Abstract
The complete chloroplast DNA sequence (122 890 bp) of the moss Physcomitrella patens has been determined. The genome contains 83 protein, 31 tRNA and four rRNA genes, and a pseudogene. Four protein genes (rpoA, cysA, cysT and ccsA) found in the liverwort Marchantia polymorpha and the hornwort Anthoceros formosae are absent from P.patens. The overall structure of P.patens chloroplast DNA (cpDNA) differs substantially from that of liverwort and hornwort. Compared with its close relatives, a 71 kb region from petD to rpoB of P.patens is inverted. To investigate whether this large inversion and the loss of rpoA usually occur in moss plants, we analyzed amplified cpDNA fragments from four moss species. Our data indicate that the large inversion occurs only in P.patens, whereas the loss of the rpoA gene occurs in all mosses. Moreover, we have isolated and characterized the nuclear rpoA gene encoding the α subunit of RNA polymerase (RNAP) from P.patens and examined its subcellular localization. When fused to green fluorescent protein, RpoA was observed in the chloroplasts of live moss protonemata cells. This indicates that chloroplast RNAP is encoded separately by chloroplast and nuclear genomes in the moss. These data provide new insights into the regulation and evolution of chloroplast transcription.
INTRODUCTION
Chloroplasts evolved from cyanobacteria through endosymbiosis, and possess independent genomes (1). To date, complete chloroplast DNA (cpDNA) sequences from two dozen land plants and algae have been determined (2,3). Algal cpDNAs range from 89 (Codium fragile) to over 1500 kb (Acetabularia), and their gene content and organization vary from species to species (2). In contrast, land plant cpDNAs are relatively uniform in size, from 120 to 160 kb, and their gene content and organization are well conserved (3). Bryophytes consist of three classes (liverworts, hornworts and mosses) and have been identified as the earliest land plants (4–6). Among the bryophytes, the cpDNA (121 024 bp) of the liverwort Marchantia polymorpha was the first to be sequenced (7), and the hornwort Anthoceros formosae cpDNA sequence (161 162 bp) has recently been published (8). The larger size of the cpDNA genome of A.formosae results from a larger inverted repeat (IR) sequence compared with the liverwort M.polymorpha. The overall gene organization is highly conserved between the two bryophytes. Single introns, however, are inserted into the 23S rRNA genes (rrn23) of A.formosae (8) but not into those of M.polymorpha (7), the charophyte Chaetosphaeridium globosum (9) or vascular plants (3). Over 900 RNA editing sites have been identified in the hornwort A.formosae chloroplast (10). In contrast, no RNA editing site has been found in the cpDNA of the liverwort M.polymorpha (11). Therefore, RNA editing events differ substantially between bryophyte species compared with the high degree of conservation of editing sites in the chloroplasts of vascular plants (12). Significant divergence in RNA editing events among the bryophytes supports bryophyte paraphyly, as suggested by molecular phylogenetic studies (13–15).
The remaining bryophyte group, the mosses, has been reported to be most closely related to the early lycopod land plants, the ferns, and the vascular plant lineage (4–6,15). The exact phylogenetic status of this group, however, remains unresolved. To answer this question, molecular data on moss chloroplast genes are required. There is little information on the gene structure (16–18) or RNA editing (19) of the moss P.patens. Therefore, we have determined the complete nucleotide sequence of the P.patens cpDNA. Moreover, we have isolated a nuclear rpoA gene encoding an α subunit of RNA polymerase (RNAP) and discuss the relocation of rpoA from the chloroplast to the nucleus.
MATERIALS AND METHODS
Accession numbers
The sequences reported in this paper have been deposited in the DDBJ/GenBank/EMBL databases with accession numbers: P.patens cpDNA, AP005672; cpDNA fragments amplified from four mosses, AB098724–AB098727; PpRpoA gene, AB110071; PpRpoA cDNA, AB110072.
Plant materials
The moss P.patens subspecies patens was grown at 25°C under continuous illumination. Total cellular DNA and RNA were isolated from P.patens protonema, as described previously (20).
Preparation of cpDNA fragments
A P.patens genomic library was screened using the tobacco ndhD sequence (21) as a probe, as described previously (22), and the genomic clone λ-D5 was isolated. The λ-D5 insert DNA was amplified by polymerase chain reaction (PCR) using primers designed from λfix-II vector sequences, AGCTCTAATACGACTCACTATAGGGCGTCGA and GAGCTCAATTAACCCTCACTAAAGGGAGTCGA. The reaction of long and accurate (LA) PCR was performed with 30 cycles of 10 s denaturation at 98°C and 15 min annealing and extension at 68°C using LA Taq DNA polymerase (Takara Shuzo). The amplified DNA contained a 12 640 bp region from rpl32 to trnV-GAC. To further amplify cpDNA fragments, total cellular DNA (90 ng) was subjected to LA PCR under the same conditions as above using primers designed from either nucleotide sequences deposited in DNA databases or sequences determined in this study as follows: for amplification of a 19 379 bp DNA region from psbD to rbcL, LA1 (TTAGGAGGTCTATGGACTTTCGTTGCTCTT, AB013655) and LA2 (AGTCATCACGAAGTAAGTCAACAAACCCTA, AB066297); for a 14 473 bp DNA region from psbB to rbcL, LA3 (TAAGTAAAAAAAAAAGATGATGGAAAA, this study) and LA4 (AGGTTCTGTTACTAACTTATTTACTTCTATTG, AB066297); for a 13 774 bp DNA region from psbB to rrn16, LA5 (AGTATTGCTGCTGTATTGTTTGCTTTT, this study) and LA6 (TTTGAGTTTCATTCTTGCGAACGTACTCCC, this study); for a 17 834 bp DNA region from rpl21 to rrn4.5, LA7 (TGTACGCTATAATTGAAACCGGAGGTGAAC, this study) and LA8 (TTTATCFATCACGATAGGTGCCAAGTGGAAG, this study); for a 15 083 bp DNA region from rpoC1 to rrn16, LA9 (GATGATTTTTAATTGTTAGTATGTATAGTCC, AB013657) and LA10 (TTTGAGTTTCATTCTTGCGAACGTACTCCC, this study); for a 9799 bp DNA region from psbA to psbD, LA11 (TGTAGGTATTTGGTTTACTGCTTTAGGTATC, X04465) and LA12 (ACCAACTACTCCAATAGCACTCATCCATAAA, AB013655); for a 19 156 bp DNA region from trnH to rpoC1, LA13 (AAATAATAAAAAATGGGCGAACGACGGGAAT, this study) and LA14 (AAAATCATCAAGGTATCTATGGTAATAAAAA, AB013657); for an 11 365 bp DNA region from rps7 to chlL, LA15 (GTTTCTTCTTTTTTTCGTATTGCTTCTCCAC, this study) and LA16 (GTAAATAACATCTTCAGGCCAAACATCTTCAT, this study). The eight amplified cpDNA fragments and the λ-D5 insert DNA encompassed the entire P.patens chloroplast genome.
Sequence analysis
The amplified cpDNA fragments were sheared, cloned into pUC18, and shotgun-sequenced using Shimadzu RISA384 sequencers by the Department of Genomic Research, Shimadzu Co., Japan. Nucleotide sequence files were assembled using the Phrap program (Phil Green, University of Washington, Seattle, WA, USA) and the resulting sequences were analyzed with Genetyx-Mac v.9.0 (Software Development) and Sequencer software (Hitachi Software Engineering). When the nucleotide sequences of either protein- or RNA-coding regions contained ambiguous bases or apparent small gaps, they were corrected by sequencing of re-amplified P.patens DNA fragments (19,23). Fifteen cpDNA regions with small gaps and 33 protein- or RNA-coding regions were amplified from total cellular DNA, and subcloned into pGEM-T Easy (Promega). At least three independent clones were picked up and sequenced using the DYEnamic ET Terminator Cycle Sequencing Kit (Amersham Bioscience) and an ABI PRISM 3100 DNA sequencer (Perkin-Elmer, Applied Biosystems). Database searches and sequence data analyses were performed as described previously (19).
DNA analysis of four mosses
Total cellular DNA from the mosses Hylocomium splendens (Hedw.) Bruch and Schimp., Plagiothecium euryphyllum (Card. and Ther.) Iwats., Bartramia pomiformis Hedw. and Ceratodon purpureus (Hedw.) were used as templates for PCR. Primers P1–P4 were designed from the sequences that have been determined for the P.patens genes rps11, rpoB, petD and petN, respectively: P1, 5′-TTTTGTTCGTGATGTAACTCCTATG-3′; P2, 5′-CTACCATAGCATCCTCAGTAGATT-3′; P3, 5′-CTAAATTAGCTAAAGGTATGG GTC-3′; and P4, 5′-TAAATCTAATTTTTATAATCCGCTTC-3′. The amplified DNA fragments were subcloned into pGEM-T Easy (Promega) and sequenced as described previously (19,24)
Isolation and sequence analysis of cDNA and genomic DNA clones
Full-length cDNA encoding PpRpoA was prepared by 5′-rapid amplification of cDNA ends using primers designed from an expressed sequence tag (EST) clone as described (24). Genomic clones were isolated using the cDNA probe from a P.patens genomic library (19). Sequencing was performed with an ABI PRISM 3100 sequencer and the DYEnamic ET Terminator Cycle Sequencing Kit (Amersham Bioscience) using appropriate sequencing primers.
Construction of fusion genes and microscopic observation
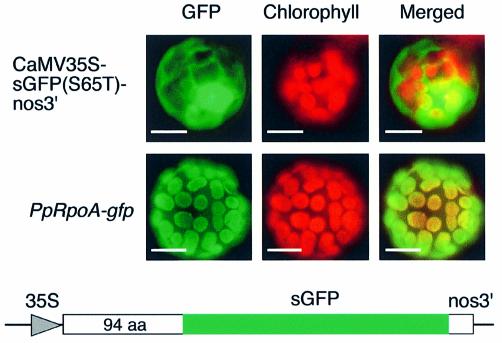
A DNA fragment encoding the N-terminal 94 amino acid residues of PpRpoA was amplified from the cDNA as above with oligonucleotides containing restriction sites (underlined), 5′-ATAGCCATGGCCTCGTCGACAGGAGCATC-3′ and 5′-AGCTTCTAGAATGGCAGCAGTCATGAGCGC-3′. SalI– NcoI-digested amplified DNA fragments were introduced into SalI–NcoI-cleaved CaMV35S-sGFP(S65T)-nos3′ (23) to create the PpRpoA-gfp construct. The reporter construct was introduced into the protoplasts prepared from P.patens protonemata by polyethyleneglycol-mediated transformation (20). One day after transformation, the fluorescence of green fluorescent protein (GFP) was monitored using a BX-50 fluorescence microscope (Olympus).
RESULTS
Overall structure of Physcomitrella cpDNA
The circular cpDNA of P.patens is 122 890 bp long, and its gene map is shown in Figure 1. Overall GC content of the cpDNA is 28.5%, which is closer to those of the liverwort M.polymorpha (28.8%) (7) and the hornwort A.formosae (32.9%) (8), the charophyte Chaetosphaeridium (29.6%) (9) and the algae (30.4–33%) (2), than to the GC content of the vascular plants (38–39%) (3). Two identical copies of a large sequence (9589 bp) containing the rDNA operon (rrn16–rrn23–rrn4.5–rrn5) are present in an inverted orientation (IRA and IRB). A large single-copy (LSC) region of 85 212 bp and a small single-copy (SSC) region of 18 501 bp separate these sequences from one another. This quadripartite structure of cpDNA is well conserved in the bryophytes (7,8), a charophyte (9) and most vascular plants (3). BglII, ClaI and SacI restriction maps predicted from the complete nucleotide sequence coincide with the physical maps constructed previously (25).
Figure 1.
Gene map of the moss P.patens chloroplast genome. IR sequences (IRA and IRB) are separated by the SSC and LSC regions. Genes on the inside of the map are transcribed clockwise and genes on the outside are transcribed counter-clockwise. Genes with related functions are shown in the same color. Asterisks denote intron-containing genes or split genes. Arrows indicate positions of the endpoint of large inversion relative to liverwort M.polymorpha and hornwort A.formosae cpDNA.
Relative to hepatic bryophytes (M.polymorpha and A.formosae), an extensive rearrangement of the P.patens cpDNA is evident within the LSC region. A large inversion is present that encompasses a 71 kb region from petD to rpoB, its apparent endpoints lying between rps11 and rpoB at one end and petD and trnC-GCA at the other (Fig. 1, arrows). To investigate whether this large inversion occurs in other moss species, we used PCR with different combinations of primers on the DNAs of C.purpurea, P.euryphyllum, H.splendens and B.pomiformis. The combinations of primers P1 and P2, and primers P3 and P4 amplified the expected DNA fragments of 1002 and 1235 bp, respectively, from P.patens DNA (data not shown). In contrast, these primer combinations failed to amplify any DNA fragments from the DNA of other mosses. However, DNA fragments containing rps11 and petD (Fig. 2B), or rpoB and trnC (data not shown), were amplified when different primer combinations (P1 and P3 or P2 and P4) were used. This indicates that the large inversion occurs only in P.patens.
Figure 2.
(A) Representation of the strategy used to detect the orientation of the 71 kb inversion. The P.patens (top), M.polymorpha and A.formosae (bottom) genomes are shown in the region of the 71 kb inversion. The positions of primers P1–P4, located adjacent to the inversion endpoints, are indicated. (B) DNA regions amplified with either the P1 and P2 pair or the P1 and P3 pair are illustrated, with the length (bp) of the intergenic spacer.
Gene content
One hundred and eighteen identified genes are listed in Table 1. Eighty-three protein-coding genes, including the hypothetical chloroplast reading frame (ycfs), were identified. The differences in genes in the cpDNAs of the three classes of bryophyte are summarized in Table 2. Four protein genes, rpoA, cysA, cysT and ccsA (ycf5) present in the liverwort M.polymorpha (7) and the hornwort A.formosae cpDNA (8), are absent from P.patens cpDNA. Two genes, cysA and cysT, that encode sulfate transport proteins, are present in the green algae Chlorella (26) and Nephroselmis (27), but are absent in charophytes (9) and vascular plants (4). ccsA is usually found in land plants (28–30) and algae (2,26), except Euglena gracilis (31). Furthermore, two open reading frames (ORFs) encoding 197 and 40 amino acid residues are located downstream of petA. The 70 N-terminal amino acid residues predicted from ORF 197 show ∼30% identity with those of Chaetosphaeridium ORF 231 (9) and Mesostigma ORF 167 (32), which also occur downstream of petA. However, no sequences similar to ORF 197 are found in the cpDNAs of other plants or algae. A sequence homologous to ORF 40 (∼40% amino acid identity) has been found in the hornwort A.formosae (8).
Table 1. Genes encoded by P.patens cpDNA.
| Gene products | Genes |
|---|---|
| Photosystem I | psaA, B, C, I, J, M |
| Photosystem II | psbA, B, C, D, E, F, H, I, J, K, L, M, N, T, Z |
| Cytochrome b6/f | petA, Ba, Da, G, L, N |
| ATP synthase | atpA, B, E, Fa, H, I |
| Chlorophyll biosynthesis | chlB, L, N |
| Rubisco | rbcL |
| NADH oxidoreductase | ndhAa, Ba, C, D, E, F, G, H, I, J, K |
| Large subunit ribosomal proteins | rpl2a, 14, 16a, 20, 21, 22, 23, 32, 33, 36 |
| Small subunit ribosomal proteins | rps2, 3, 4, 7, 8, 11, 12a,b, 14, 15, 18, 19 |
| RNAP | rpoB, C1a, C2 |
| Translation factor | infA |
| Other proteins | accD, clpPc, matK |
| Proteins of unknown function | ycf1, 2, 3c, 4, 10, 12, 66a |
| Ribosomal RNAs | rrn16d, 23d, 4.5d, 5d |
| Transfer RNAs | trnA(UGC)a,d, C(GCA), D(GUC), E(UUC), F(GAA), G(GCC), G(UCC)a, H(GUG), I(CAU), I(GAU)a,d, K(UUU)a, L(CAA), L(UAA)a, L(UAG), fM(CAU), M(CAU), N(GUU)d, P(UGG), Q(UUG), R(ACG)d, R(CCG), R(UCU), S(GCU), S(GGA), S(UGA), T(GGU), T(UGU), V(GAC)d, V(UAC)a, W(CCA), Y(GUA) |
aGene containing a single intron.
bGene divided into two independent transcription units.
cGene containing two introns.
dTwo gene copies due to the IR.
Table 2. Gene content of cpDNAs from green alga, charophyte, bryophytes and land plants.
| Plants | rpoA | ccsA | cysA | cysT | ycf66 | matK | rps15 | trnP-GGG |
|---|---|---|---|---|---|---|---|---|
| Chlorella (26) | + | + | + | + | – | – | – | – |
| Chaetosphaeridim (9) | + | + | – | – | + | + | + | + |
| Marchantia (7) | + | + | + | + | + | + | + | ψ |
| Anthoceros (8) | + | + | + | + | – | ψ | ψ | + |
| Physcomitrella | – | – | – | – | + | + | + | – |
| Psilotum (28) | + | + | – | – | – | + | + | + |
| Pinus (29) | + | + | – | – | – | + | + | + |
| Arabidopsis (30) | + | + | – | – | – | + | + | – |
The presence (+) or absence (–) of each molecular character, and pseudogene (ψ) are shown. References are shown in parenthesis.
Thirty-five RNA genes were identified, of which 31 are tRNA genes and four are rRNA genes. A tRNA-like sequence is present between rpl32 and trnL-UAG in the SSC region (Fig. 1). The nucleotide sequence shows 71.6% identity with that of A.formosae trnP-GGG (8) and 62.1% with that of M.polymorpha pseudo trnP-GGG (7), which are located in similar positions to the P.patens tRNA-like sequence. How ever, the P.patens sequence encodes a tRNA with an AAC anticodon, which is complementary to the GTT codon (encodes valine). No tRNAs or tRNA genes with AAC anticodons have been found in other organelles or cyanobacteria. Therefore, we tentatively classified this tRNA-like sequence as a pseudogene. Genes encoding stable RNAs other than rRNAs and tRNAs have been identified for tscA in Chlamydomonas reinhardtii (33), for rnpB and tmRNA genes in the non-green algae (34), and for sprA in tobacco (35), but sequences homologous to these genes are not present in this moss.
Intron content
Eighteen genes for six tRNAs and 12 proteins contain introns, as shown in Figure 1 and Table 1. The intron of the trnL gene encoding tRNALeu-UAA is classified into the group I-type intron and the remaining introns of the 17 genes are categorized into the group II intron (8,21). A maturase-like polypeptide is encoded within the longest intron (2168 bp) of trnK coding for tRNALys-UUU. The gene for ribosomal protein S12 is divided into 5′-rps12 and 3′-rps12, and each gene segment can be transcribed independently and the transcripts trans-spliced. clpP and ycf3 contain two introns, as in the corresponding tobacco and rice genes, and the other intron-containing genes have a single intron. Anthoceros formosae ycf3 also has two introns, whereas M.polymorpha ycf3 has only the first of these introns. ycf66 is absent in A.formosae, but present in P.patens and M.polymorpha. The intron of rrn23 is present in A.formosae as well as in the green algae Chlorella (26) and Chlamydomonas (36), but not in P.patens, M.polymorpha (8) or the prasinophytes Mesostigma (32) and Nephroselmis (27), which constitute the most ancient green-plant lineage (5).
Loss of rpoA from moss cpDNA
The rpoA gene is normally present immediately downstream from a ribosomal-protein gene cluster (rpl23 to rps11) and is located between rps11 and petD in most land plants. In contrast, P.patens rpoB is located downstream from rps11, and rpoA is completely absent from the cpDNA (Fig. 1). Interestingly, in the other moss species, H.splendens, P.euryphyllum, B.pomiformis and C.purpureus, petD is located downstream from rps11, and there is no rpoA gene located between them (Fig. 2B). This observation strongly suggests that the loss of the rpoA gene is a general occurrence in mosses. However, the possibility that the rpoA gene is present at some other locus on the cpDNA cannot be ruled out for other mosses.
Identification of the nuclear rpoA gene
The absence of rpoA in the moss cpDNA strongly suggests the transfer of the rpoA gene from the chloroplast to the nuclear genome. To isolate the nuclear rpoA counterpart, we searched the DNA databases and identified an EST (GenBank accession no. BI740521) encoding the C-terminal 57 amino acid residues of the α subunit. We next obtained and sequenced the full-length cDNA. The predicted protein contains 450 amino acid residues with the characteristic arrangement of protein domains identified in the Escherichia coli α subunit of RNAP (37) and shows 50% amino acid identity with chloroplast-encoded liverwort M.polymorpha RpoA and 29% identity with the E.coli α subunit (Fig. 3). Therefore, we designated this protein and gene PpRpoA and PpRpoA, respectively. The P.patens α homolog contains an N-terminal extension, which could be a transit peptide for targeting this protein to chloroplasts. The PpRpoA gene consists of two exons, which are separated by a 215 bp intron. The first exon encodes only the 94 amino acid N-terminal extension that is predicted to target PpRpoA to the chloroplast. Genomic Southern analysis showed that PpRpoA is encoded by a single copy gene (data not shown).
Figure 3.
Schematic diagram showing the domain structure of the α subunit of E.coli RNAP, P.patens nuclear PpRpoA and M.polymorpha chloroplast RpoA. αNTD and αCTD denote α subunit N-terminal and C-terminal domains, respectively. The gray boxes denote regions conserved in sequence between α homologs of prokaryotic, archaebacterial, chloroplast and eukaryotic RNAPs.
Chloroplast localization of the moss PpRpoA
The N-terminal sequence of PpRpoA was predicted to specify chloroplast targeting (a score of 0.911) by the TargetP program for protein sorting (38). To localize PpRpoA in cells we further investigated the cellular localization in protoplasts of P.patens protonemata through the use of a chimeric protein comprised of the PpRpoA N-terminal 94 amino acid residues fused to GFP. The PpRpoA-gfp construct produced GFP fluorescence associated with chloroplasts (Fig. 4). This clearly demonstrated that the N-terminal extension of the PpRpoA functions as a chloroplast-targeting signal. This strongly suggests that the nuclear-encoded PpRpoA is a component of chloroplast RNAP.
Figure 4.
Localization of fusion protein encoded by the PpRpoA-gfp construct to chloroplasts. The PpRpoA-gfp was introduced into the P.patens protoplasts of protonemata. The localization of GFP and chloroplast pigments (chlorophyll) in transformed cells was detected by fluorescent microscopy. Merged images are shown on the right. Bars are 10 µm.
DISCUSSION
The size and quadripartite structure of the chloroplast genome of P.patens is very similar to that of M.polymorpha. The cpDNA of P.patens, like the vascular plants, however, lacks cysA and cysT and contains a ycf3 gene with two introns, whereas that of the liverwort does not. This implies that the moss P.patens diverged from the hepatic bryophytes and is more closely related to the vascular plants.
The most striking feature of the cpDNA of the moss P.patens is the absence of rpoA. After the divergence of the mosses from the hepatic bryophytes, rpoA was lost from the cpDNA, together with ycf5, cysA and cysT. The loss of rpoA gene appears to be typical in mosses, and poses the question as to whether moss chloroplasts possess a transcription system unique among the green land plants. rpoA is already known to be absent from cpDNAs of the photosynthetic plant geranium (Pelargonium hortorum) (39), the parasitic vascular plant Epifagus virginiana (40), and from the plastid-like genome of the malaria parasite Plasmodium falciparum (41). The retained 60% of rpoA in the geranium cpDNA is extensively fragmented but an intact rpoA gene was not detected in the nuclear DNA by Southern hybridization analysis (39). In contrast, rpoB, C1 and C2 are most likely intact genes in P.hortorum (39) and P.falciparum (41). This suggests that rpoA gene was preferentially lost from the cpDNA, rather than rpoB, C1 and C2. The loss of single genes or groups of functionally related genes occurred similarly in specific clades, e.g. the loss of ycf1, ycf2 and accD in monocotyledonous plants (42,43), the loss of all ndh genes in the gymnosperm Pinus (29) and the loss of infA during angiosperm evolution (44).
In this study we have identified a nuclear rpoA gene encoding a protein predicted to contain 450 amino acids with high sequence identity to the α subunit of E.coli RNAP and the cpDNA-encoded RpoA (Fig. 3). Our findings clearly indicate that a functional rpoA has been transferred from the chloroplast to the nucleus. This is the first evidence for the presence of a nuclear rpoA. Nuclear rpoA genes with a chloroplast transit sequence are not found in Arabidopsis thaliana (45) and rice (46), whose cpDNAs retain an intact rpoA.
There are 171 identical amino acid residues between PpRpoA and liverwort M.polymorpha RpoA, of which 98 are encoded by codons that differ at synonymous positions. For example, PpRpoA preferentially uses GC residues at the third position in Leu, and generally uses the codons TTC for Phe, AAG for Lys and GAG for Glu. These codons are also preferentially used in P.patens ESTs (18). In contrast, A or T residues are preferentially used for the M.polymorpha rpoA as well as P.patens chloroplast protein genes. This indicates that the transfer of the moss rpoA to the nucleus was not a recent event and may have occurred immediately after the divergence of mosses from the hepatic bryophytes. There is also an EST (AW098196) in the databases from the moss C.purpureus which is predicted to encode a polypeptide similar to the P.patens PpRpoA. This indicates that mosses probably possess in general a transferred nuclear rpoA gene.
The α subunit is well characterized to be essential for RNAP assembly and basal transcription in E.coli (47,48). Therefore, in the moss chloroplast, the α subunit is indispensable for the function of plastid-encoded plastid RNAP (PEP), and the PpRpoA probably functions as the α subunit of PEP. Many P.patens chloroplast genes have canonical –10 and –35 sequences similar to those of E.coli promoter sequences (49). Moreover, cDNAs encoding σ-like transcription initiation factor for PEP have been isolated from P.patens, and the encoded transit peptide of σ-like factor has been shown to function as a chloroplast-targeting signal (20,50). Therefore, there is no doubt that PEP also transcribes photosynthetic genes in the P.patens chloroplasts. Our data indicate that the biosynthesis and assembly of the core subunits of PEP are controlled cooperatively by the chloroplast and nuclear genomes, and a relatively more complicated transcription system operates in mosses than in higher plants.
SUPPLEMENTARY MATERIAL
The amino acid sequence alignment of PpRpoA with the α subunit from E.coli, Synechococcus PCC6301, tobacco and liverwort M.polymorpha chloroplasts is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs M. Higuchi and T. Omata for providing us with DNAs of mosses, Dr T. Tsudzuki for drawing the gene map of cpDNA, and Drs K. Yoshinaga and M. Hasebe for valuable discussions. We also thank Drs A. C. Cuming and S. Bashiardes for the Physcomitrella genomic library as part of the Physcomitrella EST Programme at the University of Leeds and Washington University in St Louis. This work was supported by Grants-in-Aid from the Ministry of Education, Science, Sports, Culture and Technology (12874107 and 13206027 to M.S.) and of the Ministry of Agriculture, Forestry and Fisheries (Bio-Design Project).
DDBJ/EMBL/GenBank accession nos+ AP005672, AB110071, AB110072, AB098724–AB098727
REFERENCES
- 1.Martin W., Rujan,T., Richly,E., Hansen,A., Cornelsen,S., Lins,T., Leister,D., Stoebe,B., Hasegawa,M. and Penny,D. (2002) Evolutionary analysis of Arabidopsis, cyanobacterial and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl Acad. Sci. USA, 99, 12246–12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simpson C.L. and Stern,D.B. (2002) The treasure trove of algal chloroplast genomes. Surprises in architecture and gene content and their functional implications. Plant Physiol., 129, 957–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wakasugi T., Tsudzuki,T. and Sugiura,M. (2001) The genomics of land plant chloroplasts: gene content and alteration of genomic information by RNA editing. Photosyn. Res., 70, 107–118. [DOI] [PubMed] [Google Scholar]
- 4.Kenrick P. and Crane,P.R. (1997) The origin and early evolution of plants on land. Nature, 389, 33–39. [Google Scholar]
- 5.Qiu Y.L. and Palmer,J.D. (1999) Phylogeny of early land plants: insights from genes and genomes. Trends Plant Sci., 4, 26–30. [DOI] [PubMed] [Google Scholar]
- 6.Qiu Y.L., Cho,Y., Cox,J.C. and Palmer,J.D. (1998) The gain of three mitochondrial introns identifies liverworts as the earliest land plants. Nature, 394, 671–674. [DOI] [PubMed] [Google Scholar]
- 7.Ohyama K., Fukuzawa,H., Kohchi,T., Shirai,H., Sano,T., Sano,S., Umezono,K., Shiki,Y., Takeuchi,M., Chang,Z. et al. (1986) Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature, 322, 572–574. [Google Scholar]
- 8.Kugita M., Kaneko,A., Yamamoto,Y., Takeya,Y., Matsumoto,T. and Yoshinaga,K. (2003) The complete nucleotide sequence of the hornwort (Anthoceros formosae) chloroplast genome: insight into the earliest land plants. Nucleic Acids Res., 31, 716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turmel M., Otis,C. and Lemieux,C. (2002) The chloroplast and mitochondrial genome sequences of the charophyte Chaetosphaeridium globosum: insights into the timing of the events that restructured organelle DNAs within the green algal lineage that led to land plants. Proc. Natl Acad. Sci. USA, 99, 11275–11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kugita M., Yamamoto,Y., Fujikawa,T., Matsumoto,T. and Yoshinaga,K. (2003) RNA editing in hornwort chloroplasts makes more than half the genes functional. Nucleic Acids Res., 31, 2417–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malek O., Lättig,K., Hiesel,R., Brennicke,A. and Knoop,V. (1996) RNA editing in bryophytes and a molecular phylogeny of land plants. EMBO J., 15, 1403–1411. [PMC free article] [PubMed] [Google Scholar]
- 12.Tsudzuki T., Wakasugi,T. and Sugiura,M. (2001) Comparative analysis of RNA editing sites in higher plant chloroplasts. J. Mol. Evol., 53, 327–332. [DOI] [PubMed] [Google Scholar]
- 13.Samigullin T.H., Valiejo-Roman,K.M., Troitsky,A.V., Bobrova,V.K., Filin,V.R., Martin,W. and Antonov,A.S. (1998) Sequences of rDNA internal transcribed spacers from the chloroplast DNA of 26 bryophytes: properties and phylogenetic utility. FEBS Lett., 422, 47–51. [DOI] [PubMed] [Google Scholar]
- 14.Pruchner D., Beckert,S., Muhle,H. and Knoop,V. (2002) Divergent intron conservation in the mitochondrial nad2 gene: signatures for the three bryophyte classes (mosses, liverworts and hornworts) and the lycophytes. J. Mol. Evol., 55, 265–271. [DOI] [PubMed] [Google Scholar]
- 15.Pruchner D., Nassal,B., Schindler,M. and Knoop,V. (2001) Mosses share mitochondrial group II introns with flowering plants, not with liverworts. Mol. Genet. Genomics, 266, 608–613. [DOI] [PubMed] [Google Scholar]
- 16.Kasten B., Wehe,M., Reski,R. and Abel,W.O. (1991) trnR-CCG is not unique to the plastid DNA of the liverwort Marchantia: gene identification from the moss Physcomitrella patens. Nucleic Acids Res., 19, 5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasten B., Wehe,M., Kruse,S., Reutter,K., Abel,W.O. and Reski,R. (1992) The plastome-encoded zfpA gene of a moss contains procaryotic as well as eukaryotic promoter consensus sequences and its RNA abundance is modulated by cytokinin. Curr. Genet., 22, 327–333. [DOI] [PubMed] [Google Scholar]
- 18.Machuka J., Bashiardes,S., Ruben,E., Spooner,K., Cuming,A., Knight,C. and Cove,D. (1999) Sequence analysis of expressed sequence tags from an ABA-treated cDNA library identifies stress response genes in the moss Physcomitrella patens. Plant Cell Physiol., 40, 378–387. [DOI] [PubMed] [Google Scholar]
- 19.Miyata Y., Sugiura,C., Kobayashi,Y., Hagiwara,M. and Sugita,M. (2002) Chloroplast ribosomal S14 protein transcript is edited to create a translation initiation codon in the moss Physcomitrella patens. Biochim. Biophys. Acta, 1576, 346–349. [DOI] [PubMed] [Google Scholar]
- 20.Hara K., Sugita,M. and Aoki,S. (2001) Cloning and characterization of the cDNA for a plastid σ factor from the moss Physcomitrella patens. Biochim. Biophys. Acta, 1517, 302–306. [DOI] [PubMed] [Google Scholar]
- 21.Wakasugi T., Sugita,M., Tsudzuki,T. and Sugiura,M. (1998) Updated gene map of tobacco chloroplast DNA. Plant Mol. Biol. Rep., 16, 231–241. [Google Scholar]
- 22.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 23.Kobayashi Y., Dokiya,Y., Kumazawa,Y. and Sugita,M. (2002) Non-AUG translation initiation of mRNA encoding plastid-targeted phage-type RNA polymerase in Nicotiana sylvestris. Biochem. Biophys. Res. Commun., 299, 57–61. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi Y., Dokiya,Y. and Sugita,M. (2001) Dual targeting of phage-type RNA polymerase to both mitochondria and plastids is due to alternative translation initiation in single transcripts. Biochem. Biophys. Res. Commun., 289, 1106–1113. [DOI] [PubMed] [Google Scholar]
- 25.Calie P.J. and Hughes,K.W. (1987) The consensus land plant chloroplast gene order is present, with two alterations, in the moss Physcomitrella patens. Mol. Gen. Genet., 208, 335–341. [Google Scholar]
- 26.Wakasugi T., Nagai,T., Kapoor,M., Sugita,M., Ito,M., Ito,S., Tsudzuki,J., Nakashima,K., Tsudzuki,T., Suzuki,Y. et al. (1997) Complete nucleotide sequence of the chloroplast genome from the green alga Chlorella vulgaris: the existence of genes possibly involved in chloroplast division. Proc. Natl Acad. Sci. USA, 94, 5967–5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turmel M., Otis,S. and Lemieux,C. (1999) The complete chloroplast DNA sequence of the green alga Nephroselmis olivacea: insights into the architecture of ancestral chloroplast genomes. Proc. Natl Acad. Sci. USA, 96, 10248–10253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wakasugi T. (2002) DDBJ accession no. AP004638.
- 29.Wakasugi T., Tsudzuki,J., Ito,S., Nakashima,K., Tsudzuki,T. and Sugiura,M. (1994) Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc. Natl Acad. Sci. USA, 91, 9794–9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato S., Nakamura,Y., Kaneko,T., Asamizu,E. and Tabata,S. (1999) Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res., 6, 283–290. [DOI] [PubMed] [Google Scholar]
- 31.Hallick R.B., Hong,L., Drager,R.G., Favreau,M.R., Monfort,A., Orsat,B., Spielmann,A. and Stutz,E. (1993) Complete sequence of Euglena gracilis chloroplast DNA. Nucleic Acids Res., 21, 3537–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemieux C., Otis,C. and Turmel,M. (2000) Ancestral chloroplast genome in Mesostigma viride reveals an early branch of green plant evolution. Nature, 403, 649–652. [DOI] [PubMed] [Google Scholar]
- 33.Goldschmidt-Clermont M., Choquet,Y., Girard-Bascou,J., Michel,F., Schirmer-Rahire,M. and Rochaix,J.D. (1991) A small chloroplast RNA may be required for trans-splicing in Chlamydomonas reinhardtii. Cell, 65, 135–143. [DOI] [PubMed] [Google Scholar]
- 34.Zweib C., Gorodkin,J., Knudsen,B., Burks,J. and Wower,J. (2003) tmRDB (tmRNA database). Nucleic Acids Res., 31, 446–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugita M., Svab,Z., Maliga,P. and Sugiura,M. (1997) Targeted deletion of sprA from the tobacco plastid genome indicates that the encoded small RNA is not essential for pre-16S rRNA maturation in plastids. Mol. Gen. Genet., 257, 23–27. [DOI] [PubMed] [Google Scholar]
- 36.Rochaix J.D. and Malnoe,P. (1978) Anatomy of the chloroplast ribosomal DNA of Chlamydomonas reinhardtii. Cell, 15, 661–670. [DOI] [PubMed] [Google Scholar]
- 37.Zhang G. and Darst,S.A. (1998) Structure of the Escherichia coli RNA polymerase α subunit amino-terminal domain. Science, 281, 262–266. [DOI] [PubMed] [Google Scholar]
- 38.Emanuelsson O., Nielsen,H., Brunak,S. and von Heijne,G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol., 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- 39.Palmer J.D., Calie,P.J., dePamphilis,C.W., Logsdon,J.M., Kats-Downie,D.S. and Downie,S.R. (1990) An evolutionary genetic approach to understanding plastid gene function: lessons from photosynthetic and nonphotosynthetic plants. In Baltscheffsky,M. (ed.), Current Research in Photosynthesis. Kluwer Academic Publishers, the Netherlands, Vol. III, pp. 475–482. [Google Scholar]
- 40.Wolfe K.H., Morden,C.W. and Palmer,J.D. (1992) Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl Acad. Sci. USA, 89, 10648–10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson R.J.M., Denny,P.W., Preiser,P.R., Rangachari,K., Roberts,K., Roy,A., Whyte,A., Strath,M., Moore,D.J., Moore,P.W. et al. (1996) Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J. Mol. Biol., 261, 155–172. [DOI] [PubMed] [Google Scholar]
- 42.Hiratsuka J., Shimada,H., Whittier,R., Ishibashi,T., Sakamoto,M., Mori,M., Kondo,C., Honji,Y., Sun,C.R., Meng,B.Y. et al. (1989) The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol. Gen. Genet., 217, 185–194. [DOI] [PubMed] [Google Scholar]
- 43.Maier R.M., Neckermann,K., Igloi,G.L. and Kössel,H. (1995) Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol., 251, 614–628. [DOI] [PubMed] [Google Scholar]
- 44.Millen R.S., Olmstead,R.G., Adams,K.L., Palmer,J.D., Lao,N.T., Heggie,L., Kavanagh,T.A., Hibberd,J.M., Gray,J.C., Morden,C.W. et al. (2001) Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell, 13, 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 46.Goff S., Ricke,D., Lan,T., Presting,G., Wang,R., Dunn,M., Glazebrook,J., Sessions,A., Oeller,P., Varma,H. et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science, 296, 92–100. [DOI] [PubMed] [Google Scholar]
- 47.Igarashi K., Fujita,N. and Ishihama,A. (1991) Identification of a subunit assembly domain in the alpha subunit of Escherichia coli RNA polymerase. J. Mol. Biol., 218, 1–6. [PubMed] [Google Scholar]
- 48.Igarashi K. and Ishihama,A. (1991) Bipartite functional map of the E. coli RNA polymerase α subunit: involvement of the C-terminal region in transcription activation by cAMP–CRP. Cell, 65, 1015–1022. [DOI] [PubMed] [Google Scholar]
- 49.Hess W.R. and Börner,T. (1999) Organellar RNA polymerases of higher plants. Int. Rev. Cytol., 190, 1–59. [DOI] [PubMed] [Google Scholar]
- 50.Hara K., Morita,M., Takahashi,R., Sugita,M., Kato,S. and Aoki,S. (2001) Characterization of two genes, Sig1 and Sig2, encoding distinct plastid σ factors in the moss Physcomitrella patens: phylogenetic relationships to plastid σ factors in higher plants. FEBS Lett., 499, 87–91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.