Abstract
A potential means to improve the efficacy of steric-blocking antisense oligonucleotides (ON) is to increase their affinity for a target RNA. The grafting of cationic amino groups to the backbone of the ON is one way to achieve this, as it reduces the electrostatic repulsion between the ON and its target. We have examined the duplex stabilising effects of introducing cationic phosphoramidate internucleoside linkages into ON with a non-natural α-anomeric configuration. Cationic α-ON bound with high affinity to single-stranded DNA and RNA targets. Duplex stabilisation was proportional to the number of cationic modifications, with fully cationic ON having particularly high thermal stability. The average stabilisation was greatly increased at low ionic strength. The duplex formed between cationic α-ON and their RNA targets were not substrates for RNase H. The penalty in Tm inflicted by a single mismatch, however, was high; suggesting that they are well suited as sequence-specific, steric-blocking, antisense agents. Using a well-described target sequence in the internal ribosome entry site of the human hepatitis C virus, we have confirmed this potential in a cell-free translation assay as well as in a whole cell assay. Interestingly, no vectorisation was necessary for the cationic α-ON in cell culture.
INTRODUCTION
Antisense (AS) oligonucleotides (ON) are ligands of choice to target RNA sequences and diminish, or abrogate, the expression of specific genes (1,2). Indeed, over 42 AS-ON are currently being screened as potential drugs and one is currently an FDA-approved drug (3).
The inherent instability of phosphodiester ON in biological media, and the problems often encountered with the widely used phosphorothioate analogue, have led to the elaboration of a large panoply of novel DNA analogues that possess vastly augmented nuclease resistance (4,5). The potential pharmacological benefits of such analogues are, however, tempered by their incapacity to form a substrate for RNase H, a cellular enzyme that degrades the RNA strand of the DNA:RNA heteroduplex formed between the AS-ON and its target RNA. Despite this, a number of studies have shown that RNase H-incompetent ON can efficiently down regulate specific gene expression by steric hindrance of a specific stage in the processing of information from DNA to protein (6).
An obvious means to increase the efficacy of these so-called ‘steric-blocking’ ON is to improve their affinity for the RNA target sequence. The grafting of cationic amino groups to the ON is a potential means to increase the thermal stability of the duplex formed between the ON and the RNA target, as it can reduce the electrostatic repulsion between the phosphate moieties of the two strands of the duplex.
Cationic groups can be introduced via nucleobase (7–10), sugar (11–13) or backbone modifications (14–23). However, cationisation of the internucleoside linkage has the clear advantage that it can be achieved with only minor adaptations to existing automated DNA synthesis protocols. Furthermore, whilst the grafting of cationic groups on nucleobases or sugars allows the formation of zwitterionic ON, backbone modifications can lead to both zwitterionic as well as fully modified cationic ON.
Although backbone-modifications generally confer a good resistance to nuclease degradation, in most cases, modifications of phosphate groups induce a destabilisation of the duplex. This presumably results from the diastereoisomery due to phosphorus chirality (24), compared with unmodified phosphodiester (PO)-ON. Such stereoisomeric constraints also apply to cationic backbone-modified ON (21). For example, Letsinger has shown that an ON with alternating anionic phosphodiester and cationic stereo-uniform phosphoramidate linkages formed a stable duplex with its DNA target whereas a similar ON containing the other stereoisomer did not bind efficiently (16). Despite this chirality problem, zwitterionic ON, containing several terminal cationic phosphoramidate linkages (as diastereoisomeric mixtures) and central phosphodiester linkages, have been reported to bind to their mRNA target and show a greater antisense activity than unmodified ON (22).
In addition, we have demonstrated that, unlike ON with the natural β-configuration, the stereoisomery of a single cationic dimethylaminopropyl phosphoramidate linkage (PNHDMAP) introduced into an ON with a non-natural α-anomeric configuration was not detrimental to the stability of duplexes formed with either DNA or RNA, regardless of the isomer considered (25). These α-anomeric ON, which were pioneered by our group, form stable parallel-stranded duplexes with complementary DNA and RNA (26). Interestingly, we have demonstrated that backbone modified α-ON (including methylphosphonates and non-ionic phosphoramidates) form more stable duplexes than their β-analogues (25,27). This has prompted us to investigate the possibility of combining the potential duplex stabilizing effects of cationic PNHDMAP linkages and an α-configuration for the sugar moieties.
In this paper, we focus on the effect of the cationic phosphoramidate internucleotide linkage on the hybridisation properties of the α-ON with single-stranded (ss)DNA and RNA. Of special interest, with respect to potential therapeutic and diagnostic purposes, we demonstrate that these cationic ON bind with high affinity to their single-stranded targets. The effects on duplex stability of the number and distribution of cationic linkages within an anionic α-ON have been investigated. We have also examined the base pairing specificity of the cationic α-ON and the influence of ionic strength on their hybridisation as compared to natural anionic β-ON.
Finally, we have demonstrated that neither a fully cationic PNHDMAP α-ON, nor chimeric PNHDMAP/PO α-ON, were able to elicit Escherichia coli RNase H hydrolysis of a complementary RNA target. Despite this, they were capable of specifically inhibiting translation initiated at the internal ribosome entry site (IRES) of the human hepatitis C virus (HCV) in vitro and in intact cells.
MATERIALS AND METHODS
Oligonucleotides
It is well established that α-ON, and their backbone modified phosphorothioate, methylphosphonate and phosphoramidate analogues, hybridise to their nucleic acid targets with a parallel orientation (25–30). For this reason, all the PNHDMAP α-ON described here were designed with a parallel orientation with respect to the target.
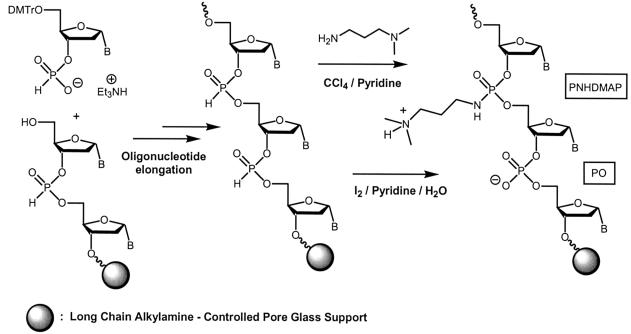
Modified α-ON 3–5 and 11 (Table 1) were synthesized (1 µmol scale) with an ABI Model 381A DNA synthesizer using phosphoramidite chemistry (31). Protected deoxy-2′-α-ribonucleoside-3′-phosphoramidites (α-dT, α-dCBz and α-dABz) were prepared according to published procedures (32). The phosphoramidate linkages were introduced into the α-analogues 3–5 through 3′-phosphoramidite dinucleoside building blocks containing a PNHDMAP linkage (T+T, C+T, T+C), prepared according to a procedure described in the literature for the synthesis of β-analogues (17) and further used by us for α-analogues (25). Oligomers 3–5 were synthesised following standard phosphoramidite chemistry except that extended coupling times (3 × 3 min) were applied for the coupling step of the α-dinucleotides. The elongation of chimeric α-ON 6, 7, 12, 12*, 13, 16 and 17 (Table 1), containing mixed PO and PNHDMAP domains, was performed in blocks on an ABI Model 394 DNA synthesizer using hydrogen phosphonate chemistry (33) with protected α-nucleoside 3′-H-phosphonates (25,34) (Scheme 1). The oxidation of each block was carried out manually by the syringe technique outside the synthesizer (22). The phosphodiester bonds were generated by oxidation of the corresponding H-phosphonate diester block with standard iodine–pyridine–water treatment, whereas the PNHDMAP linkages were obtained by oxidation with anhydrous CCl4 (450 µl) in the presence of pyridine (450 µl) and N-dimethylaminopropylamine (100 µl) (Aldrich) (Scheme 1). In the same way, fully backbone-modified ON 18, 19 and 20 were prepared via H-phosphonate-chemistry. The H-phosphonate diester linkages were then manually oxidized by CCl4 in the presence of 2-methoxyethylamine for ON 19 or N-dimethylaminopropylamine for ON 18 and 20. After deprotection with concentrated aqueous ammonia (30%) at 40°C for 5 h, α-ON were purified by preparative HPLC. For ON 3–6, 12, 12*, 16 and 17, purification was performed with a Nucleogel SAX 1000-8 column (50 × 4.6 mm; Macherey Nagel) using a 30 min linear gradient of 0–300 mM NaCl in 5 mM NaOH (pH 12) at a flow rate of 1.5 ml/min. ON 7, 13, 14, 18 and 20 were purified with a Nucleogel SCX 1000-8 column (50 × 4.6 mm; Macherey Nagel) using a 25 min linear gradient of 0–1 M NaCl in 20 mM KH2PO4 (pH 6) containing 20% CH3CN at a flow rate of 1.5 ml/min. Purification of ON 19 was performed by preparative RP–HPLC as previously described (25). All ON were further desalted with Chromafix PS-RP cartridges (Macherey-Nagel). Their final purity was confirmed by HPLC and their characterization was performed by MALDI-TOF mass spectrometry (data not shown). Unmodified ON 1, 2, 8, 8a–c, 9, 9a–c, 10, 15 (Table 1) were purchased from Eurogentec (Seraing, Belgium).
Table 1. Oligonucleotides synthesized.
| No. | Oligonucleotides | Sequence 5′→3′ | PNHDMAP linkages(+) | Net charge |
|---|---|---|---|---|
| 1 | DNA target | β-d(AAGAGGAAGAAA) | 0 | –11 |
| 2 | β-PO | β-d(TTTCTTCCTCTT) | 0 | –11 |
| 3 | α-PO/PNHDMAP | α-d(T+TCTCCT+TCT+TT) | 3 | –5 |
| 4 | α-PO/PNHDMAP | α-d(TT+CT+CCTT+CT+TT) | 4 | –3 |
| 5 | α-PO/PNHDMAP | α-d(TT+CT+CC+TT+CT+TT) | 5 | –1 |
| 6 | α-PO/PNHDMAP | α-d(TTCT+C+C+T+T+CTTT) | 5 | –1 |
| 7 | α-PO/PNHDMAP | α-d(TT+C+T+C+C+T+T+C+T+TT) | 9 | +7 |
| 8 | DNA target | β-d(AGAATTGGGTGT) | 0 | –11 |
| 8a | β-d(AGAACTGGGTGT) | 0 | –11 | |
| 8b | β-d(AGAAATGGGTGT) | 0 | –11 | |
| 8c | β-d(AGAAGTGGGTGT) | 0 | –11 | |
| 9 | RNA target | β-r(AGAAUUGGGUGU) | 0 | –11 |
| 9a | β-r(AGAACUGGGUGU) | 0 | –11 | |
| 9b | β-r(AGAAAUGGGUGU) | 0 | –11 | |
| 9c | β-r(AGAAGUGGGUGU) | 0 | –11 | |
| 10 | β-PO | β-d(ACACCCAATTCT) | 0 | –11 |
| 11 | α-PO | α-d(TCTTAACCCACA) | 0 | –11 |
| 12 | α-PO/PNHDMAP | α-d(TCTT+A+A+C+C+CACA) | 5 | –1 |
| 12* | α-PO/PNHDMAP | α-d(TCTT+A*+A+C+C+CACA) | 5 | –1 |
| 13 | α-PO/PNHDMAP | α-d(T+C+T+TAACCC+A+C+A) | 6 | +1 |
| 14 | α-PNHDMAP | α-d(T+C+T+T+A+A+C+C+C+A+C+A) | 11 | +11 |
| 15 | IRES RNA target | β-r(UAGUGUUGGGUCGCGA) | 0 | –15 |
| 16 | α-PO/PNHDMAP | α-d(ATCACA+A+C+C+C+A+G+C+G+C+T) | 10 | +5 |
| 17 | α-PO/PNHDMAP | α-d(ATC+A+C+A+A+C+C+C+A+G+C+GCT) | 11 | +7 |
| 18 | α-PNHDMAP | α-d(A+T+C+A+C+A+A+C+C+C+A+G+C+G+C+T) | 15 | +15 |
| 19 | α-PNHME | α-d(A•T•C•A•C•A•A•C•C•C•A•G•C•G•C•T) | 0 | 0 |
| 20 | α-PNHDMAP | α-d(C+A+A+C+T+C+A+G+A+C+C+C+T+C+G+A) | 15 | +15 |
+, catio nic PNHDMAP linkage; •, neutral PNHME linkage; A* = 2-amino-α-2′-deoxyadenosine.
Scheme 1. Synthesis of chimeric α-ON with PO/PNHDMAP internucleoside linkages on solid support via H-phosphonate chemistry.
UV melting experiments
The concentration of each ON was determined spectrophotometrically at 260 nm and at 80°C assuming that the molar extinction coefficient of each ON is the sum of the molar extinction coefficients of the constitutive deoxynucleotides. Optical measurements were carried out on a Uvikon 943 spectrophotometer (Kontron). The temperature of the cell was controlled by a Huber temperature programmer (Ministat) connected to a refrigerated ethyleneglycol/water bath. The cell compartment was continuously flushed with dry nitrogen when the temperature was below room temperature. Prior to the experiments, the ON, each at a final concentration of 3 µM, were mixed in 100 mM NaCl, 10 mM sodium cacodylate (pH 7) and allowed to incubate at 90°C for 30 min. During the melting experiments the heating rate was 0.5°C/min. Digitised absorbance and temperature values were stored in a computer for subsequent plotting and analysis. Tm values were determined from the maxima of the first derivative plots of absorbance versus temperature.
Hydrolysis of RNA heteroduplexes with E.coli RNase H
RNA target 15 was 5′-end labeled using [γ 32P]ATP (ICN 3000 Ci/mmol) and T4 Polynucleotide Kinase (Invitrogen) following the manufacturer’s protocol. Control PO β-ODN or modified PNHDMAP ON 16–18 (200 pmol) and RNA 15 (0.2 pmol) were incubated in 10 µl of assay buffer (20 mM Tris–Cl, pH 8, 50 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol) for 30 min at 37°C in the presence of 2 U of E.coli RNase H (Sigma). Reactions were stopped by heating to 90°C for 2 min and adding 2 µl of 80% formamide gel loading buffer containing tracking dyes. Cleavage products were separated on a 20% polyacrylamide/7M urea sequencing gel. Radioactive bands were visualised by autoradiography.
Plasmids
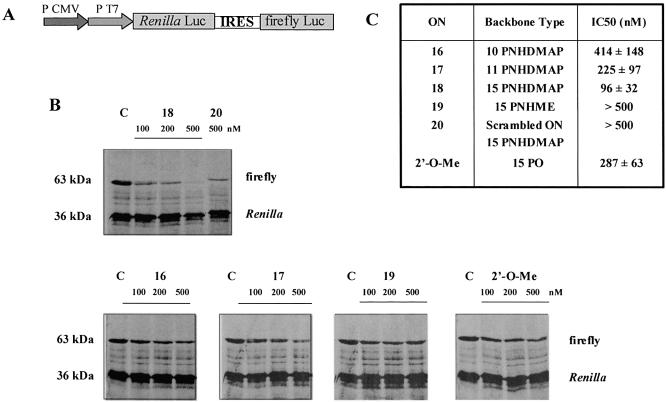
The bi-cistronic plasmid pRL-HL was kindly provided by S. Lemon (Galveston, Texas), and is outlined in Figure 2. Cistrons 1 and 2 code for Renilla and firefly luciferases, respectively, and are separated by the HCV IRES. The plasmid is more fully described by Honda et al. (35).
Figure 2.
Inhibition of HCV IRES driven cell-free translation by oligonucleotides. (A) Schematic representation of the bi-cistronic reporter cassette from plasmid pRL-HL (35). (B) Dose-dependent inhibition of HCV IRES driven cell-free translation by ON. Specificity is demonstrated by the lack of activity on Renilla luciferase translation and the lack of activity of a scrambled ON 20 on firefly luciferase translation. No ON were added in control reactions (C). Representative gels from three independent experiments are presented. (C) Calculated IC50 values for the specific inhibition of IRES-mediated translation by ON on capped RNA. For each concentration, signal from the firefly luciferase band is normalized to signal from the Renilla luciferase band and compared to the control lane. The mean values of three independent experiments are given.
RNA production
5′-capped RNA (to prime in vitro translation assays) was transcribed in vitro from the T7 promoter of pRL-HL using the Ribomax-T7 kit (Promega) following the manufacturer’s protocol. The plasmid was linearised immediately after the 3′-end of the required transcript, using HindIII restriction enzyme, purified on 1% agarose gels and electro-eluted from the excised gel bands. Five micrograms of purified plasmid were used as a template in 100 µl transcription reactions. DNase-treated RNA transcripts were purified using RNeasy mini columns (Qiagen). Purity and integrity were verified on 1% agarose gels.
In vitro translation
Translation assays were performed as described by Robbins and LeBleu (36), using nuclease-treated rabbit reticulocyte lysate (Promega) and [35S]methionine (Amersham-Pharmacia redivue; 1000 Ci/mmol) in 10 µl reactions. The translation reactions were primed with 200 ng of capped RNA in all cases. ON were added, on ice, prior to the addition of the lysate, with no specific prehybridisation step. Translated products were resolved on 12% running/4% stacking polyacrylamide–SDS gels. Vacuum-dried gels were analysed by autoradiography using Kodak Biomax films. Protein bands were quantified using NIH Image software (version 1.62).
Cell culture and establishment of stable transfectants
The hepatocyte cell line HepG2 was maintained in DMEM supplemented with non-essential amino acids (Gibco BRL), 20 mM HEPES pH 7.5 (Gibco BRL) and 10% fetal calf serum, at 37°C and 5% CO2.
pRL-HL was purified using an endotoxin-free plasmid maxiprep column (Qiagen) and transfected into HepG2 cells using FuGENE-6 (Roche) following the manufacturer’s instructions. Stably transfected clones were selected using G418 (1 mg/ml).
Intact cell assays
pRL-HL-expressing HepG2 cells (5.105) were plated to each well of six-well plates and allowed to grow overnight, giving a confluency of between 60 and 80% at time of loading. Cells were incubated with ON and harvested after 24 h, by scraping into Passive Lysis Buffer (Promega). Luciferase activities were assayed using dual-luciferase chemistry (Promega) and an EG & G Berthold MicroLumat LB96P luminometer.
RESULTS
Cationic internucleoside linkages stabilise hybrids
The stability of the double-stranded helix formed between an ON and a complementary ssDNA or RNA target is a compromise between hydrogen bonding and the electrostatic repulsion between the negatively charged phosphate moieties of the two strands. To investigate the extent to which the introduction of cationic internucleoside bonds (which reduce the net negative charge) into the backbone could stabilize duplexes, we evaluated melting temperatures (Tm) by thermal denaturation and renaturation experiments. We observed no differences between the thermal association and dissociation curves of the ON/DNA complementary strands tested. The introduction of cationic PNHDMAP linkages into homopyrimidine α-ON 3–7 (Tables 1 and 2), as well as into α-heteropolymers 12–14 (Tables 1 and 3), greatly increased the thermal stability of the duplexes, see for example unmodified phosphodiester β-ON 2 or 10 and α-ON 11. This stabilisation is proportional to the number of modifications. The average stabilisation was 4.4°C per modification in duplexes obtained with the homopyrimidine ON (Table 2) and 2°C in duplexes with the heteropolymer (Table 3). The average stabilisation was greatly increased at low ionic strength (6.7°C per modification) (Table 2).
Table 2. Thermal stability (Tm and ΔTm) of duplexes formed between homopyrimidines ON 2–7 and their DNA target 1 and salt concentration dependence of the duplexes stability.
| DNA target 1 | ||||
|---|---|---|---|---|
| 100 mM NaCl | No NaCl | |||
| ON | Tm (°C) | ΔTm (°C) | Tm (°C) | ΔTm (°C) |
| 2 | 35.5 | 20.6 | ||
| 3 | 48.0 | +12.5 | 39.0 | +18.4 |
| 4 | 54.5 | +19.0 | 47.5 | +26.9 |
| 5 | 60.0 | +24.5 | 58.2 | +37.6 |
| 6 | 58.5 | +23.0 | 55.5 | +34.9 |
| 7 | 69.0 | +33.5 | 78.5 | +57.9 |
Experiments w ere carried out at 3 µM concentration for each strand in a buffer containing 10 mM sodium cacodylate, pH 7 at the indicated NaCl concentration. Δ Tm = Tm modified ON – Tm 2.
Table 3. Thermal stability (Tm and ΔTm) of duplexes formed between tat sequence ON 10–14 and their DNA 8 and RNA 9 targets.
| DNA target 8 | RNA target 9 | |||
|---|---|---|---|---|
| ON | Tm (°C) | ΔTm (°C) | Tm (°C) | ΔTm (°C) |
| 10 | 45.5 | 44.6 | ||
| 11 | 43.5 | –2.0 | 41.8 | –2.8 |
| 12 | 55.0 | +9.5 | 43.0 | –1.6 |
| 12* | 55.0 | +9.5 | 46.0 | +1.4 |
| 13 | 54.0 | +8.5 | 43.0 | –1.6 |
| 14 | 72.5 | +27.0 | 55.0 | +10.4 |
Experiments were carried out at 3 µM concentration for each strand in a buffer containing 10 mM sodium cacodylate, pH 7, 100 mM NaCl. Δ Tm = Tm modified ODN – Tm 10.
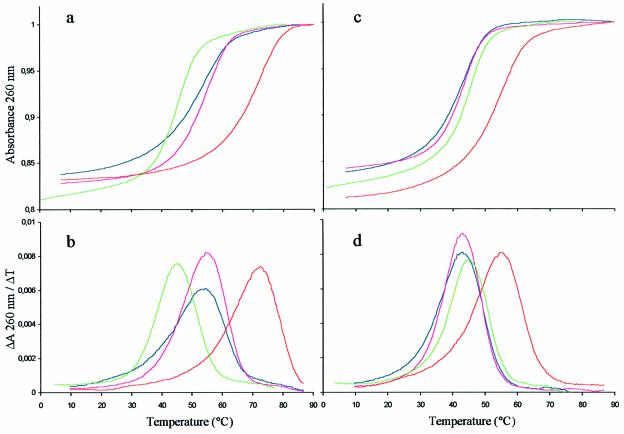
The effect of cationic charge distribution on duplex formation was evaluated by comparison of the ΔTm per modification calculated for each modified ON. The homopyrimidine α-ON 6, harbouring an internal stretch of five contiguous PNHDMAP linkages, formed a slightly less stable hybrid (ΔTm/mod + 4.6°C) than an isosequential ON 5 that harbours five PNHDMAP alternated with phosphodiester linkages (ΔTm/mod + 4.9°C) (Table 2). On the other hand, ON 12, which also has a central window of five PNHDMAP linkages, formed a more stable duplex on DNA target 8 than ON 13, which has the cationic linkages in the 5′ and 3′ wings, even though there is one additional PNHDMAP linkage in this ON (Table 3). The sharpness of the melting curves with the DNA targets was slightly affected with increasing modifications. The higher the number of cationic linkages, the broader the temperature range over which the duplexes melted (Fig. 1A and B).
Figure 1.
(a) UV-melting curves and (b) first derivative plots for the duplexes formed with DNA target 8 and β-PO 10 (green line), ON 12 (blue line), ON 13 (pink line) or ON 14 (red line). (c) UV-melting curves and (d) first derivative plots for the duplexes formed with RNA target 9 and β-PO 10 (green line), ON 12 (blue line), ON 13 (pink line) or ON 14 (red line).
As with DNA targets, no differences between the thermal association and dissociation curves were observed with RNA targets. The stability of the hybrids formed between the RNA target 9 and α-ON 12 and 13, containing respectively five and six cationic linkages, was only slightly higher than that with the corresponding PO α-ON 11 (ΔTm + 1.2°C), but somewhat lower than the duplex formed with the natural PO β-ON 10 (ΔTm –1.6°C) (Table 3). The fully modified ON 14, however, showed a substantial enhancement of duplex stability, compared with both PO β-ON 10 and α-ON 11 (ΔTm + 10.4°C and + 13.2°C, respectively). The decrease in Tm observed between α-ON 12 and 13 hybridised to an RNA target when compared with a DNA target was considerably greater (ΔTm –12°C and –11°C, respectively) than the decrease observed with either PO α-ON 11 (ΔTm –1.7°C) or with PO β-ON 10 (ΔTm –0.9°C) (Table 3). In contrast with the DNA targets, all duplexes formed with RNA exhibited the same cooperativity of melting, regardless of the number of modifications and their distribution (Fig. 1C and D).
The replacement of one α-2′-deoxyadenosine, in the central cationic linkages window of α-ON 12, with a 2-amino-α-2′-deoxyadenosine (ON 12*) (37) had no effect on the duplex stability with DNA, but increased the Tm with an RNA target by 3°C.
A single mismatch in a cationic α-ON/target duplex is highly destabilising
As it has been reported that the enhancement of duplex stability by cationic polyamines can be sufficient to counter-balance the instability caused by mismatched bases (38), we evaluated the influence of a single mismatch AX in the DNA 8 or RNA 9 targets on the stability of the hybrids.
Introduction of one AC, AA or AG mismatch in the hybrids between the cationic α-ON 14 and DNA targets 8a–c decreased the Tm by 15.5, 14.0 and 12.5°C, respectively (Table 4). The average destabilisation per mismatch was higher for cationic α-ON 14 (ΔTm/mismatch –14.0°C) than for β-ON 10 (ΔTm/mismatch –12.0°C). Nevertheless, it should be noted that the increased stability of the duplex formed by the cationic α-ON compared with the β-ON with the perfect duplex is reflected by more stable hybrids with the corresponding mismatched duplexes (Table 4). In addition, the cationic α-ON 12 and 14 discriminate AG mismatches with either DNA or RNA targets better than the β-ON 10. Surprisingly, with the RNA target, the CA mismatch is less destabilising with the cationic α-ON 14 (ΔTm –8.5°C) than for the regular β-ON (ΔTm –11.3°C). The average destabilisation per mismatch for both cationic α-ON 14 and β-ON was identical (ΔTm/mismatch –10.0°C). Here again, the mismatched duplexes formed between the cationic α-ON 14 and the mismatched RNA targets 9a–c were more stable than the corresponding duplexes with the β-ON 10 and, indeed, greater than that of the perfectly matched RNA 9/β-ON 10 duplex (Table 4).
Table 4. Influence of a single mismatch on the Tm of duplexes with ON 10 and 14.
| Tm (°C) | ||||||||
|---|---|---|---|---|---|---|---|---|
| DNA targets | rRNA targets | |||||||
| ON | 8 (AT) | 8a (AC) | 8b (AA) | 8c (AG) | 9 (AT) | 9a (AC) | 9b (AA) | 9c (AG) |
| 10 | 45.5 | 30.5 | 32.3 | 37.7 | 44.6 | 33.3 | 33.8 | 36.8 |
| 14 | 72.5 | 57.0 | 58.5 | 60.0 | 55.0 | 46.5 | 45.0 | 43.5 |
Cationic α-ON/RNA duplexes are not substrates for RNase H
Under conditions (37°C for 30 min) where the control PO ON stimulated RNA 15 degradation (data not shown), the chimeric α-ON 16–18/RNA 15 duplexes were not substrates for E.coli RNase H.
Cationic α-ON arrest HCV IRES-mediated translation in an acellular assay
ON complementary to the IIId loop of the HCV IRES were tested in dose-response in an in vitro translation assay programmed with a bi-cistronic construction that allows the simultaneous monitoring of translation initiated at the HCV IRES (firefly luciferase) and from a classical, eukaryote, cap-dependent translation initiation site (Renilla luciferase) (Fig. 2A). Cationic α-ON were capable of arresting IRES-dependent translation, whilst having no effect on cap-dependent translation initiation (Fig. 2B). The IC50 was inversely proportional to the number of cationic charges for ON 16–18 (Fig. 2C). The fully cationic ON 18 has a significantly lower IC50 than an isosequential 2′-O-methyloligoribonucleotide (2′-O-Me ON). Neither neutral backboned methoxyethylphosphoramidate (PNHME) ON 19 nor a scrambled, fully cationic α-ON 20 affected IRES-mediated translation (Fig. 2C).
Cationic α-ON arrest HCV IRES-mediated translation in cell culture
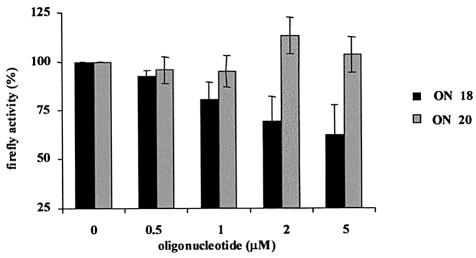
In order to establish whether cationic α-ON are capable of blocking IRES-mediated translation in a cellular environment, the bi-cistronic construction used for the cell-free translation experiments was stably transfected into a HepG2 hepatoma cell line. The cells were incubated with the cationic α-ON and the activities of the two luciferase (firefly and Renilla) reporter genes were determined by luminometry. α-ON 18 inhibited firefly (compared with Renilla) luciferase activity in a dose-dependent manner in the whole cell assay while the scrambled α-ON 20 showed no significant activity (Fig. 3). Interestingly, this effect was observed without the use of specific transfection agents. Under similar conditions, neither the isosequential 2′-O-Me ON nor the neutral backboned PNHME ON had any effect (data not shown).
Figure 3.
Dose-dependent inhibition of HCV-IRES driven translation by ON 18 in intact cells. HepG2 cells stably transfected with the pRL-HL construct, were incubated with ON at the given doses. Protein extracts were made 24 h later and firefly and Renilla luciferase activities assayed by luminometry. Firefly luciferase activity (IRES driven) was normalized with respect to Renilla luciferase activity. Comparative efficiencies of ON 18 with respect to a scrambled control ON 20 are shown. Representative values from three independent experiments are presented.
DISCUSSION
Although several antisense ON are currently being developed as drugs, their large size (usually 20mer), low stability and poor membrane passage reduce their potential as pharmaceutical agents. We hypothesised that by combining the potential duplex-stabilising effects of the non-natural α-anomeric DNA configuration with PNHDMAP cationic inter-nucleotide linkages, we could produce antisense ON with high affinity for target nucleic acid sequences and improved cellular uptake.
The incorporation of cationic modifications, into both an oligopyrimidine α-ON and a mixed purine/pyrimidine ON, increased the melting temperatures of the duplexes formed with a DNA target in comparison with that of an unmodified phosphodiester β-ON. The increase in Tm was positively correlated to the number of cationic modifications. Curiously, the increased Tm observed with DNA targets was only observed with an RNA target when the ON was fully cationic. The lower melting cooperativity of modified duplexes with the DNA target 8 compared to PO duplex accounts for the effect of increasing mixtures of diastereoisomers, when the number of modifications increases, due to the chirality of the phosphorus atom. Indeed, this difference in hybridization abilities between isomers was previously observed in backbone-modified β-ON as well as α-ON (25). In contrast, we observed only a minor difference between isomers with α-ON when the target was RNA.
At low salt concentration, the differences in the affinity of the cationic ON 7 and the natural ON 2 is striking. This reflects the charge repulsion between the two negatively charged PO backbones of 2 and its target 1 and in contrast, the charge attraction between the cationic ON 7 and its anionic target 1. In comparison, hybrids with ON 5 and 6, with a lower net charge (–1), were less sensitive to a change of ionic strength. Compared to previously studied non-ionic PNHME α-ON (25), which induced a slight stabilisation effect, anionic or zwitterionic α-ON 12 and 13 do not improve affinity for RNA target. We have found, however, that the affinity of a PNHDMAP α-ON for the RNA target could be increased by replacing an α-2′-deoxyadenosine by a 2-amino-α-2′-deoxyadenosine (A*) that can potentially form three hydrogen bonds (37) with a complementary thymine and uracil, unlike adenine, which forms only two hydrogen bonds.
Cationic PNHDMAP α-ON exhibit the highest affinity for DNA and RNA targets of all the backbone-modified α-ON studied. This is particularly true of the fully modified cationic α-ON 14 and probably results from electrostatic interactions between cationic PNHDMAP linkages and the anionic PO backbone of the targets.
For the partially cationic α-ON, the distribution of cationic charges appears to influence the affinity for both DNA and RNA targets. A 12mer oligopyrimidine ON with five cationic linkages, for example, has a substantially higher Tm when the charged linkages are evenly distributed compared with when they are bunched into the central region. Centrally grouped cationic linkages in the mixed purine/pyrimidine α-ON, on the other hand, are more favourable than an ON with cationic linkages restricted to the wings.
It is clear that, as with virtually all second-generation (i.e. not phosphorothioate) analogues described to date, cationic PNHDMAP α-ON are incapable of forming a suitable substrate for RNase H. This is of little surprise as unmodified α-ON are themselves RNase H incompetent (39). This has a major implication for their use as antisense reagents as, unless they are coupled to an RNase activator (40) or contain an RNase H-competent central window (41), their activity is restricted to steric blockade. However, RNase H competence, although potentially increasing activity is also a source of decreased specificity, as only a few contiguous nucleotides need to hybridise to provoke target cleavage and this reduces their ability to distinguish between a bona fide target sequence and a partially complementary by-stander sequence (42). The advantage of steric blocking ON is that specificity is a function of affinity. We demonstrate here that a single mismatch within a 12mer fully cationic α-ON has a large impact on Tm. This is in keeping with similar mismatches in the corresponding duplexes formed with a neutral PNHME α-ON (25) (ΔTm/mismatch –15.4°C), indicating that base pairing fidelity is not overwhelmed by the electrostatic attraction between the cationic ON and the anionic backbone of the target. As reported for anionic β-ON (43), for cationic α-ON hybridised to DNA targets, the least stable mismatches are those containing cytosine and the most stable mismatches are those containing guanine.
Cationic α-ON, therefore, fulfill a number of criteria critical to their use as antisense agents; namely high affinity, high stability and good selectivity. We therefore tested their capacity to selectively arrest the formation of a translational pre-initiation complex on the HCV internal ribosome entry site, in a cell-free assay. Cationic α-ON were capable of effectively and specifically abrogating IRES-mediated translation, with an IC50 that was inversely correlated to the number of cationic charges. The fully cationic 16mer α-ON 18 had a lower IC50 than a 2′-O-methyl analogue or a PNHME α-ON 19 targeting the same site, and a similar IC50 to that of a 12mer peptide nucleic acid (Martinand-Mari,C., Markarian,A., Burm,B.E.A., Meeuvenoord,N.J., van Boom,J.H., Lebleu,B. and Robbins,I., in revision). Tallet-Lopez et al. (44) have also demonstrated the efficacity of 14–17mer 2′-O-methyl analogues targeted to this site, and Jubin et al. (45) that of longer (20–24mer) morpholino analogues.
The poor uptake of anionic or neutral oligonucleotides by cells has long proved a major obstacle to their utilisation. A number of strategies used to increase ON uptake rely on their complexation or conjugation with oligo/polycationic moieties, such as lipids or peptides (46,47). The incorporation of cationic linkages within the backbone of the ON itself could potentially overcome the need to use cationic vectors. We therefore tested the fully cationic α-ON 18 in a whole cell assay. A significant, dose-dependent reduction in firefly luciferase activity (IRES-mediated translation) was observed. Although the antisense effect was only moderate, it does demonstrate the capacity of cationic ON to be taken up by the cells and to bind to target in a physiologically relevant context. The degree of inhibition is similar to that observed with transfected 2′-O-methyl analogues or PNA (Martinand-Mari,C., Markarian,A., Burm,B.E.A., Meeuvenoord,N.J., van Boom,J.H., Lebleu,B. and Robbins,I., in revision).
Taken together, these results demonstrate the potential applicability of cationic phosphoramidate α-ON as simple, non-vectorised antisense agents.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the Association pour la Recherche contre le Cancer for financial support (J.-J.V. and B.L.) and the Ligue Nationale pour la Recherche contre le Cancer for a post-doctoral fellowship (C.M.-M.).
REFERENCES
- 1.Opalinska J.B. and Gewirtz,A.M. (2002) Nucleic-acid therapeutics: basic principles and recent applications. Nature Rev. Drug Discov., 1, 503–513. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal S. and Zhao,Q. (1998) Antisense therapeutics. Curr. Opin. Chem. Biol., 2, 519–528. [DOI] [PubMed] [Google Scholar]
- 3.Stephens A.C. and Rivers,R.P.A. (2003) Antisense oligonucleotide therapy in cancer. Curr. Opin. Mol. Therapeutics, 5, 118–122. [PubMed] [Google Scholar]
- 4.Altmann K.-H., Cuenoud,B. and Von Matt,P. (1998) Novel chemistry. In Stein,C.A. and Krieg,A.M. (eds), Applied Antisense Oligonucleotide Technology. Wiley-Liss Inc., New York, NY, pp. 73–107. [Google Scholar]
- 5.Seeberger P.H. and Caruthers,M.H. (1998) Modified oligonucleotides as antisense therapeutics. In Stein,C.A. and Krieg,A.M. (eds), Applied Antisense Oligonucleotide Technology. Wiley-Liss Inc., New York, NY, pp. 51–71. [Google Scholar]
- 6.Tidd D.M. and Giles,R.V. (2000) Mechanisms of action of antisense oligonucleotides. In Couvreur,P. and Malvy,C. (eds), Pharmaceutical aspects of oligonucleotides. Taylor & Francis, London, UK, pp. 3–31. [Google Scholar]
- 7.Manoharan M., Ramasamy,K.S., Mohan,V. and Cook,P.D. (1996) Oligonucleotides bearing cationic groups: N2-(3-aminopropyl)deoxyguanosine. Tetrahedron Lett., 37, 7675–7678. [Google Scholar]
- 8.Soto A.M., Kankia,B.I., Dande,P., Gold,B. and Marky,L.A. (2001) Incorporation of a cationic aminopropyl chain in DNA hairpins: thermodynamics and hydration. Nucleic Acids Res., 29, 3638–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bijapur J., Keppler,M.d., Bergqvist,S., Brown,T. and Fox,K.R. (1999) 5-(1-propargylamino)-2′-deoxyuridine (Up): a novel thymidine analogue for generating DNA triplexes with increased stability. Nucleic Acids Res., 27, 1802–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajeev K.G., Jadhav,V.R. and Ganesh,K.N. (1997) Triplex formation at physiological pH: comparative studies on DNA triplexes containing 5-Me-dC tethered at N4 with spermine and tetraethyleneoxyamine. Nucleic Acids Res., 25, 4187–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prakash T.P., Manoharan,M., Fraser,A.S., Kawasaki,A.M., Lesnik,E.A. and Owens,S.R. (2000) Zwitterionic oligonucleotides with 2′-O-[3-(N,N-dimethylamino)propyl]-RNA modification: synthesis and properties. Tetrahedron Lett., 41, 4855–4859. [Google Scholar]
- 12.Puri N., Majumdar,A., Cuenoud,B., Natt,F., Martin,P., Boyd,A., Miller,P.S. and Seidman,M.M. (2002) Minimum number of 2′-O-(2-aminoethyl) residues required for gene knockout activity by triplex helix forming oligonucleotides. Biochemistry, 41, 7716–7724. [DOI] [PubMed] [Google Scholar]
- 13.Kanazaki M., Ueno,Y., Shuto,S. and Matsuda,A. (2000) Highly nuclease-resistant phosphodiester-type oligodeoxynucleotides containing 4′α-C-aminoalkylthymidines form thermally stable duplexes with DNA and RNA. A candidate for potent antisense molecules. J. Am. Chem. Soc., 122, 2422–2432. [Google Scholar]
- 14.Letsinger R.L., Singman,C.N., Histand,G. and Salunkhe,M. (1988) Cationic oligonucleotides. J. Am. Chem. Soc., 110, 4470–4471. [Google Scholar]
- 15.Jung P.M., Histand,G. and Letsinger,R.L. (1994) Hybridization of alternating cationic/anionic oligonucleotides to RNA segments. Nucl. Nucl., 13, 1597–1605. [Google Scholar]
- 16.Horn T., Chaturvedi,S., Balasubramaniam,T.N. and Letsinger,R.L. (1996) Oligonucleotides with alternating anionic and cationic phosphoramidate linkages: synthesis and hybridization of stereo-uniform isomers. Tetrahedron Lett., 37, 743–746. [Google Scholar]
- 17.Chaturvedi S., Horn,T. and Letsinger,R.L. (1996) Stabilization of triple-stranded oligonucleotide complexes: use of probes containing alternating phosphodiester and stereo-uniform cationic phosphoramidate linkages. Nucleic Acids Res., 24, 2318–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dagle J.M. and Weeks,D.L. (1996) Positively charged oligonucleotides overcome potassium-mediated inhibition of triplex DNA formation. Nucleic Acids Res., 24, 2143–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bailey C.P., Dagle,J.M. and Weeks,D.L. (1998) Cationic oligonucleotides can mediate specific inhibition of gene expression in Xenopus oocytes. Nucleic Acids Res., 26, 4860–4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kore A. and Salunke,M. (1998) Fluoresceine-labelled cationic oligonucleotide probes: detection of hybrid formation in solution using fluorescence polarization spectroscopy. Indian J. Chem., Section A, 37B, 536–539. [Google Scholar]
- 21.Robles J., Ibanez,V., Grandas,A. and Pedroso,E. (1999) Synthesis and triple helix-forming ability of oligonucleoties with N,N-dimethylaminoethyl phosphoramidate linkages. Tetrahedron Lett., 40, 7131–7134. [Google Scholar]
- 22.Dagle J.M., Littig,J.L., Sutherland,L.B. and Weeks,D.L. (2000) Targeted elimination of zygotic messages in Xenopus laevis embryos by modified oligonucleotides possessing terminal cationic linkages. Nucleic Acids Res., 28, 2153–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vasquez K.M., Dagle,J.M., Weeks,D.L. and Glazer,P.M. (2001) Chromosome targeting at short polypurine sites by cationic triplex-forming oligonucleotides. J. Biol. Chem., 276, 38536–38541. [DOI] [PubMed] [Google Scholar]
- 24.Lebedev A.V. and Wickstrom,E. (1996) The chirality problem in P-substituted oligonucleotides. Perspect. Drug Disc. Des., 4, 17–40. [Google Scholar]
- 25.Laurent A., Naval,M., Debart,F., Vasseur,J.-J. and Rayner,B. (1999) Chiral and steric effects in the efficient binding of [alpha]-anomeric deoxyoligonucleoside N-alkylphosphoramidates to ssDNA and RNA. Nucleic Acids Res., 27, 4151–4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rayner B., Malvy,C., Paoletti,J., Lebleu,B., Paoletti,C. and Imbach,J.-L. (1989) α-Oligodeoxynucleotide Analogues. In Cohen,J.S. (ed.), Topics in Molecular and Structural Biology, Vol.12: Oligodeoxynucleotides, Antisense Inhibitors of Gene Expression. MacMillan Press, London, UK, pp. 119–136. [Google Scholar]
- 27.Debart F., Meyer,A., Vasseur,J.-J. and Rayner,B. (1998) Anomeric inversion (from β to α) in methylphosphonate oligonucleosides enhances their affinity for DNA and RNA. Nucleic Acids Res., 26, 4551–4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morvan F., Porumb,H., Degols,G., Lefebvre,I., Pompon,A., Sproat,B.S., Rayner,B., Malvy,C., Lebleu,B. and Imbach,J.L. (1993) Comparative evaluation of 7 oligonucleotide analogues as potential antisense agents. J. Med. Chem., 36, 280–287. [DOI] [PubMed] [Google Scholar]
- 29.Peyrottes S., Vasseur,J.-J., Imbach,J.-L. and Rayner,B. (1996) Oligodeoxynucleoside phosphoramidates (P-NH2): synthesis and thermal stability of duplexes with DNA and RNA target. Nucleic Acids Res., 24, 1841–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peyrottes S., Vasseur,J.-J., Imbach,J.-L. and Rayner,B. (1996) Dramatic effect of the anomeric configuration on the thermal stability of duplex formed between novel dodecathymidine phosphoramidate (P-NH2) and complementary DNA and RNA strands. Tetrahedron Lett., 37, 5869–5872. [Google Scholar]
- 31.Beaucage S.L. and Iyer,R.P. (1992) Advances in the synthesis of oligonucleotides by the phosphoramidite approach. Tetrahedron, 48, 2223–2311. [Google Scholar]
- 32.Morvan F., Rayner,B. and Imbach,J.-L. (1993) α-Oligodeoxynucleotides. In Agrawal,S. (ed.), Methods in Molecular Biology: Protocols for Oligonucleotides and Analogs, Vol. 20. Humana Press, Totowa, NJ, pp. 261–285. [DOI] [PubMed] [Google Scholar]
- 33.Froehler B.C. (1993) Oligodeoxynucleotide synthesis. H-phosphonate approach. In Agrawal,S. (ed.), Methods in molecular biology. Protocols for oligonucleotides and analogs, Vol. 20. Humana Press, Totowa, NJ, pp. 63–80. [DOI] [PubMed] [Google Scholar]
- 34.Ozola V., Reese,C.B. and Song,Q. (1996) Use of ammonium aryl H-phosphonates in the preparation of nucleoside H-phosphonate building blocks. Tetrahedron Lett., 37, 8621–8624. [Google Scholar]
- 35.Honda M., Kaneko,S., Matsushita,E., Kobayashi,K., Abell,G.A. and Lemon,S.M. (2000) Cell cycle regulation of hepatitis C virus internal ribosomal entry site-directed translation. Gastroenterology, 118, 152–162. [DOI] [PubMed] [Google Scholar]
- 36.Robbins I. and Lebleu,B. (2000) Vesicular stomatitis virus as model system for studies of antisense oligonucleotide translation arrest. Methods Enzymol., 313, 189–203. [DOI] [PubMed] [Google Scholar]
- 37.Naval M., Michel,T., Vasseur,J.-J. and Debart,F. (2002) 2-amino-α-2′-deoxyadenosine increased duplex stability of methoxyethylphosphoramidate α-oligodeoxynucleotides with RNA target. Bioorg. Med. Chem. Lett., 12, 1435–1438. [DOI] [PubMed] [Google Scholar]
- 38.Hou M.-H., Lin,S.-B., Yuann,J.-M.P., Lin,W.-C., Wang,A.H.-J. and Kan,L.-S. (2001) Effects of polyamines on the thermal stability and formation kinetics of DNA duplexes with abnormal structure. Nucleic Acids Res., 29, 5121–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gagnor C., Bertrand,J.-R., Thenet,S., Lemaître,M., Morvan,F., Rayner,B., Malvy,C., Lebleu,B., Imbach,J.-L. and Paoletti,C. (1987) Alpha-ADN VI: Comparative study of alpha- and beta-anomeric oligodeoxyribonucleotides in hybridization to mRNA and in cell free translation inhibition. Nucleic Acids Res., 15, 10419–10436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robbins I., Mitta,G., Vichier-Guerre,S., Sobol,R., Ubysz,A., Rayner,B. and Lebleu,B. (1998) Selective mRNA degradation by antisense oligonucleotide-2,5A chimeras: involvement of RNase H and RNase L. Biochimie, 80, 1–10. [DOI] [PubMed] [Google Scholar]
- 41.Malchere C., Verheijen,J., Van der Laan,S., Bastide,L., Van Boom,J., Lebleu,B. and Robbins,I. (2000) A short phosphodiester window is sufficient to direct RNase H-dependent RNA cleavage by antisense peptide nucleic acid. Antisense Nucleic Acid Drug Dev., 10, 463–468. [DOI] [PubMed] [Google Scholar]
- 42.Branch A. (1998) A good antisense molecule is hard to find. Trends Biochem. Sci., 23, 45–50. [DOI] [PubMed] [Google Scholar]
- 43.Aboul-ela. F., Koh,D., Tinoco,I. and Martin,F.H. (1985) Base-base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A,C,G,T). Nucleic Acids Res., 13, 4811–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tallet-Lopez B., Aldaz-Carroll,L., Chabas,S., Dausse,E., Staedel,C. and Toulme,J.J. (2003) Antisense oligonucleotides targeted to the domain IIId of the hepatitis C virus IRES compete with 40S ribosomal subunit binding and prevent in vitro translation. Nucleic Acids Res., 31, 734–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jubin R., Vantuno,N.E., Kieft,J.S., Murray,M.G., Doudna,J.A., Lau,J.Y. N. and Baroudy,B.M. (2000) Hepatitis C virus internal ribosome entry site (IRES) stem loop IIId contains a phylogenetically conserved GGG triplet essential for translation and IRES folding. J. Virol., 74, 10430–10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thierry A.R., Vives,E., Richard,J.-P., Prevot,P., Martinand-Mari,C., Robbins,I. and Lebleu,B. (2003) Cellular uptake and intracellular fate of antisense oligonucleotides. Curr. Opin. Mol. Therapeutics, 5, 133–138. [PubMed] [Google Scholar]
- 47.Juliano R.L. (2000) Mechanisms of transmembrane transport of oligonucleotides. In Couvreur,P. and Malvy,C. (eds), Pharmaceutical Aspects of Oligonucleotides. Taylor & Francis, London, UK, pp. 213–225. [Google Scholar]