at the 24B/C boundary. (B) and (C) Tip of the left arm of the third chromosome stained with the anti-Bab2 antibody (B) or with a digoxigenin labeled bab2 cDNA (C).
The BabCD is highly conserved between the Bab proteins of D.melanogaster, Drosophila virilis, Anopheles gambiae and Apis mellifera
The bab locus is composed of two paralogous genes, bab1 and bab2. The corresponding proteins Bab1 and Bab2 are nuclear and have a BTB/POZ domain in their N-terminal region (10). However, in contrast to many other known BTB/POZ domains containing nuclear proteins, neither Bab1 nor Bab2 contain a zinc-finger motif. The Bab1 and Bab2 proteins have a second domain in common, the BabCD that is 183 amino acids long and shows 78% identity between the two Bab proteins (10). A search of the genome of A.gambiae (26) and of a library of EST sequences of A.mellifera (Honey Bee Brain EST Project, http://titan.biotec.uiuc.edu/bee/honeybee_project.htm) revealed that a bab gene is present in these species. The presence of only one bab gene in the genome of A.gambiae suggests that the duplication of the bab gene might have occurred after the divergence of mosquitoes and flies. We have also identified part of the genomic sequence of a bab gene from D.virilis from a genomic library (a gift from Ron Blackman, University of Illinois) (C.Lours, unpublished data). A comparison of the BabCD of these different insect species shows that this domain is highly conserved as at least 85% of the amino acids are identical and most of the differences represent conservative exchanges (see Supplementary Material), indicating that the BabCD must have functional importance in Bab proteins.
The BabCD contains two known motifs, a psq motif and an AT-Hook motif (10). The psq motif has been defined by comparison of the Bab proteins with the Psq domains of the Drosophila proteins Tkr (27), Piefke (28), Ribbon (29) and Psq (16). The sequence similarity between the psq motifs of these different proteins is low compared with the psq motifs of Bab proteins of different insect species that are almost identical (D.melanogaster, A.gambiae, A.mellifera and D.virilis). The psq domains of Bab 1 and Bab2 of D.melanogaster differ by only two amino acids (see Supplementary Material). The sequence of the 10 amino acid long Bab AT-Hook domain is perfectly conserved between these different insect species and contains the consensus core RGRP sequence surrounded by several basic amino acids (four of six). These motifs have been shown to be involved in the DNA binding activity of many proteins (17–18). We therefore speculated that the BabCD could function as a DNA binding domain.
The BabCD specifically binds to DNA in vitro
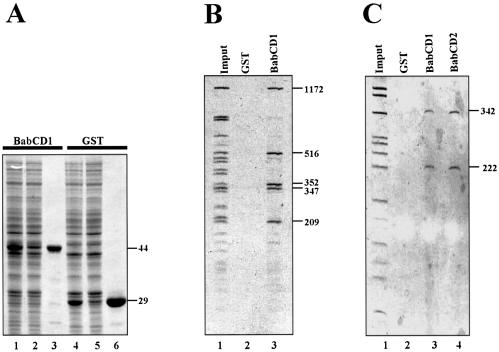
Since the Bab2 protein is localized to the bab locus on polytene chromosomes, we decided to scan several genomic subclones covering the bab locus to test for direct binding of the BabCD to DNA. To test DNA binding activity, the BabCD of Bab1 protein (BabCD1) was expressed as a GST fusion protein in E.coli (Fig. 2A, lane 1) as well as GST alone (Fig. 2A, lane 4). These two proteins were purified using glutathione–agarose beads (Fig. 2, lanes 3 and 6). The purified GST–BabCD1 and GST were used for IVPO experiments. The results obtained with HaeIII fragments from either an 8 kb subclone of the bab1 intron 1 or a 2.5 kb subclone of the bab2 intron 1 are shown in Figure 2B and C, respectively. Five fragments of the bab1 subclone ranging from 209 to 1172 bp are specifically retained by the BabCD1 (Fig. 2B, lane 3), and two fragments of the bab2 subclone, which are 222 and 342 bp in size, are retained by the BabCD1 (Fig. 2C, lane 3). No plasmid sequences are retained, and not all the bab genomic fragments are retained under these conditions. The GST protein alone did not retain any of the DNA fragments (Fig. 2B, lane 2 and C, lane 2). Together, these data show that the BabCD1 binds specifically in vitro to DNA sequences of the bab locus and that the BabCD is sufficient for this specific binding.
Figure 2.
The BabCD specifically binds to DNA in vitro. (A) GST–BabCD1 fusion protein and GST were produced in E.coli, purified and analyzed by SDS–PAGE electrophoresis, and stained with Coomassie blue. Total protein extracts were analyzed after induction (lanes 1 and 4) or without induction (lanes 2 and 5), and after purification (lanes 3 and 6). The molecular mass of the GST–BabCD1 fusion protein and GST are indicated in kDa on the right. (B) An 8 kb bab1 subclone was digested with HaeIII and the digestion products were incubated with either GST or GST–BabCD1 fusion protein bound to glutathione–agarose beads. The HaeIII digestion products (lane 1) and the fragments retained by GST (lane 2) or GST–BabCD1 (lane 3) were analyzed on 5% polyacrylamide gel. (C) A 2.5 kb bab2 subclone was digested with HaeIII and the digestion products were incubated with either GST, GST–BabCD1 or GST–BabCD2 fusion proteins bound to glutathione–agarose beads. The experiment is as in (B). Lane 1 shows the HaeIII digestion products, lanes 3 and 4 the fragments retained by the GST–BabCD1 and GST–BabCD2 proteins, respectively, and the control experiment with GST is shown in lane 2. The length of the DNA fragments retained by both the GST–BabCD1 and GST–BabCD2 protein are indicated in bp on the right.
The BabCDs of Bab1 and Bab2 are 78% identical (10). To test whether both have the same binding specificity, the fusion protein GST–BabCD2 was expressed in E.coli and IVPO experiments were performed with the 2.5 kb bab2 subclone. BabCD2 specifically retains the 222 and 342 bp fragments (Fig. 2C, lane 4), that are also bound by the BabCD1 protein (Fig. 2C, lane 3). This result shows that the BabCD is a DNA binding domain and suggests that the BabCDs of Bab1 and Bab2 have the same DNA binding specificity. We have concluded that the BabCD is a DNA binding domain, and is responsible for specific DNA binding.
The BabCD binds to TA- or TAA-rich sequences
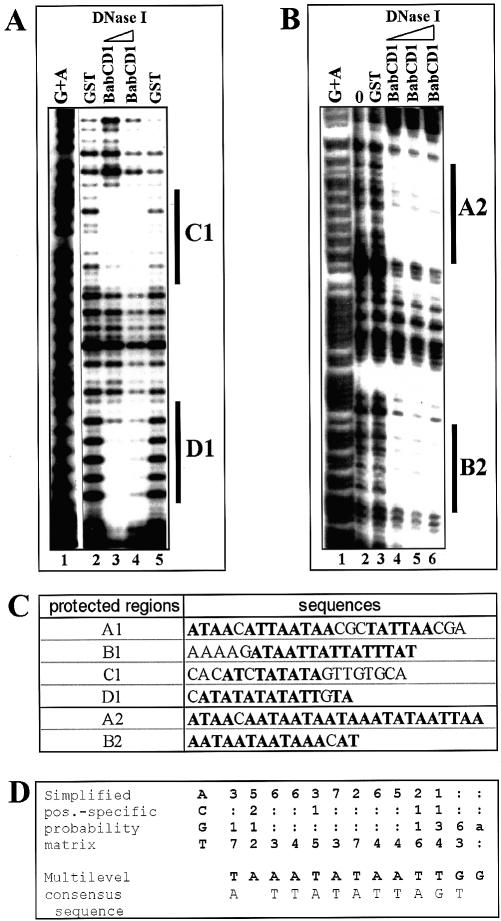
To examine in detail which sequences are bound by the BabCD, DNase I footprinting experiments were carried out using the BabCD1 protein and some of the retained genomic fragments identified. The 209 and the 347 bp bab1 fragments and the 222 bp bab2 fragment were used as probes (see Fig. 2B and C). Two regions (C1 and D1) of the 209 bp bab1 fragment were protected from DNase I digestion (Fig. 3A), and two footprints, called A1 and B1 were detected on the 347 bp bab1 fragment (data not shown). The 222 bp bab2 fragment showed two footprints, called A2 and B2 (Fig. 3B). The protected sites are ∼20 bp in length and T/A-rich containing several TA or TAA repeats (Fig. 3C). Footprints like C1/D1 and A2/B2 are separated by <20 bp. Thus, the 209 and the 347 bp bab1 and the 222 bp bab2 fragments contain several contiguous Bab binding sites.
Figure 3.
The BabCD binds to TA/TAA rich sites. A DNase I footprinting analysis was done with the 209 bp bab1 fragment (A) or the 220 bp bab2 fragment (B) as probes and the GST or GST–BabCD1 fusion proteins. ‘0’ indicates that the experiment was done without protein (Fig. 4B, lane 2). Two or three increasing dilutions of DNase I enzyme were used as indicated by the triangle above the lanes. Products were analyzed on a 7% polyacrylamide gel in parallel with a G + A sequence reaction (lane 1). (C) Sequences of the protected regions on the 209 bp bab1 fragment (C1 and D1), on a 347 bp bab1 fragment (A1 and B1, not shown) and on the 220 bp bab2 fragment (A2 and B2). The T and A bases are in bold. (D) CAST: oligonucleotides retained by the BabCD1 fusion protein were selected by IVPO. DNA from 27 plasmids were sequenced and the internal variable sequences were aligned using MEME version 3.0. The position-specific probability matrix and the multilevel consensus sequence calculated by MEME are shown.
A different approach, CAST, was used to identify a consensus binding site for the BabCD. The amplified DNA was selected by IVPO. After 10 rounds of binding site selection, 27 independent fragments were sequenced. The same data were obtained in a second CAST experiment in which the amplified fragments were selected by gel-shift. These experiments show that selected targets of the BabCD have a very high A/T content. Analysis using the motif elicitation method (MEME) (25) revealed similarities between selected targets (Fig. 3D): almost all the sequences contain repeats of TAA or TA with the possibility of permutations between A and T. These results are consistent with our footprinting data from bab genomic fragments. The analysis of footprint sequences with MEME gave similar results, revealing that the BabCD binds specifically to DNA containing TA or TAA repeats.
DNA binding specificity of the BabCD domain
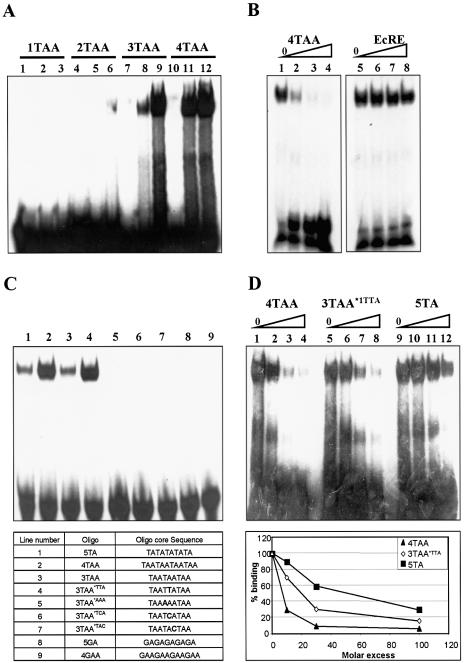
Repeats of TA and TAA motifs are found in BabCD cognate binding sites. In order to better define the binding specificity of the BabCD, we conducted EMSAs with GST–BabCD1 protein and synthetic double-stranded oligonucleotides containing one, two, three or four TAA repeats (Fig. 4A). The BabCD1 does not bind to an oligonucleotide containing a single TAA motif (1TAA) (Fig. 4A, lanes 2 and 3) and binds very weakly to an oligonucleotide containing two TAA motifs (2TAA) (Fig. 4A, lanes 5 and 6). The presence of a retarded complex indicates that the BabCD1 binds strongly to oligonucleotides containing three or four TAA motifs (3TAA or 4TAA) (Fig. 4A, lanes 8, 9 and 11, 12). The GST protein alone does not bind to any of these oligonucleotides (Fig. 4A, lanes 1, 4, 7 and 10). The BabCD1 shows a higher affinity for 4TAA than for 3TAA oligonucleotides (Fig. 4A, lanes 8 and 11). A 30-fold molar excess of an unlabeled 4TAA oligonucleotide completely abolished the labeled retarded complex (Fig. 4B, lane 3), whereas addition of a 100-fold excess of an oligonucleotide containing an ecdysone binding site (Fig. 4B, lane 8) did not significantly compete with the 4TAA oligonucleotides for BabCD binding. These results confirm that the BabCD binds to TAA repeats and that more than two repeats are required for efficient binding in vitro. Furthermore, the BabCD1 binds to an oligonucleotide containing five TA repeats (5TA) with the same affinity as to a 3TAA oligonucleotide (Fig. 4C, lanes 1–3). Oligonucleotides containing less than five TA repeats are not recognized (data not shown). In our gel-shift assays, the BabCD thus equally recognizes TA or TAA repeats if there is a minimal length of at least 9 nt.
Figure 4.
GST–BabCD fusion proteins bind to repeats of TA or TAA. (A) EMSAs with GST (lanes 1, 4, 7 and 10) or GST–BabCD1 (lanes 2, 3, 5, 6, 8, 9, 11 and 12) purified proteins and oligonucleotides containing one (lanes 1–3), two (lanes 4–6), three (lanes 7–9) or four (lanes 10–12) repeats of TAA. Lanes 3, 6, 9 and 12 contain 5-fold more protein than lanes 2, 5, 8 and 11. (B) GST–BabCD1 protein was incubated with radiolabeled 4TAA oligonucleotide, without (lanes 1 and 5) or with (lanes 2–4) increasing amounts of cold 4TAA oligonucleotide or EcRE binding site (lanes 6–8). Each competitor was added in an molar excess of 10-, 30- and 100-fold. (C) EMSAs with GST–BabCD1 proteins and different oligonucleotides in each lane. The name and the sequence of the corresponding oligonucleotides are listed in the table. (D) GST–BabCD1 protein was incubated with radiolabeled 4TAA double-stranded oligonucleotide without (lanes 1, 5 and 9) or with increasing amounts of cold 4TAA (lanes 2–4), 3TAA*TTA (lanes 6–8) or 5TA (lanes 10–12) oligonucleotides. The amounts of competitors used are 10, 30 and 100 molar excess. A graphical representation of the percentage of bound radiolabeled oligonucleotide 4TAA, quantitated by PhosphorImager analysis, is shown below. The amount of probe bound in the absence of a competitor was given the value 100%.
Mutations were introduced into oligonucleotides containing 3TAA repeats. Introducing one cytosine in the center as in oligonucleotides 3TAA*1TCA or 3TAA*1TAC completely abolishes binding by the BabCD1 (Fig. 4C, lanes 6 and 7) as does the replacement of only one T by an A as in the oligonucleotide 3TAA*1AAA (Fig. 4C, lane 5). In oligonucleotide 3TAA*1TTA, the central TAA is replaced by the complementary sequence TTA, a change that increases the affinity of the BabCD (Fig. 4C, lane 4) to a similar level as for the 4TAA oligonucleotide. Thus, the BabCD does not bind to A/T-rich sites that are interrupted by a C or a G and does not recognize a monotonous stretch of A or T. Moreover, BabCD binds neither to a 5GA- nor to a 4GAA-containing oligonucleotide (Fig. 4C, lanes 8 and 9). A series of competition EMSAs were done in order to compare the affinity of BabCD1 for the 4TAA, 5TA and 3TAATTA oligonucleotides (Fig. 4D). The increase of cold 4TAA (lanes 2–4), 3TAATTA (lanes 6–8) or 5TA (lanes 10–12) oligonucleotides leads to a decrease of labeled 4TAA oligonucleotides bound by the BabCD. Each shift was quantified and the percentage of labeled 4TAA oligonucleotides bound by the BabCD1 was calculated (Fig. 4D). A 30-fold molar excess of unlabeled 4TAA oligonucleotide is sufficient to almost completely abolish the retarded complex containing the labeled oligonucleotide, showing the specificity of the interaction. A 10-fold molar excess of unlabeled 4TAA oligonucleotide drastically reduced the labeled retarded complex; a 30- and 100-fold molar excess of 3TAATTA oligonucleotide and 5TA oligonucleotide, respectively, were required to obtain the same reduction. These results reveal that the affinity of the BabCD for oligonucleotides decreases from 4TAA to 3TAATTA to 5TA oligonucleotides confirming that the BabCD binds to different target sites with different affinities. Together, these data indicate that the BabCD binds specifically to uninterrupted repeats of TA or TAA and that the optimal binding site is a repeat of three or four TAA motifs.
DNA binding is mediated by both the Psq motif and AT-Hook motif of the BabCD
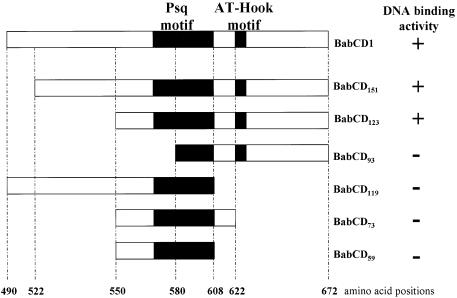
The BabCD, as defined by sequence comparison, is a large domain that consists of 183 amino acids. We wanted to determine whether the entire domain is involved in the DNA binding activity, and more specifically, whether the psq motif and/or the AT-Hook motif might be involved in DNA binding. To address this issue a series of BabCD1 deletion constructs were produced and tested with IVPO using the HaeIII fragments of the 2.5 kb bab2 subclone (Fig. 5). Deletions BabCD151 and BabCD123 that truncate the N-terminal region of the BabCD, but keep the psq and AT-Hook motifs intact, retain DNA binding activity. BabCD93, which lacks part of the psq motif, has lost DNA binding ability. BabCD119, BabCD73 and BabCD59 which have an intact psq motif but lack the AT-Hook motif are also unable to bind DNA. Thus, only a BabCD that contains both the psq and AT-Hook motif is able to bind DNA, indicating that both motifs are required for the DNA binding function of the BabCD. Point mutations were introduced in either one or the other motif and the ability of the mutated BabCD to bind to DNA was tested.
Figure 5.
Structure and DNA binding activity of full-length or truncated versions of the BabCD. Designations of the truncated derivatives indicate the number of amino acid residues. The positions of the deletions in the BabCD1 amino acid sequence are indicated at the bottom. The psq and AT-Hook motif are indicated in each protein by black boxes. The DNA binding activity of each protein, determined by IVPO analysis of the 2.5 kb bab2 subclone that was digested with HaeIII, is indicated on the right.
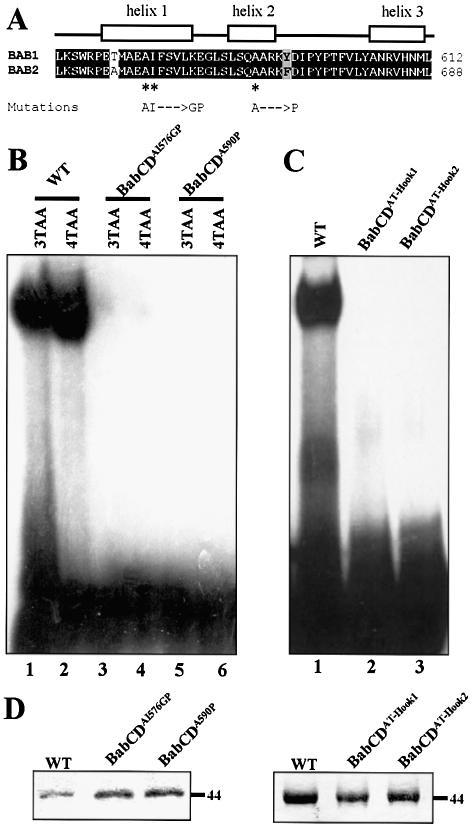
In the Psq protein, the DNA binding domain consists of four tandem repeats of the psq motif (16). At least three psq motifs are required for the DNA binding activity (18). The single psq motif of the BabCD appears to be required for DNA binding, since BabCD92, which has the AT-Hook but lacks a part of the psq domain, is not able to bind to DNA; whereas BabCD123, which contains both the psq and the AT-Hook motifs, is able to bind to DNA (Fig. 5). The psq motif of Bab1 and Bab2 is likely to be composed of three α helices (Fig. 6A). Mutations disrupting the first and the second helix of the BabCD1 psq motif were made (Fig. 6A). These mutated proteins were tested in EMSAs using labeled 3TAA and 4TAA oligonucleotides (Fig. 6B). Although BabCD1 protein strongly binds to these oligonucleotides, neither BabCDAI576GP which destroys helix1 nor BabCDA590P which destroys helix2 are able to bind DNA. These data indicate that the integrity of the unique psq domain of the BabCD is important for specific binding to DNA.
Figure 6.
The AT-Hook and the psq motif are required for the DNA binding activity of the BabCD. (A) Indicated above the sequence alignment of the psq motif of Bab1 and Bab2 are the positions of three predicted alpha helices. The mutations made in helix 1 or in helix 2 of the psq domain of BabCD1 are shown below the sequences. (B) EMSAs with the 3TAA (lanes 1, 3 and 5) or the 4TAA (lanes 2, 4 and 6) oligonucleotides and the wild-type BabCD1 (lanes 1 and 2), or two psq domain mutants: BabCDAI576GP (lanes 2 and 4) and BabCDA590P (lanes 5 and 6). (C) EMSAs with the 4TAA oligonucleotide and the wild-type BabCD1 (lane 1), the BabCDAT-Hook1 (lane 2) or the BabCDAT-Hook2 (lane 3) mutants. (D) Coomassie blue-stained SDS–PAGE containing the indicated purified wild-type or mutant BabCD1 proteins. Their molecular mass is indicated in kDa on the right.
The AT-Hook motif is composed of a short basic amino acid sequence containing the motifs GRGRP or PRGRP (17,20). The conserved amino acids RGR in the AT-Hook core motif have been changed to DGR or DGD in the proteins BabCDAT-Hook1 and BabCDAT-Hook2, respectively (Fig. 6D). These mutated BabCD1 proteins were not able to bind and shift a target site in EMSAs using the labeled 4TAA oligonucleotide (Fig. 6C), indicating that the AT-Hook motif is necessary for the DNA binding activity of the BabCD and that the Psq motif is not sufficient. The anti-tumor drug distamycin selectively binds to AT-rich DNA via minor groove interactions and effectively competes with AT-Hook proteins such as High Motility Group protein A (HMGA) proteins for DNA binding to AT-rich regions (30). Binding of the BabCD to the 4TAA oligonucleotide is abolished by distamycin in EMSA experiments (data not shown) indicating that the BabCD binds to this AT-rich target site through the minor groove. It is presumably the AT-Hook of the BabCD that is involved in DNA binding to the minor groove of DNA. In summary, our data show that the psq and AT-Hook motifs are both necessary for the DNA binding activity of the BabCD.
DISCUSSION
Bab1 and Bab2 belong to the family of nuclear BTB/POZ domain proteins and are required during metamorphosis for proper development and morphogenesis of ovaries (8–10), legs (8,10,11), antennae (11,12) and abdomen (10,13) of D.melanogaster. Many BTB/POZ proteins are transcriptional regulators and a large number contain, in addition to this N-terminal protein–protein interaction motif, a C2H2 zinc-finger DNA binding domain (6,31–33). There is no detectable zinc-finger motif in the two Bab proteins, although the Bab2 protein binds to several discrete sites on the polytene chromosomes suggesting that it can bind to DNA. We show here that the BabCD, a second conserved domain in the Bab proteins, is a sequence-specific DNA binding domain. The BabCDs of Bab1 and Bab2 can bind sequences that contain several TA or TAA repeats. This domain contains an AT-Hook and a psq motif which are both necessary for DNA binding. It had been previously determined that zinc-finger (1,6,31–33), basic leucine zipper (bZip) (34) or Psq domains [in the case of Psq and Pfk (19)] function as DNA binding domains of BTB/POZ proteins. Here we reveal a new type of DNA binding domain which is a combination between a psq motif and an AT-Hook motif.
The AT-Hook motif is primarily found in HMGA proteins, which, like other HMG proteins, are thought to function as architectural elements that modify the structure of DNA and chromatin to facilitate various DNA-dependent activities (17). The AT-Hook motifs of HMGA proteins bind specifically to the minor groove of stretches of A/T-rich DNA with the optimal DNA binding site centered at the sequence AA(T/A)T (17,20). High-affinity HMGA binding sites have two or three appropriately spaced A/T tracts allowing at least two AT-Hooks to bind to DNA, indicating that multiple AT-Hooks are required in HMGA proteins for high-affinity binding to DNA (21). We have shown that in the BabCD, a single AT-Hook, in combination with a psq domain, is required for DNA binding. In several proteins a single AT-Hook is associated with an additional DNA binding domain, such as a homeodomain, ETS domain or zinc finger (20). It has been postulated that in these proteins the AT-Hook modulates the specificity and the affinity of the associated domain through its interaction in the minor groove. We also favor this hypothesis for the BabCD as we have shown that distamycin prevents the binding of the BabCD to its DNA target sites.
The Psq domain of the Psq protein has been shown to specifically recognize the sequence GAGAG, with three of the four psq motifs being required for binding (18). There is only one psq motif in the BabCD, which we have shown to be required for DNA binding activity. However, the BabCD has a binding specificity that is different from Psq, as it binds to TA- or TAA-rich sites and is not able to bind to GA or GAA sites. In Drosophila there are nine BTB/POZ proteins that do not contain a zinc-finger motif but do contain one or more psq motifs (19). Two of these proteins, Psq and Pfk, have three or more psq motifs, the seven others have only a single psq motif. We have shown that an AT-Hook motif is required in addition to a psq motif for the DNA binding activity of the BabCD. We believe that in BTB/POZ proteins containing a single psq motif, this motif is not sufficient to bind to a specific DNA sequence, but requires an additional motif for specific binding. Of the seven Drosophila proteins that contain a single psq motif, only the two Bab proteins and the protein encoded by the CG3726 gene (35) contain an AT-Hook motif. Whether the four remaining proteins contain alternative DNA binding domains remains to be determined.
The psq motif shows significant sequence similarity to the DNA binding domain of eukaryotic and prokaryotic recombinases such as the Pogo and the Hin recombinases, respectively (19). The secondary structure of the Hin recombinase, determined by X-ray crystallography (36,37), indicates that its DNA binding domain is composed of three α helices forming a helix–turn–helix motif. Protein secondary structure prediction programs indicate that the psq motif of the BabCD also forms three α helices (Fig. 6A). Point mutations disrupting one of these helices abolish the DNA binding activity of the BabCD. Specific binding of Hin recombinases requires both major groove interactions by the helix–turn–helix motif and minor groove interactions by a part of the protein containing the sequence GRPR. The two residues Gly–Arg (GR) are invariant among all DNA invertases and are required for DNA binding (37). This GRPR sequence is similar to the AT-Hook core motif that is present in the BabCD. Therefore, the BabCD could bind DNA in the same manner as the Hin recombinase. We speculate that the BabCD binds to target sites with its AT-Hook motif (RGRP) interacting with the minor groove and with one of the α helices of the psq motif recognizing specific bases in the major groove, providing the binding specificity of the BabCD. Determining the secondary structure of the BabCD complexed to target sites will be required to confirm this model.
We have shown that the BabCD binds specifically to several different DNA fragments in the regulatory regions of the bab locus. In two of these fragments (2.5 kb bab2 and 8.0 kb bab1) several smaller sub-fragments are recognized by the BabCD. DNase1 footprinting analysis of three of these sub-fragments (222, 209 and 347 bp) reveals that there are at least two binding sites per sub-fragment. Both bab genes contain a large number of contiguous sites that are specifically recognized by the BabCD. The BTB/POZ domain is a protein–protein interacting domain that allows oligomerization (3,38), suggesting that the Bab proteins that are bound to the bab locus might form large aggregates. Such aggregates might play an architectural role by bringing together regulatory elements that are scattered over the large bab locus. Moreover, the BTB domain of the Bab proteins interacts with a component of the basal transcriptional machinery, the TAF3 protein (14,15). These results suggest a model in which Bab proteins directly modulate the transcription of target genes by bringing regulatory sequences close to the basal transcriptional machinery.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank V. Calco for excellent technical assistance, R. Blackman for the D.virilis genomic library, Dominique Leprince, Muriel Grammont and Adrienne Blair for comments on the manuscript. C.L. was supported by a fellowship from the Ministère de la Recherche et de la Technologie, the Association pour la Recherche contre le Cancer. Funding was provided by INSERM (to J.L.C.), NSERC (to D.G.) and NIH (to F.A.L.).
REFERENCES
- 1.Bardwell V.J. and Treisman,R. (1994) The POZ domain: a conserved protein–protein interaction motif. Genes Dev., 8, 1664–1677. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi A., Yamagiwa,H., Hoshino,H., Muto,A., Sato,K., Morita,M., Hayashi,N., Yamamoto,M. and Igarashi,K. (2000) A combinatorial code for gene expression generated by transcription factor Bach2 and MAZR (MAZ-related factor) through the BTB/POZ domain. Mol. Cell. Biol., 20, 1733–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmad K.F., Engel,C.K. and Prive,G.G. (1998) Crystal structure of the BTB domain from PLZF. Proc. Natl Acad. Sci. USA, 95,12123–12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melnick A., Carlile,G., Ahmad,K.F., Kiang,C.L., Corcoran,C., Bardwell,V., Prive,G.G. and Licht,J.D. (2002) Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol. Cell. Biol., 22, 1804–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsukiyama T., Becker,P.B. and Wu,C. (1994) ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature, 367, 525–532. [DOI] [PubMed] [Google Scholar]
- 6.Dong S., Zhu,J., Reid,A., Strutt,P., Guidez,F., Zhong,H.J., Wang,Z.Y., Licht,J., Waxman,S., Chomienne,C., Chen,Z., Zelent,A. and Chen,S.J. (1996) Amino-terminal protein–protein interaction motif (POZ-domain) is responsible for activities of the promyelocytic leukemia zinc finger–retinoic acid receptor-α fusion protein. Proc. Natl Acad. Sci. USA, 93, 3624–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huynh K.D. and Bardwell,V.J. (1998) The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene, 17, 2473–2484. [DOI] [PubMed] [Google Scholar]
- 8.Godt D. and Laski,F.A. (1995) Mechanisms of cell rearrangement and cell recruitment in Drosophila ovary morphogenesis and the requirement of bric à brac. Development, 121, 173–187. [DOI] [PubMed] [Google Scholar]
- 9.Sahut-Barnola I., Godt,D., Laski,F.A. and Couderc,J.L. (1995) Drosophila ovary morphogenesis: analysis of terminal filament formation and identification of a gene required for this process. Dev. Biol., 170, 127–135. [DOI] [PubMed] [Google Scholar]
- 10.Couderc J.L., Godt,D., Zollman,S., Chen,J., Li,M., Tiong,S., Cramton,S.E., Sahut-Barnola,I. and Laski,F.A. (2002) The bric à brac locus consists of two paralogous genes encoding BTB/POZ domain proteins and acts as a homeotic and morphogenetic regulator of imaginal development in Drosophila. Development, 129, 2419–2433. [DOI] [PubMed] [Google Scholar]
- 11.Chu J., Dong,P.D. and Panganiban,G. (2002) Limb type-specific regulation of bric à brac contributes to morphological diversity. Development, 129, 695–704. [DOI] [PubMed] [Google Scholar]
- 12.Godt D., Couderc,J.L., Cramton,S.E. and Laski,F.A. (1993) Pattern formation in the limbs of Drosophila: bric à brac is expressed in both a gradient and a wave-like pattern and is required for specification and proper segmentation of the tarsus. Development, 119, 799–812. [DOI] [PubMed] [Google Scholar]
- 13.Kopp A., Duncan,I., Godt,D. and Carroll,S.B. (2000) Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature, 408, 553–559. [DOI] [PubMed] [Google Scholar]
- 14.Pointud J.C., Larsson,J., Dastugue,B. and Couderc,J.L. (2001) The BTB/POZ domain of the regulatory proteins bric à brac 1 (BAB1) and bric à brac 2 (BAB2) interacts with the novel Drosophila TAF(II) factor BIP2/dTAF(II)155. Dev. Biol., 237, 368–380. [DOI] [PubMed] [Google Scholar]
- 15.Gangloff Y.G., Pointud,J.C., Thuault,S., Carre,L., Romier,C., Muratoglu,S., Brand,M., Tora,L., Couderc,J.L. and Davidson,I. (2001) The TFIID components human TAF(II)140 and Drosophila BIP2 (TAF(II)155) are novel metazoan homologues of yeast TAF(II)47 containing a histone fold and a PHD finger. Mol. Cell. Biol., 21, 5109–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horowitz H. and Berg,C.A. (1996) The Drosophila pipsqueak gene encodes a nuclear BTB-domain-containing protein required early in oogenesis. Development, 122, 1859–1871. [DOI] [PubMed] [Google Scholar]
- 17.Reeves R. and Nissen,M.S. (1990) The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J. Biol. Chem., 265, 8573–8582. [PubMed] [Google Scholar]
- 18.Lehmann M., Siegmund,T., Lintermann,K.G. and Korge,G. (1998) The pipsqueak protein of Drosophila melanogaster binds to GAGA sequences through a novel DNA-binding domain. J. Biol. Chem., 273, 28504–28509. [DOI] [PubMed] [Google Scholar]
- 19.Siegmund T. and Lehmann,M. (2002) The Drosophila Pipsqueak protein defines a new family of helix–turn–helix DNA-binding proteins. Dev. Genes Evol., 212, 152–157. [DOI] [PubMed] [Google Scholar]
- 20.Aravind L. and Landsman,D. (1998) AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res., 26, 4413–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maher J.F. and Nathans,D. (1996) Multivalent DNA-binding properties of the HMG-1 proteins. Proc. Natl Acad. Sci. USA, 93, 6716–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reeves R. (2001) Molecular biology of HMGA proteins: hubs of nuclear function. Gene, 277, 63–81. [DOI] [PubMed] [Google Scholar]
- 23.Zink B. and Paro,R. (1989) In vivo binding pattern of a trans-regulator of homoeotic genes in Drosophila melanogaster. Nature, 337, 468–471. [DOI] [PubMed] [Google Scholar]
- 24.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G. and Struhl,K. (1993) Current Protocols in Molecular Biology. John Wiley and Sons, Canada. [Google Scholar]
- 25.Bailey T.L. and Elkan,C. (1995) The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol., 3, 21–29. [PubMed] [Google Scholar]
- 26.Holt R.A., Subramanian,G.M., Halpern,A., Sutton,G.G., Charlab,R., Nusskern,D.R., Wincker,P., Clark,A.G., Ribeiro,J.M., Wides,R. et al. (2002) The genome sequence of the malaria mosquito Anopheles gambiae. Science, 298, 129–149. [DOI] [PubMed] [Google Scholar]
- 27.Haller J., Cote,S., Bronner,G. and Jackle,H. (1987) Dorsal and neural expression of a tyrosine kinase-related Drosophila gene during embryonic development. Genes Dev., 1, 862–867. [DOI] [PubMed] [Google Scholar]
- 28.Schwendemann A., Siegmund,T. and Lehmann,M. (2001) Piefke encodes a new member of the family of Psq-domain DNA-binding proteins. A. Dros. Res. Conf., 42, 245B. [Google Scholar]
- 29.Shim K., Blake,K.J., Jack,J. and Krasnow,M.A. (2001) The Drosophila ribbon gene encodes a nuclear BTB domain protein that promotes epithelial migration and morphogenesis. Development, 128, 4923–4933. [DOI] [PubMed] [Google Scholar]
- 30.Wegner M. and Grummt,F. (1990) Netropsin, distamycin and berenil interact differentially with a high-affinity binding site for the high mobility group protein HMG-I. Biochem. Biophys. Res. Commun., 166, 1110–1117. [DOI] [PubMed] [Google Scholar]
- 31.DiBello P.R., Withers,D.A., Bayer,C.A., Fristrom,J.W. and Guild,G.M. (1991) The Drosophila broad-complex encodes a family of related proteins containing zinc fingers. Genetics, 129, 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrison S.D. and Travers,A.A. (1990) The tramtrack gene encodes a Drosophila finger protein that interacts with the ftz transcriptional regulatory region and shows a novel embryonic expression pattern. EMBO J., 9, 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deweindt C., Albagli,O., Bernardin,F., Dhordain,P., Quief,S., Lantoine,D., Kerckaert,J.P. and Leprince,D. (1995) The LAZ3/BCL6 oncogene encodes a sequence-specific transcriptional inhibitor: a novel function for the BTB/POZ domain as an autonomous repressing domain. Cell Growth Differ., 6, 1495–1503. [PubMed] [Google Scholar]
- 34.Oyake T., Itoh,K., Motohashi,H., Hayashi,N., Hoshino,H., Nishizawa,M., Yamamoto,M. and Igarashi,K. (1996) Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol. Cell. Biol., 16, 6083–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams M.D., Celniker,S.E., Holt,R.A., Evans,C.A., Gocayne,J.D., Amanatides,P.G., Scherer,S.E., Li,P.W., Hoskins,R.A., Galle,R.F. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 36.Hughes K.T., Gaines,P.C., Karlinsey,J.E., Vinayak,R. and Simon,M.I. (1992) Sequence-specific interaction of the Salmonella Hin recombinase in both major and minor grooves of DNA. EMBO J., 11, 2695–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng J.A., Johnson,R.C. and Dickerson,R.E. (1994) Hin recombinase bound to DNA: the origin of specificity in major and minor groove interactions. Science, 263, 348–355. [DOI] [PubMed] [Google Scholar]
- 38.Li X., Lopez-Guisa,J.M., Ninan,N., Weiner,E.J., Rauscher,F.J.,III and Marmorstein,R. (1997) Overexpression, purification, characterization and crystallization of the BTB/POZ domain from the PLZF oncoprotein. J. Biol. Chem., 272, 27324–27329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.