Abstract
Animal proteins that contain a methyl-CpG-binding domain (MBD) are suggested to provide a link between DNA methylation, chromatin remodelling and gene silencing. However, some MBD proteins reside in chromatin remodelling complexes, but do not have specific affinity for methylated DNA. It has recently been shown that the Arabidopsis genome contains 12 putative genes encoding proteins with domains similar to MBD, of which at least three bind symmetrically methylated DNA. Using a bioinformatics approach, we have identified additional domains in a number of these proteins and, on this basis and extended sequence similarity, divided the proteins into subgroups. Using RT–PCR we show that 10 of the AtMBD genes are active and differentially expressed in diverse tissues. To investigate the biological significance of AtMBD proteins, we have transformed Arabidopsis with a construct aimed at RNA interference with expression of the AtMBD11 gene, normally active in most tissues. The resulting 35S::AtMBD11-RNAi plants displayed a variety of phenotypic effects, including aerial rosettes, serrated leaves, abnormal position of flowers, fertility problems and late flowering. Arabidopsis lines with reduced expression of genes involved in chromatin remodelling and transgene silencing show similar phenotypes. Our results suggest an important role for AtMBD proteins in plant development.
INTRODUCTION
DNA methylation, one of the most abundant epigenetic modifications in higher plants and animals, plays an important role in regulating development and developmental processes (1,2). In animal systems, DNA methylation is essential for normal embryonic development. Mouse embryos with significantly reduced DNA methylation levels abort spontaneously during gestation. DNA methylation is known to be crucial for genomic imprinting as well as X chromosome inactivation and to be important in controlling the activity of genomic parasites like transposable elements, retrotransposons and retroviruses (3).
In plants, recent studies show that genome-wide demethylation has pleiotropic effects on morphology. Plants with a general reduction in DNA methylation have been generated either by 5-azacytidine treatment (4), mutation of the Arabidopsis DDM1 gene encoding a member of the SWI2/SNF2 family of chromatin remodelling proteins (5) or by the use of antisense technology to suppress the Arabidopsis MET1 gene, which encodes a DNA methyltransferase (6,7). The hypomethylated background is associated with the onset of developmental abnormalities, including loss of apical dominance, stunting and changes in flowering time (6–8).
Evidently there is a strong correlation between DNA methylation and repression of gene expression. An understanding of the underlying molecular mechanisms has emerged in recent years from the discovery of mammalian proteins, MeCP2 and MBD1–MBD4, which contain a common methyl-CpG-binding domain (MBD) (9,10). A general notion is that MBD proteins bind methylated CpG in vitro and in vivo, function in transcriptional repression and are associated with histone deacetylases: when bound to methylated DNA, MeCP2 recruits the SIN3 histone deacetylase- containing complex, while MBD2 and MBD3 interact with the NuRD/Mi2 deacetylase complex (11) and MBD1 transcriptional repression has been shown to be deacetylase-dependent (12). Thus, MBD proteins are suggested to link DNA methylation to gene silencing via chromatin remodelling events where deacetylation of histone tails is essential. However, all data do not fit into this model (13), e.g. the human MBD3 is not capable of binding methylated DNA (14) and the human MBD4 is involved in mismatch repair of methylated DNA (15).
Recently, it was shown that MeCP2 can recruit histone H3K9 methyltransferase activity (16). In addition, mammalian proteins, e.g. ESET (17), with a MBD motif and a histone methyltransferase (HMT) type of bifurcated SET domain, have been identified. Although so far the MBD of ESET has not been shown to bind methylated DNA, ESET has been suggested to mediate a coupling of histone methylation with DNA methylation where MBD-containing HMTs may bind directly to methylated nucleosomal DNA to methylate histone tails (18). ESET can also recruit histone deacetylases (17). An MBD that binds DNA, but without preference for methylated DNA, has been found in combination with a number of other domains of nuclear proteins in the mouse TTF-I-interacting protein 5 (TIP5), a protein in the nucleolar remodelling complex (NoRC) involved in silencing of ribosomal gene transcription (19,20).
So far, the information on proteins binding methylated DNA in plants is limited. One such protein (DBPm) has been partially purified from pea and a similar binding activity was shown to be present in several higher plant species (21). In addition, two classes of methylated DNA-binding proteins, dcMBP1 and dcMBP2, have been identified in carrot (22). The amino acid sequences of these proteins are unknown and the genes encoding them have not been identified.
By a bioinformatics approach, we and others (23; ChormBD, http://chromdb.biosci.arizona.edu/) have identified 12 MBD genes in Arabidopsis thaliana. Three of these were recently shown to bind symmetrically methylated CpGs (23). Although the evidence in general is that proteins with MBD are involved in epigenetic control of gene expression, it is important to emphasise that MBDs, defined as amino acid regions with sequence similarity to the MBDs of MeCP2 and MBD1–MBD4, do not necessarily bind methylated DNA. Sequence comparisons suggest that a number of the MBD proteins in Arabidopsis are unlikely to bind methylated CpGs. Therefore we are focusing our work on the biological function of AtMBDs. In the present paper we show that of the 12 putative Arabidopsis AtMBD genes, at least 10 are active in diverse tissues. As a first approach to investigate the biological significance of plant MBD genes, Arabidopsis was transformed with an AtMBD11 RNA interference construct, resulting in plants with pleiotropic developmental alterations.
MATERIALS AND METHODS
Bioinformatics tools
Database searches were performed using BLASTP, TBLASTN and PSI-BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
Protein domains were identified using the programs RPS-BLAST (NCBI) and ProfileScan (http://hits.isb-sib.ch/cgi-bin/PFSCAN) searching the Pfam-A, Prosite profiles and Smart databases (NCBI).
Amino acid sequence alignments were created using ClustalX 1.8 (http://www-igbmc.u-strasbg.fr/BioInfo/ClustalX) with default parameters and manual adjustments from GeneDoc 2.6.001 (http://www.psc.edu/biomed/genedoc/).
RT–PCR expression and rapid amplification of cDNA ends (RACE) analyses
mRNA was isolated from different Arabidopsis tissues using magnetic oligo(dT) beads (GenoPrep™ mRNA beads, Qiagen). First strand cDNA was made by incubation at 42°C for 1 h of bead-bound mRNA in a 30 µl reaction containing 127 µM dNTP mixture, 30 U RNasin (Promega) and 8 U AMV reverse transcriptase (RT) (Promega). The bead-bound first strand cDNA was resuspended in 20 µl of 10 mM Tris–HCl pH 8. Control reactions were run without RT to reveal DNA contamination in the mRNA. Five per cent of the first strand cDNA generated, and a corresponding amount of the control reaction, was used as template in subsequent PCRs using gene-specific primers annealing to different exons.
5′-RACE and 3′-RACE were performed using a Marathon™ cDNA Amplification Kit (Clontech) with 1 µg of poly(A)+ RNA isolated from floral buds as template and Advantage® 2 Polymerace Mix in accordance with the manufacturer’s recommendations. RT–PCR and RACE PCR products were sequenced using a MegaBACE 1000 sequencer.
Sequences of primers used for RT–PCR and RACE analyses are available on request.
Nuclear localisation assay
For onion epidermis nuclear localisation assays (24), a cDNA–GFP fusion construct was made using Gateway cloning technology (Invitrogen). The vector pAVA319 (25) was converted into a Gateway destination vector by ligating a blunt-ended Gateway cassette A into a blunted NcoI site between the translational enhancer sequence and the DNA fragment encoding GFP. The resulting vector was called pAVAGAW-GFP. The pKEx-327 vector (24) was similarly converted to a Gateway vector (pKEGAW-smGFP) by inserting the C.1 Gateway cassette into a blunted SalI site.
The AtMBD11 cDNA was PCR amplified by the proof-reading Platinum® Pfx DNA polymerase (Invitrogen) using the primers 5′-GGGAAAGGAGAGAGATGGGTGGTGAAGA-3′ and 5′-TCCGGTAGCTTCTCCTTCTGCTGT-3′ with additional attB1 and attB2 recombinase attachment sites designed according to the manufacturer’s instructions to ensure in-frame integration of the cDNA into the vectors. The cDNA was first recombined into pDONR201, and thereafter recombined into pAVAGAW-GFP and pKEGAW-smGFP. The resultant constructs pAVAMBD11-GFP and pKEMBD11-GFP were introduced into onion cells by microprojectile bombardment using a Biolistic PDS-1000/He Particle Delivery System (Bio-Rad) (26). As a positive control for nuclear localisation, the cDNA of heterochromatin protein 1 of Drosophila melanogaster (NM_057407) fused to GFP in pKEx-137 was used.
Generation of transgenic Arabidopsis plants
A fragment of 853 bp encompassing the putative promoter sequence of the AtMBD11 gene (pMBD11) was amplified with the primers 5′-GGCATCAAGCTTCCATATATGAAGAGGCTTTGGTG-3′ and 5′-GCTACGGATCCTCTCCTTTCCCTTTAGCAACC-3′, which have HindIII and BamHI restriction endonuclease sites in their 5′ ends, respectively, and cloned into the vector pPZP211G, using the BamHI and HindIII sites upstream of the GUS reporter gene. The pPZP211G vector is a pPZP Agrobacterium binary vector (27) with a spectinomycin bacterial selectable marker. Between the T-DNA right and left borders, pPZP211G carries the nptII gene as a plant selectable marker and a promoterless GUS gene with a nos terminator inserted between the SmaI and EcoRI sites of the polylinker.
A 459 bp fragment of the second exon of the AtMBD11 gene, with no significant similarity to other Arabidopsis genes, was amplified with the primers 5′-ATGCAGAAGGTGAGAAATCA-3′ and 5′-ACAGAAGCAGAGCAGAACAG-3′, with appropriate additional restriction endonuclease recognition sites at the ends, and inserted in inverse orientation in corresponding cloning sites of the pKANNIBAL vector (28). A NotI fragment containing the RNAi construct driven by the 35S promoter was introduced into the pART27 Ti-vector.
The two constructs were transferred to the Agrobacterium tumefaciens strain C58C1 pGV2260 and used to transform Arabidopsis (ecotype C24) by the floral dip method (29). Transformants were selected on MS2 medium with 50 µg/ml kanamycin. PCR with construct-specific primers was used to confirm that kanamycin-resistant plants were true transformants.
GUS assay
Transformants containing the pMBD11-GUS construct were tested for GUS expression according to a standard procedure (30).
Northern analysis
For northern analyses 1.2 µg mRNA, isolated using Genoprep mRNA beads, was loaded in each lane on a denaturing RNA gel, blotted onto Hybond N+ membrane and hybridised as described (31). Single-stranded 32P-labelled antisense probes were prepared as previously described (32). The non-specific adherence of rRNA to magnetic beads during isolation of poly(A)+ RNA allows the use of an Arabidopsis 18S rRNA probe (1803 bp), amplified with the primers 5′-GCAAGTCGTGTGCCAGCAGCC-3′ and 5′-CTTCCGTCAATTCCTTTAAG-3′, as a control for the relative amounts of RNA loaded in different lanes.
RESULTS
Arabidopsis contains 12 putative genes encoding proteins with a methyl-CpG-binding domain
The MBD motif from the human protein MeCP2 (XP_010162.2) was used for a BLASTP search against the Arabidopsis non-redundant sequence database. The MBDs of putative proteins thus identified were used in further BLASTP, TBLASTN and PSI-BLAST searches. Twelve putative proteins containing the MBD motif were found (E-value inclusion threshold <0.001). The corresponding genes encoding these proteins were identified from the annotations in the databases. These genes, AtMBD1–AtMBD12, have also been identified by the ChromDB (http://www.chromdb.org/), as well as by Zemach and Grafi (23).
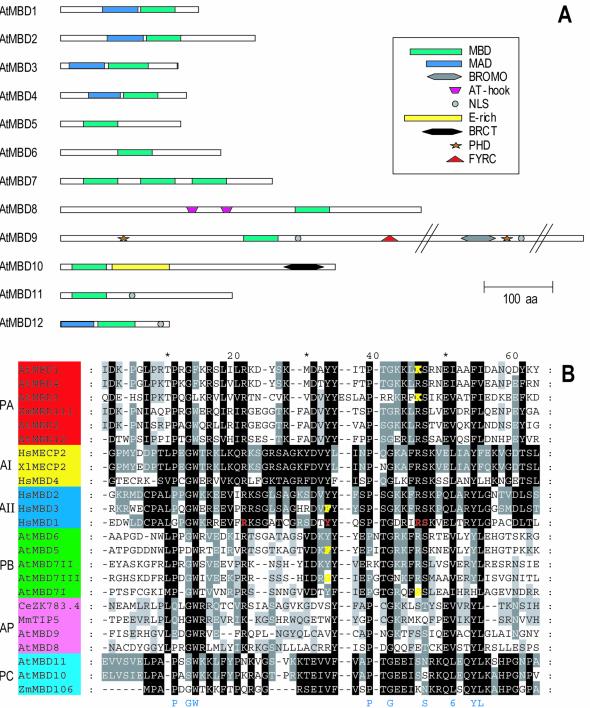
The majority of the predicted proteins are small (155–300 amino acids; Fig. 1A and Table 1). AtMBD9 distinguishes itself from the others, being 10-fold larger (2167 amino acids) than the smallest of the proteins. We identified three MBD motifs in AtMBD7, which dominate the entire protein. Two of these (II and III) were recognised by Zemach and Grafi (23).
Figure 1.
Structure of putative Arabidopsis MBD proteins. (A) Protein sequences obtained from annotations in the EMBL and MIPS databases, adjusted by sequences of RT–PCR and RACE products or cDNAs were analysed for conserved domains (see Materials and Methods). Lengths of proteins and positions of domains are shown to scale except when indicated by //. MBD, methyl-CpG-binding domain; MAD, MBD-associated domain; BROMO, BROMO domain; NLS, nuclear localisation signal; E-rich, region rich in glutamic acid residues; BRCT, BRCA1 C-terminal domain; PHD, PHD finger; FYRC, C-terminal FY-rich domain. (B) Alignment of MBD domains. The degree of conservation is distinguished in different plant and animal subgroups at four levels (100, 80 and 60% and not conserved), where 100% has the darkest shade of grey. Amino acids conserved in at least 17 of the 24 motifs are indicated in blue below the alignment (6, hydrophobic residues). Residues of human MBD1 important for binding methylated CpG are shown in red. Residues of human MBD3 and plant subgroups A and C deviating at these positions are shown in yellow. PA, red, plant subgroup A; PB, green, plant subgroup B; PC, light blue, plant subgroup C; AI, yellow, animal MeCP2 subgroup; AII, dark blue, animal MBD1 subgroup; AP, pink, deviating animal and plant MBDs. Protein accession numbers: HsMBD1, NP_056671; HsMBD2, NP_003918; HsMBD3, NP_003917; HsMBD4, NP_003916; HsMeCP2, P51608; XlMeCP2, AAD03736; MmTIP5, Q91YE5; CmZK783.4, NP_498673. For accession nos of the Arabidopsis proteins see Table 1. For accession nos for the Zm proteins, see text. Hs, Homo sapiens; Xl, Xenopus laevis; At, Arabidopsis thaliana; Zm, Zea mays; Dm, Drosophila melanogaster; Mm, Mus musculus; Ce, Caenorhabditis elegans.
Table 1. Putative genes encoding methyl-CpG-binding domain proteins in A.thaliana.
| Gene name | Protein accession no. | BAC clone | MIPS code (AGI-TIGR code) | No. of introns | Protein (no. of amino acids) | EST cDNA RT |
|---|---|---|---|---|---|---|
| AtMBD1 | CAA16559a | T12H17.130 | (At4g22745) | 2 | 198 | er |
| AtMBD2 | BAB11480 | MVP2.2 | At5g35330 | 4 | 272 | er |
| AtMBD3 | NP_567177 | IG005I10 F5I10 | (At4g00416) | 0 | 163 | |
| AtMBD4 | CAB87748 | T20O10.130 | At3g63030 | 1 | 186 | ec |
| AtMBD5 | CAB62328 | F12A12.100 | At3g46580 | 2 | 182 | er |
| AtMBD6 | BAA97474 | F2O15.40 | At5g59380 | 2 | 225 | er |
| AtMBD7 | BAB08347 | MMN10.2 | At5g59800 | 4/5 | 121/208/306 | r |
| AtMBD8 | AAF87260 | T16E15.7 | At1g22310 | 1/2 | 524/425 | ec |
| AtMBD9 | AAF24616a | T13O15.10 | At3g01460 | 9/10 | 2167/2257 | er |
| AtMBD10 | AAD39661 | F9L1.28 | At1g15340 | 1 | 384 | er |
| AtMBD11 | BAB02310 | MSJ11.19 | At3g15790 | 1 | 254 | ec |
| AtMBD12 | BAB11482 | MVP2.10 | 2 | 155 |
The table lists the EMBL accession numbers of the proteins, BAC or P1 clone annotations, MIPS gene localisation codes (for AtMBD1 and AtMBD3 the AGI-TIGR code). Intron numbers (in cases of alternative splicing in both transcripts), the number of amino acids, RT–PCR and RACE products (r) or cDNAs (c) generated for the present study and the presence of matching cDNA or ESTs (e) in the databases.
aProteins have been modified based on sequencing of RT–PCR and RACE products.
Alignment of the conserved 54–60 residue MBD region of the putative Arabidopsis MBD proteins, in addition to MBD regions from two annotated maize proteins, ZmMBD106 (AAK40308.1) and ZmMBD111 (AAK40310.1), was performed using ClustalX and manual adjustment with GeneDoc (Fig. 1B). MBD motifs from selected known MBD proteins from animals were included for comparison; human MeCP2 and MBD1–MBD4 and Xenopus laevis MeCP2, as well as mouse TIP5 and its Caenorhabditis elegans homologue ZK783.4.
The best conserved residues in the whole set of MBDs are the hydrophobic V, L or I in position 51, in our alignment present in all 24 motifs, and P11, G14W15, P39, G42, S47 and Y54L55, present in at least 17 of 24 motifs (Fig. 1B). With the exception of S47, these conserved residues are not identical to those found to be crucial for binding of the human MBD1 to methylated cysteines in a CpG context (m5CpG) (33).
Plant MBDs constitute distinct subgroups
Compared to the animal MBD1–MBD4 and MeCP2 proteins, the plant MBDs make up distinct subgroups. The ChromDB has divided the proteins into subclasses and this numbering has been adopted by Zemach and Grafi (23). We have, on the other hand, divided the proteins not only based on the sequence similarity within the MBD domain, but in addition on the presence of additional domains not identified by Zemach and Grafi (23; see below).
In subgroup A (groups II and III in 23), comprising the MBDs of AtMBD1–AtMBD4 and AtMBD12 together with ZmMBD111, one third of the amino acids, evenly spaced along the motif, are completely conserved. AtMBD12 and, even more so, AtMBD3 deviate at some positions conserved among the other members. The MBD of AtMBD3 has two additional amino acids in the middle of the motif compared to all other MBDs.
In subgroup B (AtMBD5–AtMBD7) (groups IV and VI in 23), a distinctive feature is the additional amino acid at position 40, the hydrophobic amino acid (Val or Ile) at position 30 and a conserved glutamic acid (E37). Each of the three AtMBD7 motifs deviate at certain otherwise conserved positions.
Subgroup C (group I in 23) is represented by the MBDs of AtMBD10 and AtMBD11 as well ZmMBD106. More than half of the amino acids of the motif are completely conserved.
The MBDs of AtMBD8 and AtMBD9 may be said to be unique. They do, however, share features with TIP5 and ZK783.4, in that they have some common conserved amino acids, e.g. L10, G13W14R15, E50V51 and Y54L55, while at the same time they lack conservation of a number of amino acids well conserved in the other subgroups (Fig. 1B).
Residues crucial for binding of m5CpG are best conserved in subgroups I and II
The solution structure and mutational analysis of the MBD of the human MBD1 protein in complex with methylated DNA (33) have shown the importance of the residues depicted in red in our alignment: Arg, Tyr, Arg and Ser in positions 21, 33, 46 and 47, respectively (Fig. 1B). Substitutions of the Arg residues with either alanine or lysine, Tyr with alanine or phenylalanine or Ser with alanine abolish or reduce DNA binding. Human MBD3, which does not bind m5CpG, has a tyrosine/phenylalanine substitution. In the plant subgroups I and II the crucial residues are well conserved. One should, however, note the phenylalanine (F33) in AtMBD5, the serine (S33) in motif III of AtMBD7, the serine (S46) in motif I of AtMBD7 and the lysine (K46) in AtMBD1 and AtMBD3 (yellow residues, Fig. 1B) changes compared to human MBD1 that indicate a reduced ability of binding of methyl-CpG. However, Zemach and Grafi (23) have shown that AtMBD5 and motif III of AtMBD7 bind methylated CpG.
Members of subgroup III, as well as AtMBD8 and AtMBD9, differ at several important positions, suggesting that these proteins are not capable of binding m5CpG.
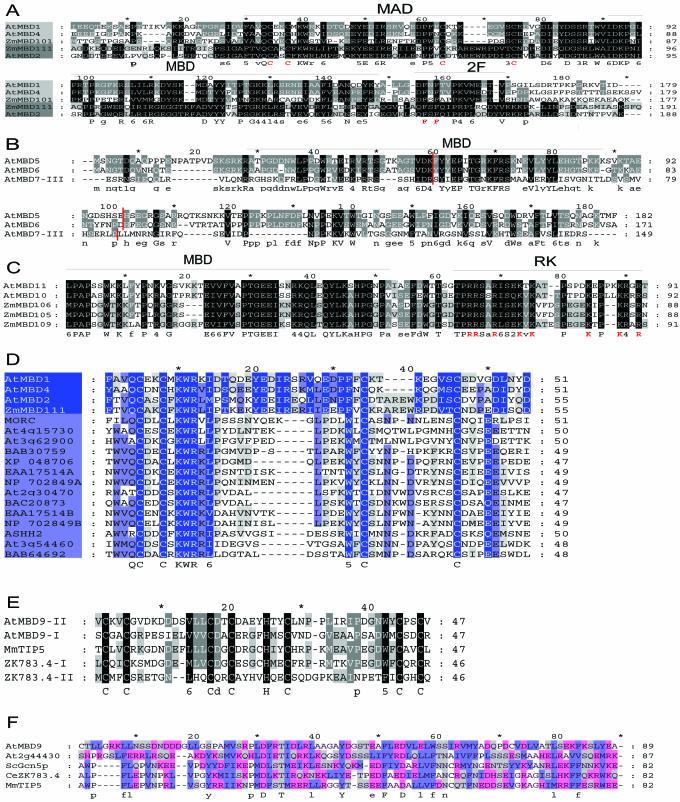
A novel domain was identified in subgroup A
No sequence similarity outside the MBD motif was detected between the AtMBD proteins and the animal MBD1–MBD4 and MeCP2 proteins. However, within each plant MBD subgroup extended similarity was found (Fig. 2A–C).
Figure 2.
Alignments of domains found in AtMBD proteins. (A) The central parts of plant subgroup A proteins. Proteins in subgroup A can be further divided into two subclasses (light and dark grey). AtMBD3 and AtMBD12 have been omitted since we have not been able to confirm that these two putative genes are active (cf. text). The four cysteine residues of MAD and the two phenylalanine residues of the 2F motif found in this subgroup are indicated in red. (B) The C-terminus of plant subgroup B proteins. Note the conserved C-terminal region and the conserved position of introns (red lines). (C) The N-terminus of plant subgroup C proteins. Note the conserved RK-rich motif (in red) next to the MBD. (D) MAD in plant subgroup A (light blue) and in other proteins (grey blue). MORC, NP_034946; EAA17514A and EAA17514B, two MAD motifs of the EAA17514 hypothetical protein of Plasmodium yoelii yoelii; NP_702849A and NP_702849B, two MAD motifs of the NP_702849 hypothetical protein of Plasmodium falciparum 3D7; ASHH2, gene product of Arabidopsis gene At1g77300. (E) PHD fingers. Both AtMBD9 and CeZK783.4 have two PHD fingers (I and II). (F) Putative BROMO domain. ScGcn5p, Saccharomyces cerevisieae histone H3 acetyltransferase Gcn5p; At2g44430, Arabidopsis gene of unknown function. Blue, hydrophobic residues; red, charged polar residues; grey, uncharged polar residues.
The subgroup A proteins are similar over a stretch of 150 amino acids that includes the MBD (Fig. 2A). Just C-terminal to the MBD there is a short motif with two phenylalanine residues (2F, Fig. 2A) and N-terminally there is a region of about 50 amino acids containing four cysteine residues, which we have chosen to name the MBD-associated domain (MAD). MAD in conjunction with MBD is also found in AtMBD3 and AtMBD12 and in two additional putative subgroup A proteins from maize, ZmMBD101 (AAK40305) and ZmMBD108 (AAK40309). While ZmMBD111 and ZmMBD108 are more similar to AtMBD2, ZmMBD101 is most similar to AtMBD1 and AtMBD4 (Fig. 2A).
The MAD sequences of AtMBD1 and AtMBD4 were used in PSI-BLAST searches, resulting in the identification of this motif in proteins from mammals, Plasmodium spp., rice and Arabidopsis (Fig. 2D). MAD is characterised by the consensus sequence Q-C-x2-C-x-K-W-R-x-[LVIM]-x9–17-[FW]-x-C- x7–11-C, i.e. two signatures, each with two conserved cysteine residues, separated by a variable number of residues. The distance between the two signatures is shortest in the non-MBD proteins (Fig. 2D), which mainly are predicted proteins of unknown function, e.g. proteins encoded by the putative Arabidopsis genes At3g62900 and At4g15730 and asparagine-rich proteins from two different Plasmodium spp., each with two copies of MAD. However, the mouse MORC protein is known as a nuclear protein with an N-terminal region similar to the histidine kinase, DNA gyrase B and HSP90 ATPase (HATPase) domain (34). Mutation in MORC causes arrest in spermatogenesis (34). Human homologues of MORC (hMORC, KIAA0136 and KIAA085) (34) as well as related mammalian putative HATPases (BAB30759 and HsXP_ 048706, Fig. 2D) also contain a MAD. A third Arabidopsis gene, ASHH2, encodes a protein with a SET domain (24) and is likely to be involved in chromatin modulation. The At3g54460 protein and its rice homologue BAB64692 are putative helicases, while At2g30470 and BAC20873 from rice encode proteins with a B3 DNA-binding domain found in the VP1/ABI3 family regulatory proteins (35).
The amino acid similarities within each of subgroups B and C extend beyond the MBDs
In MBD subgroup B, almost the entire AtMBD5 (182 amino acids) is similar to AtMBD6 (Fig. 2B), which, however, has 54 additional amino acids N-terminally compared to AtMBD5. AtMBD7 is similar to these two proteins C-terminal to its last MBD domain. The position of the last two introns of the AtMBD7 gene is the same as for the two introns in AtMBD5 and AtMBD6 (Fig. 2B), indicating that all three genes have a common ancestor.
In subgroup C, AtMBD10 and AtMBD11 show high identify to ZmMBD106 and two other putative subgroup C maize MBD proteins, ZmMBD105 (AAK40305) and ZmMBD109 (AAM93219), in a region rich in lysine and arginine adjacent to the MBD (RK motif, Fig. 2C). ZmMBD106 and ZmMBD105 have an overall amino acid identity of 75%, while ZmMBD109 has 52% similarity to ZmMBD106 and 53% to ZmMBD105 (data not shown).
Additional domains known from nuclear proteins are found in some AtMBD proteins
Searches in the conserved domain databases were used to identify other domains in addition to MBD and MAD (Fig. 1A). In subgroup B proteins only the MBD domain was found. A nuclear localisation signal (NLS) was recognised in 3 of the 12 putative proteins, namely AtMBD9, AtMBD11 and AtMBD12. We cannot exclude that other undetected types of NLSs are present in the other proteins.
Just C-terminal to the MBD motif of the AtMBD10 protein, 35% of the amino acids are glutamic acid (amino acids 112–206). At the C-terminal end of this protein the SMART database identified a BRCA1 C-terminal (BRCT) domain (amino acids 242–359, E = 3e–7). This domain, first detected in the breast cancer protein BRCA1, is comprised of several distinct clusters of conserved hydrophobic amino acids (36). It is present in a number of proteins that are involved in various chromosomal events, e.g. DNA repair, transcriptional activation, chromatin remodelling and cell cycle control.
At the centre of the AtMBD8 protein, two adjacent AT-hooks, which mediate protein binding to the minor groove of AT-rich tracts in DNA (37), were found, while in the large AtMBD9 protein several chromatin-associated domains were identified. One PHD finger, a Cys4-His-Cys3 zinc finger-like motif found in a number of nuclear proteins (38), is present in the N-terminal region and another in the middle of the protein (amino acids 86–131, E = 7e–12 and 1289–1335, E = 5e–8, Fig. 2E). A sequence similar to FYRC is found about 700 amino acids from the N-terminus of the protein (amino acids 671–703, E = 0.073). FYRC is often found C-terminal to another FY-rich region (FYRN) in chromatin-associated proteins, e.g. Drosophila trithorax and its mammalian homologues (39). A putative BROMO domain, found in histone acetyltranferases and in the ATPase component of certain nucleosome remodelling complexes (40), was identified in the middle of AtMBD9 (amino acids 1180–1243, E = 0.035, Fig. 2F).
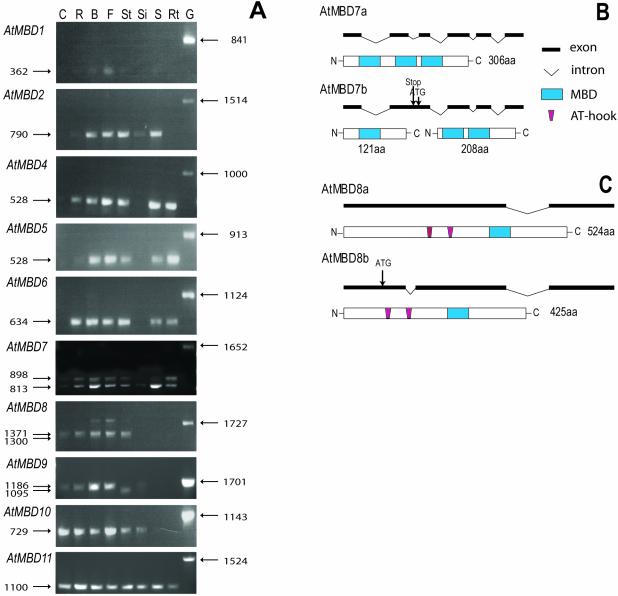
Ten AtMBD genes are active in different tissues and some are alternatively spliced
For 9 of the 12 putative AtMBD genes, matching ESTs were found in the databases (Table 1). Using RT–PCR we verified the expression of these nine genes as well as AtMBD7 (Fig. 3A). We failed to amplify RT–PCR products for two potential genes, AtMBD3 and AtMBD12.
Figure 3.
Expression analyses. (A) Agarose gels stained with ethidium bromide showing cDNA fragments of AtMBD1, AtMBD2, AtMBD4, AtMBD5, AtMBD6, AtMBD7, AtMBD8, AtMBD9, AtMBD10 and AtMBD11 amplified by RT–PCR using gene-specific primers. RT–PCR reactions were performed on mRNA, bound to magnetic beads, isolated from cauline leaves (C), rosette leaves (R), floral buds (B), flowers (F), stems (St), green siliques (Si), mature seeds (S) and roots (Rt). Positive control reactions (genomic DNA, G) are shown for each gene. Note that the RT–PCR fragments are shorter than the corresponding genomic fragments, due to introns. The sizes in base pairs of the RT–PCR and genomic fragments are given to the left and right, respectively. Note RT–PCR products of two sizes for AtMBD7, AtMBD8 and AtMBD9 due to alternative splicing. (B) Alternative splicing of AtMBD7. When the second intron is not removed shorter translational products may result from the AtMBD7b transcript due to a stop codon in that intron (121 amino acids) or the use of an in-frame ATG in the intronic sequence as an alternative start codon (208 amino acids). The AtMBD7a transcript is depicted from its start to its stop codon. (C) Alternative splicing of AtMBD8. When the first intron is spliced out a different ATG start codon must be used to translate the AtMBD8b transcript into a protein (426 amino acids) with AT-hooks and the MBD. The AtMBD8a transcript is depicted from its start to its stop codon.
The RT–PCR analysis showed that the AtMBD genes are differentially expressed (Fig. 3A). AtMBD11 was active in every tissue tested (cauline leaves, rosette leaves, buds, flowers, stems, siliques, mature seeds and roots) and similar amounts of RT–PCR products were generated from each tissue. The other subgroup C gene, AtMBD10, was not amplified from seeds and roots. All 10 transcripts were present in flowers and buds, and AtMBD1 was hardly detectable in any of the other tissues. The other two subgroup A genes, AtMBD2 and AtMBD4, seem absent from cauline leaves and have similar expression pattern in most tissues, one exception being that only AtMBD4 was amplified from roots. The subgroup B genes AtMBD5 and AtMBD6 are expressed in all tissues except siliques and cauline leaves. The relative abundance of the AtMBD6 transcript seems to be higher than that of AtMBD5 in rosette leaves. AtMBD7 of subgroup B was expressed in all tissues, but only weak RT–PCR products were amplified from cauline leaves and siliques. In contrast to the subgroup C genes, AtMBD8 and AtMBD9 RT–PCR products were not amplified from seeds and roots.
The 5′- and 3′-untranslated regions (UTRs) of the active genes were determined by rapid amplification of cDNA ends (RACE). Introns of 122, 345 and 90 bp were found in the leader sequences of AtMBD1, AtMBD2 and AtMBD11, respectively. Our analysis suggests that annotation of the gene T12H17.130 as encoding a protein of 566 amino acids (CAA16559) is incorrect and that this predicted gene is actually two genes, the AtMBD1 gene (At4g22745 in AGI-TIGR) and gene At4g22740 encoding a glycine- and phenylalanine-rich protein.
Alternative splicing was revealed for three genes, AtMBD7, AtMBD8 (Fig. 3B and C) and AtMBD9. In AtMBD9, the 90 bp intron lies downstream of the MBD encoding region. The smallest transcript seems only to be present in stems (Fig. 3A), while the longer one is present in the other tissues tested. The alternatively spliced intronic sequence of AtMBD9 does not contain any stop codon and therefore translates into a putative protein with an additional 30 amino acids.
When an 85 bp alternatively spliced intronic sequence is included in the AtMBD7 transcript (AtMBD7b, Fig. 3B), this may result in a shorter peptide (121 amino acids) due to a stop codon in the middle of the intron. This putative peptide sequence contains only the first of the three MBD domains present in the long variant of AtMBD7 (306 amino acids). Alternatively, an ATG codon present in the unspliced intron may be used as a start codon in the translation of a protein of 208 amino acids encompassing the last two MBDs. RT–PCR analysis shows that both alternatively spliced transcripts (AtMBD7a and AtMBD7b) are present in most of the tissues, but the smallest one (AtMBD7a) seems to be somewhat more abundant, especially in buds and seeds (Fig. 3A).
The 71 bp alternatively spliced intron in AtMBD8 is localised upstream of the regions encoding the AT-hooks. The largest transcript (AtMBD8a) is slightly more dominant than the smaller one (AtMBD8b) in tissues where the gene is expressed (Fig. 3A). The two alternatively spliced transcripts must be translated from different start codons if they are to be translated into two proteins having AT-hooks and MBD, but of different sizes (a difference of 99 amino acids, see Fig. 3C).
AtMBD11 was chosen for functional analysis of Arabidopsis MBD genes
We have approached the question of biological function by initiating a closer study of the in vivo function of the AtMBD11 gene, since the RT–PCR analysis indicated AtMBD11 to be expressed in all tissues (Fig. 3A) and, therefore, it could be anticipated to be of importance in several tissues and developmental stages. Although the MBD domain of AtMBD11 diverges significantly from human MBD1 (Fig. 1B), this may not exclude an important biological function. Nuclear localisation, a prerequisite for involvement in epigenetic gene control, was anticipated for AtMBD11 protein due to its predicted NLS. This was also important for our choice of the AtMBD11 gene for biological investigations.
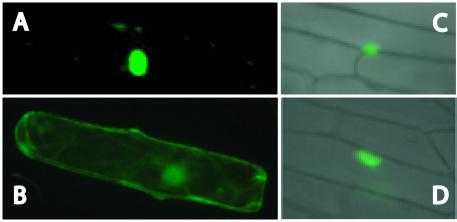
AtMBD11 is localised to the nucleus
The cDNA encoding AtMBD11 was recombined into the transient expression vectors pAVAGAW-GFP and pKEGAW-smGFP (see Materials and Methods), in in-frame fusions with the green fluorescent protein (GFP) gene, resulting in the constructs pAVAMBD11-GFP and pKEMBD11-GFP. The former construct employs the strong constitutive CaMV 35S promoter (25), while the latter has a weaker version of this promoter (24). These constructs were introduced into the inner epidermis of onion cells using particle bombardment. Regardless of promoter strength, the AtMBD11–GFP fusion protein, as well as the positive control, HP1 of D.melanogaster, could be detected in the nucleus (Fig. 4A, C and D). In contrast, the small GFP protein on its own was present in the whole cell (Fig. 4B), as reported previously (25).
Figure 4.
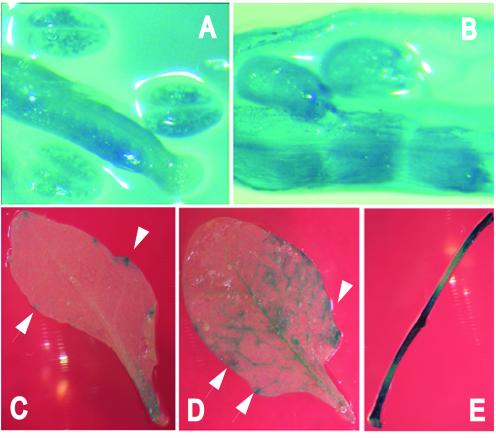
Nuclear localisation assay in onion epidermis transient expression system. Subcellular localisation of: (A) AtMBD11 fused to GFP using the pAVAMBD11-GFP construct; (B) GFP alone using the pAVA319 vector; (C) heterochromatin protein 1 of D.melanogaster fused to GFP in the pKEx-327 vector; (D) AtMBD11 fused to GFP using the pKEMBD11-GFP construct. GFP fluorescence was revealed 2 h after bombardment utilising a Nikon Microphot microscope with epifluorescence and Nomarski optics.
The AtMBD11 promoter directs distinct expression patterns in various organs
To examine the expression pattern of AtMBD11 in more detail, Arabidopsis was transformed with a promoter-AtMBD11 (pMBD11) GUS reporter gene construct. Representative GUS expression patterns are shown in Figure 5. GUS is expressed in the carpel and in the pollen grains of the anther (Fig. 5A), in seeds and in most cases also in siliques (Fig. 5B), as well as in the cauline and rosette leaves around the water excreting glands (hydrathodes) found at the points of the leaf margin to where veins extend (Fig. 5C and D). GUS expression could also be seen in the stem, especially around the nodes (Fig. 5E).
Figure 5.
GUS expression driven by the AtMBD11 promoter in transgenic Arabidopsis plants. Blue colour reflects GUS activity in (A) carpel and anthers, (B) seeds and silique, (C) cauline leaf, (D) rosette leaf and (E) stem. Note the GUS activity at the margin of the leaves around the water excreting glands (arrows).
RNA interference with AtMBD11 expression results in plants with morphological and developmental abnormalities
To investigate the function of AtMBD11 in planta, Arabidopsis was transformed with a RNA interference (RNAi) construct (35S::AtMBD11-RNAi) containing two copies of a fragment near the 3′ end of the AtMBD11 coding region in inverse orientation on each side of an intron for efficient generation of double-stranded RNA (28). Thirty-six transformants were obtained. PCR with construct-specific primers was used to confirm that these primary transformants contained the RNAi construct (data not shown).
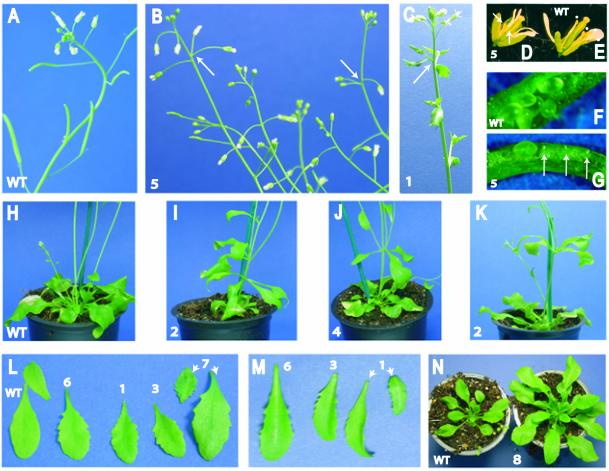
A number of aberrant phenotypic traits were observed (Table 2 and Fig. 6). Wild-type plants or pMBD11::GUS transformants did not show such phenotypes. A third of the RNAi plants displayed late flowering compared to wild-type (Table 2), i.e. they flowered from 20 to 31 days after transfer to soil, with an average of 24.8 ± 4.1 days. Wild-type plants (n = 54), on the other hand, flowered after 16.2 ± 3.2 days. Eleven RNAi plants developed a majority of siliques that were abnormally small. In two of these plants the siliques hardly developed at all (Fig. 6A and B). The infertility was most likely a result of mechanical male sterility (Fig. 6D and E). The flowers had fewer and/or shorter anthers and pollen development was in most cases aborted. The siliques from such plants showed a majority of undeveloped seeds (Fig. 6F and G). Crossings with wild-type pollen led to a higher number of developed seeds (data not shown). Interestingly, the progeny of such a cross to one of these infertile plants did not give any kanamycin-resistant seedlings, indicating that the AtMBD11 gene can also be involved in gametophyte and embryo development.
Table 2. Phenotypes of primary Arabidopsis transformants harbouring the 35S::AtMBD11-RNAi construct.
| Phenotype | No. of plants | Per cent (n = 36) |
|---|---|---|
| Reduced seed production | 11 | 31 |
| Many small siliques | 6 | 17 |
| Serrated leaves | 7 | 19 |
| Late floweringa | 12 | 33 |
| Extra rosettes | 2 | 6 |
| Reduced apical dominance | 3 | 8 |
| Combined phenotypes | 9 | 25 |
| Normal | 8 | 22 |
See text and Figure 6 for further information on phenotypes.
aPlants flowering ≥20 days after transfer to soil.
Figure 6.
Phenotypes of Arabidopsis plants transformed with a 35S::AtMBD11-RNAi construct. Eight different plant lines are depicted, cf. the numbering. (A) Inflorescence of wild-type plant. (B) Inflorescence of transformant with reduced fertility. Note tiny siliques compared to wild-type and abnormal positioning of flowers (arrows). (C) Inflorescence of plant with serrated cauline leaves. (D) Flower of transformant with reduced fertility. Note that only two short anthers are present (arrows). (E) Flower of wild-type plant where four of the six anthers are visible (dots). (F) Wild-type immature silique. (G) Immature silique of transformant with reduced fertility. Note undeveloped seeds (arrows). (H) Wild-type rosette. (I–K) Transformants with aerial rosettes. (L and M) Serrated rosette leaves of transformants. Note lack of serration in wild-type (WT) leaves. (N) Wild-type rosette and transformant with a rosette of smaller leaves inside a rosette of larger leaves. (B), (E), (G), (I), (J) and (L) show primary transformants, while (C), (K), (M) and (N) depict plants of the next generation.
Six additional primary transformants had a mixture of seemingly normal and underdeveloped siliques. The normal siliques were more frequent late in the flowering period. It is possible that this weaker phenotype is due to a weaker interference with the AtMBD11 transcript, either related to the genomic position of the inserted RNAi T-DNA or because of a gradual silencing of the expression of 35S::AtMBD11-RNAi. Three plants were very small with reduced apical dominance in addition to small siliques (not shown). In inflorescences of some plants, an abnormal positioning of the flowers was observed, i.e. more than one flower grew out at the same node on the stem (see arrows in Fig. 6B).
Two plants developed an additional rosette on the first node on the stem (Fig. 6H, compared to I and J). In contrast to wild-type plants, seven of the AtMBD11-RNAi transformants had serrated leaves (Fig. 6L). Some plants had a combination of aberrant phenotypes (Table 2), i.e. five of the plants that flowered later than wild-type had serrated leaves, one plant with an extra rosette also had serrated leaves and small siliques, and two of the tiny plants flowered later than wild-type. Eight of the 36 primary plants did not exhibit any obvious phenotype.
Northern hybridisation was used to investigate the expression level of AtMBD11 and the RNAi construct. The single-stranded antisense probe used recognises both the native transcript and RNAi transcripts. The common pattern seen was a reduced expression level of the endogenous transcript compared to wild-type plants (blue arrow, Fig. 7), while the RNAi-transcript was strongly expressed (red arrow, Fig. 7). One to two weaker additional bands were visible in the mRNA from the RNAi plants (red dots, Fig. 7). These bands may possibly represent RNAi transcripts that are unspliced or have a longer 3′-UTR.
Figure 7.
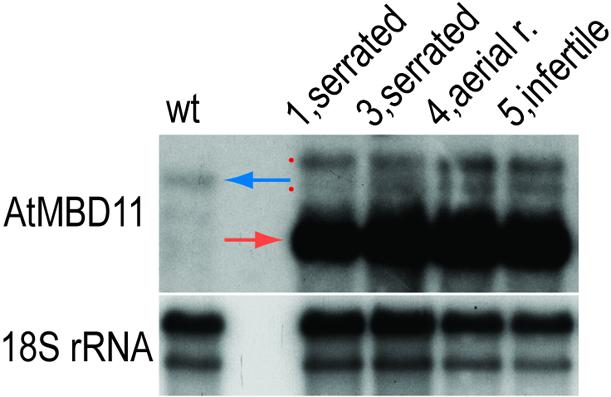
Northern hybridisation using mRNA from rosette leaves of wild-type (wt) and four independent 35S::AtMBD11-RNAi primary transformants, cf. numbering and phenotypes in Figure 6. The same blot was hybridised with a single-stranded antisense AtMBD11 probe (upper panel) and, as a loading control, an 18S rRNA probe (lower panel). Blue arrow, endogenous AtMBD11 transcript; red arrow, major RNAi transcript; red dots, minor RNAi transcripts (see text).
To investigate whether the observed phenotypes were heritable, seeds of six primary transformants were germinated on plates with kanamycin and resistant plants inspected for aberrant phenotypes. Such phenotypes were seen in siblings from five of the six lines. We observed plants with aerial rosettes (Fig. 6K), serrated rosette leaves (Fig. 6M), serrated cauline leaves (Fig. 6C) and with a rosette of 10 large leaves within which another rosette formed with leaves about a third of the length (Fig. 6N). Northern hybridisation confirmed that the RNAi construct was expressed in plants of this generation (data not shown); while Southern hybridisation showed that lines 2, 3 and 8 harboured a single insert of the RNAi construct (data not shown). Single copy lines are less susceptible to transgene silencing and will be used in future extended investigations of the RNAi phenotype.
DISCUSSION
At least 10 active genes encoding putative methyl-CpG-binding proteins are present in Arabidopsis
BLAST screenings of the Arabidopsis genome identified 12 putative genes encoding proteins with a conserved MBD (ChromDB; 23; this study). Our RT–PCR analyses and the presence of matching ESTs verified that 10 of these are active genes. Using northern hybridisation the ChromDB has also documented expression from 7 of the 10 genes we found to be expressed. Our RT–PCR analysis does not allow a rigid quantitative comparison of the expression levels between tissues, but demonstrates that all the active genes are expressed in flowers and buds. In other parts of the plant, the genes appear to be differentially expressed (Fig. 3A). Genes within the same subgroup have similar, but not identical, expression patterns, suggesting that each gene has a specialised function.
Despite the use of several primer pairs and mRNA from eight different tissues, we failed to amplify RT–PCR products of AtMBD3 and AtMBD12, which also do not have matching ESTs. Their expression may be too low to be detected or restricted to tissues and developmental stages not tested here. Alternatively, these genes represent pseudogenes, since they have accumulated mutations in their MBD motifs and thus deviate from the other members of subclass I at several conserved positions (Fig. 1B). In contrast to the other genes, AtMBD3 is without introns, indicating that it may have been generated by a retrotransposition event. AtMBD12 is most similar to AtMBD2 and positioned on the same BAC clone as this gene, suggesting that this pair represents a duplication event.
Plant MBD proteins have evolved separately from the animal proteins
The amino acids in human MBD1 shown to be crucial for efficient interaction with nucleosomal DNA (33) are conserved in the plant subgroups A and B (Fig. 1B). The three members of subgroup B (AtMBD5–AtMBD7) have recently been shown to have methyl-CpG-binding properties (23). Proteins of subgroup A, AtMBD1, AtMBD2 and AtMBD4, on the other hand, did not show any methyl-CpG-binding activity (23). A stretch of amino acids constituting the loop L1 in human MBD1 (residues 22–28 in Fig. 1B) consists of important residues (Lys, Ser, Ala and Thr) involved in hydrogen bonding to phosphate groups on one of the DNA strands, and undergoes major structural rearrangements upon binding of m5CpG (33). AtMBD5 and AtMBD6 have amino acid regions similar to L1 of human MBD1, and this can be suggested to contribute to the m5CpG binding. The L1 region is less similar for the proteins of plant subgroup A, however, also deviating in the MBDs of AtMBD7.
We would like to emphasise that plants have the ability to methylate cysteines not only in CpGs, but also in CpNpG contexts and even in asymmetric contexts (41). In pea, the protein dcMBP1 binds oligonucleotides with symmetric m5CpG and m5CpNpG sequences, but shows a higher specificity for CpG methylation, while dcMBP2 binds methylated cytosines in the CpNpG and CpNpN contexts and does not have strong requirements for strand-symmetric methylation (22). Possibly, some of the AtMBDs are involved in recognition of methylated cytosines in these settings. Others may bind unmethylated DNA, like MBD3 (14) and the TIP5 MBD, which, assisted by AT-hooks, bind the upstream control element of the murine rDNA promoters (20). These examples suggest that AtMBD proteins without general m5CpG- or m5CpNpG-binding properties can be targeted to specific, yet unknown, DNA sequences, guided by additional domains and/or interacting partners.
Most of the AtMBDs have no conspicuous conserved domains in addition to MBD. However, in AtMBD8 two AT-hooks are found. This is also the case for the mammalian MeCP2. AT-hooks are frequently associated with other domains of chromatin- and DNA-binding proteins, functioning as an auxiliary protein motif cooperating with other DNA-binding activities (37). Except for this domain and the MBD, the plant MBD proteins do not have any similarity to MBD1–MBD4 or MeCP2, indicating that specific plant and animal functions of MBD proteins have evolved.
In animals, proteins have recently been discovered with a MBD in combination with a bifurcated SET domain (e.g. ESET) (42). In Arabidopsis, about 30 SET domain proteins have been identified, some of which have split or truncated SET domains, but none of them contain a MBD domain (24). Neither is the plant MBD found in combination with the helix–hairpin–helix DNA glycosylase domain of MBD4 (15) and other DNA repair proteins. The CXXC zinc finger of human MBD1 is not present in any AtMBD protein. One should, however, note the MAD of subgroup I with four conserved cysteine residues (Fig. 2A). MAD might represent a new domain involved in DNA binding or protein–protein interactions, since several of the MAD-containing proteins we have identified (Fig. 2D) are likely nuclear proteins based on their additional domains.
In AtMBD10 a sequence similar to the BRCT domain was found. The BRCT of BRCA1 has been shown to interact with histone deacetylases (HDAC1 and HDAC2) and also RbAb46, which together with RbAp48 are components of histone deacetylase complexes involved in chromatin remodelling (43). RbAb46, RbAp48, HDAC1 and HDAC2 are also core components of the NuRD and SIN3 complexes of mammalian cells, with which MBD proteins are associated (11). One may speculate that AtMBD proteins like AtMBD10 reside in similar complexes. The MBD sequence of AtMBD10 is, however, not likely to bind m5CpG, since its MBD motif deviates more from the human MBDs than the subgroup A proteins. AtMBD6, which binds m5CpG, has recently been found to be associated with histone deacetylase activity in leaf nuclear extracts (23).
AtMBD9 is a very large protein compared to the other Arabidopsis MBDs. PHD fingers, which are thought to be involved in chromatin-mediated transcriptional regulation (38), are found both in the N-terminus and in the middle of the protein. In addition, regions with weaker similarity to FYRC and the BROMO domain were found. Interestingly, the TIP5 protein and its C.elegans homologue ZK783.4 contain MBD, PHD and BROMO domains (see Figs 1B and 2E and F). TIP5 is crucial in rDNA silencing, in that it targets the nucleolar remodelling complex, NoRC, to rDNA promoters and recruits DNA methyltransferase and histone deacetylase activities, thus establishing heterochromatin (19). Although the domain positions are not the same in AtMBD9 and TIP5 and AtMBD9 is lacking other domains (i.e. DDT, WAKZ, BAZ1 and BAZ2) present in TIP5, one may hypothesise that AtMBD9 is the functional equivalent of TIP5.
AtMBD11 is important for normal plant development
As a first approach to explore AtMBD biological function we have investigated the in planta effect of transforming Arabidopsis with a 35S::AtMBD11-RNAi construct. The majority of the primary transformants (78%) showed one or more aberrant phenotypes (Table 2) and a third of the plants flowered significantly later than wild-type plants. There are several lines of evidence for epigenetic control of flowering time. Repression of the FLC gene by cold treatment (vernalisation) accelerates flowering in late flowering ecotypes and a nuclear localised zinc finger protein (VRN2) with similarity to polycomb group proteins is involved in maintenance of the repressed state (44). In vernalisation-responsive Arabidopsis lines, earlier flowering can take place in MET1 antisense (asMET1) plants that have a reduced level of CpG methylation (45). However, hypomethylation, e.g. in a asMET1 or ddm1 mutant background, can also give rise to plants with a delay in flowering (46). This phenotype has been ascribed to ectopic expression of the FWA gene, which is normally silenced due to dense methylation of promoter repeats (47).
Late flowering has also been observed in Arabidopsis plants (CASH lines) with reduced levels of the histone deacetylase AtHD1 (also called HDA19 or AtRPD3A) (48). Immmuno logical data indicate that the maize homologue of HDA19 (zmRPD3) associates with proteins similar to human RbAp46/48 (49). We suggest that epigenetic changes required for normal timing of flowering are associated with histone deacetylation and thus heterochromatinisation and that MBD proteins may be a part of a chromatin remodelling complex. We are presently establishing late flowering RNAi lines to investigate this possibility.
Interestingly, the other 35S::AtMBD11-RNAi phenotypes (serrated leaves, aerial rosettes and reduced infertility) found in 56% of the primary transformants have also been observed in CASH lines (48). In those lines, 55% of the primary transformants showed abnormal phenotypes other than late flowering. Early senescence and homeotic transformations were observed in some CASH plants, but not in the 35S::AtMBD11-RNAi plants, suggesting that HDA19 is involved in additional epigenetic regulatory pathways. In the 35S::AtMBD11-RNAi plants, phenotypic changes were seen in rosette and cauline leaves, the first node on the stem, anthers and developing seeds (Fig. 6). AtMBD11 promoter-driven GUS expression in the same organs (Fig. 5) supports the notion that the observed phenotypes are the result of RNA interference with normal AtMBD11 expression. In particular, we will draw attention to the GUS activity around the hydrathodes of rosette and cauline leaves (arrows, Fig. 5C and D). Serration in leaves is exactly associated with the hydrathodes (50). Thus, it can be suggested that a normal function of AtMBD11 is to suppress gene activity leading to serration.
In Arabidopsis, some mutants with serrated leaves have been described. Weak alleles of ARGONAUTE1 show serrated leaves and a 1–2 week delay in the initiation of flowering, as compared to wild-type. These alleles, as well as other ago1 alleles, are deficient in post-transcriptional gene silencing and show a decrease in transgene methylation (51). fasciata1 (fas1) mutants display a pleiotropic phenotype with serrated leaves, fertility problems and defects in maintenance of meristem organisation and function. FAS1 encodes the largest subunit of chromatin assembly factor I, which directs assembly of chromatin onto newly replicated DNA (52). serrate (se) mutants have, in addition to serrated leaves, increased numbers of hydrathodes, changes in positions of leaves, abnormal clusters of flowers and up to 12 aerial rosettes (50,53). While the AtMBD11 promoter can drive expression around the hydrathodes, it is not known whether that is the case for SE, FAS1 or AGO1. SE, encoding a zinc finger protein, has been suggested to coordinate the repression of multiple genes during development, by modifying the chromatin structure surrounding these genes (53), and to be required for maintenance of integrity of the shoot apical meristem (50). The aerial rosettes and abnormal positioning of flowers in AtMBD11 RNAi plants (Fig. 6B, I, J and K) may similarly arise as the result of defective inflorescence meristems.
What are the functions of AtMBD proteins?
MBD proteins of animals have been shown to be involved in transcriptional repression, mismatch repair in methylated DNA and silencing of rRNA genes, and mutation in MBD genes affects normal development (9,13,15,19). Some, but not all, bind methylated CpGs.
Our bioinformatics analysis indicates that plants have evolved their own sets of MBD proteins that can be divided into several distinct subgroups. Differential expression and alternative splicing suggest diverse roles of these proteins during plant growth and morphogenesis. Differential biological functions can also be anticipated, since AtMBDs differ in their m5CpG-binding activity (23). Sequence comparisons show that the MBD motif in AtMBD11 deviates from those shown to bind to m5CpG. However, interference with normal AtMBD11 expression shows, for the first time, the important role of a MBD protein in plant development. The pleiotropic effects seen in 35S::AtMBD11-RNAi transformants overlap with phenotypes resulting from lowered expression of genes thought to be involved in epigenetic control of gene expression. Although circumstantial, this suggests that plant MBD proteins may reside in chromatin remodelling complexes, as do their animal counterparts.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Albrecht von Arnim (University of Tennessee, TN) for the pAVA319 vector, Prof. Gunter Reuter (Martin Luther University, Halle, Germany) for the pKEx-327 vector and the pKE-HP1 construct, Dr Wuyi Wang (University of Wisconsin, Madison, WI) for the pZPZ211G vector and CSIRO Plant Industry (Canberra, Australia) for the pKANNIBAL vector. We thank P.E. Grini for critical and constructive comments and suggestions on the manuscript and S.H. Engebretsen, R. Falleth and K.E. Rakkestad for technical assistance at the Division of Cell and Molecular Biology, University of Oslo. This work was supported by grant no. 129525/420 from the Research Council of Norway.
REFERENCES
- 1.Meehan R.R. (2003) DNA methylation in animal development. Semin. Cell Dev. Biol., 14, 53–65. [DOI] [PubMed] [Google Scholar]
- 2.Finnegan E.J., Peacock,W.J. and Dennis,E.S. (2000) DNA methylation, a key regulator of plant development and other processes. Curr. Opin. Genet. Dev., 10, 217–223. [DOI] [PubMed] [Google Scholar]
- 3.Yoder J.A., Walsh,C.P. and Bestor,T.H. (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends Genet., 13, 335–340. [DOI] [PubMed] [Google Scholar]
- 4.Sano H., Kamada,I., Youssefian,S., Katsumi,M. and Wabiko,H. (1990) A single treatment of rice seedlings with 5-azacytidine induces heritable dwarfism and undermethylation of genomic DNA. Mol. Gen. Genet., 220, 441–447. [Google Scholar]
- 5.Jeddeloh J.A., Stokes,T.L. and Richards,E.J. (1999) Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nature Genet., 22, 94–97. [DOI] [PubMed] [Google Scholar]
- 6.Finnegan E.J., Peacock,W.J. and Dennis,E.S. (1996) Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl Acad. Sci. USA, 93, 8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ronemus M.J., Galbiati,M., Ticknor,C., Chen,J. and Dellaporta,S.L. (1996) Demethylation-induced developmental pleiotropy in Arabidopsis. Science, 273, 654–657. [DOI] [PubMed] [Google Scholar]
- 8.Kakutani T., Jeddeloh,J.A., Flowers,S.K., Munakata,K. and Richards,E.J. (1996) Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc. Natl Acad. Sci. USA, 93, 12406–12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballestar E. and Wolffe,A.P. (2001) Methyl-CpG-binding proteins. Targeting specific gene repression. Eur. J. Biochem., 268, 1–6. [DOI] [PubMed] [Google Scholar]
- 10.Hendrich B. and Bird,A. (1998) Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol., 18, 6538–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahringer J. (2000) NuRD and SIN3; histone deacetylase complexes in development. Trends Genet., 16, 351–356. [DOI] [PubMed] [Google Scholar]
- 12.Ng H.H., Jeppesen,P. and Bird,A. (2000) Active repression of methylated genes by the chromosomal protein MBD1. Mol. Cell. Biol., 20, 1394–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wade P.A. (2001) Methyl CpG-binding proteins and transcriptional repression. Bioessays, 23, 1131–1137. [DOI] [PubMed] [Google Scholar]
- 14.Saito M. and Ishikawa,F. (2002) The mCpG-binding domain of human MBD3 does not bind to mCpG but interacts with NuRD/Mi2 components HDAC1 and MTA2. J. Biol. Chem., 277, 35434–35439. [DOI] [PubMed] [Google Scholar]
- 15.Wu P., Qiu,C., Sohail,A., Zhang,X., Bhagwat,A.S. and Cheng,X. (2003) Mismatch repair in methylated DNA. Structure and activity of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4. J. Biol. Chem., 278, 5285–5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuks F., Hurd,P.J., Wolf,D., Nan,X., Bird,A.P. and Kouzarides,T. (2003) The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem., 278, 4035–4040. [DOI] [PubMed] [Google Scholar]
- 17.Yang L., Mei,Q., Zielinska-Kwiatkowska,A., Matsui,Y., Blackburn,M.L., Benedetti,D., Krumm,A.A., Taborsky,G.J.,Jr and Chansky,H.A. (2003) An ERG (ets-related gene)-associated histone methyltransferase interacts with histone deacetylases 1/2 and transcription co-repressors mSin3A/B. Biochem. J., 369, 651–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y. and Reinberg,D. (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev., 15, 2343–2360. [DOI] [PubMed] [Google Scholar]
- 19.Santoro R., Li,J. and Grummt,I. (2002) The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nature Genet., 32, 393–396. [DOI] [PubMed] [Google Scholar]
- 20.Strohner R., Nemeth,A., Jansa,P., Hofmann-Rohrer,U., Santoro,R., Langst,G. and Grummt,I. (2001) NoRC—a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J., 20, 4892–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ehrlich K.C. (1993) Characterization of DBPm, a plant protein that binds to DNA containing 5-methylcytosine. Biochim. Biophys. Acta, 1172, 108–116. [DOI] [PubMed] [Google Scholar]
- 22.Pitto L., Cernilogar,F., Evangelista,M., Lombardi,L., Miarelli,C. and Rocchi,P. (2000) Characterization of carrot nuclear proteins that exhibit specific binding affinity towards conventional and non-conventional DNA methylation. Plant Mol. Biol., 44, 659–673. [DOI] [PubMed] [Google Scholar]
- 23.Zemach A. and Grafi,G. (2003) Characterization of Arabidopsis thaliana methyl-CpG-binding domain (MBD) proteins. Plant J., 34, 565–572. [DOI] [PubMed] [Google Scholar]
- 24.Baumbusch L.O., Thorstensen,T., Krauss,V., Fischer,A., Naumann,K., Assalkhou,R., Schulz,I., Reuter,G. and Aalen,R.B. (2001) The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res., 29, 4319–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Arnim A.G., Deng,X.W. and Stacey,M.G. (1998) Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene, 221, 35–43. [DOI] [PubMed] [Google Scholar]
- 26.Stacy R.A.P., Munthe,E., Steinum,T., Sharma,B. and Aalaen,R.B. (1996) A peroxiredoxin antioxidant is encoded by a dormancy-related gene, Per1, expressed during late development in the aleurone and embryo of barley grains. Plant Mol. Biol., 31, 1205–1216. [DOI] [PubMed] [Google Scholar]
- 27.Hajdukiewicz P., Svab,Z. and Maliga,P. (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol., 25, 989–994. [DOI] [PubMed] [Google Scholar]
- 28.Wesley S.V., Helliwell,C.A., Smith,N.A., Wang,M.B., Rouse,D.T., Liu,Q., Gooding,P.S., Singh,S.P., Abbott,D., Stoutjesdijk,P.A. et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J., 27, 581–590. [DOI] [PubMed] [Google Scholar]
- 29.Clough S.J. and Bent,A.F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J., 16, 735–743. [DOI] [PubMed] [Google Scholar]
- 30.Jefferson R.A., Kavanagh,T.A. and Bevan,M.W. (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J., 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galau G.A., Hughes,D.W. and Dure,I.L. (1986) Abscisic acid induction of cloned cotton late embryogenesis-abundant (Lea) mRNAs. Plant Mol. Biol., 7, 155–170. [DOI] [PubMed] [Google Scholar]
- 32.Espelund M., Stacy,R.A. and Jakobsen,K.S. (1990) A simple method for generating single-stranded DNA probes labeled to high activities. Nucleic Acids Res., 18, 6157–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohki I., Shimotake,N., Fujita,N., Jee,J.-G., Ikegami,T., Nakao,M. and Shirakawa,M. (2001) Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell, 105, 487–497. [DOI] [PubMed] [Google Scholar]
- 34.Inoue N., Hess,K.D., Moreadith,R.W., Richardson,L.L., Handel,M.A., Watson,M.L. and Zinn,A.R. (1999) New gene family defined by MORC, a nuclear protein required for mouse spermatogenesis. Hum. Mol. Genet., 8, 1201–1207. [DOI] [PubMed] [Google Scholar]
- 35.Ezcurra I., Wycliffe,P., Nehlin,L., Ellerstrom,M. and Rask,L. (2000) Transactivation of the Brassica napus napin promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box. Plant J., 24, 57–66. [DOI] [PubMed] [Google Scholar]
- 36.Huyton T., Bates,P.A., Zhang,X., Sternberg,M.J. and Freemont,P.S. (2000) The BRCA1 C-terminal domain: structure and function. Mutat. Res., 460, 319–332. [DOI] [PubMed] [Google Scholar]
- 37.Aravind L. and Landsman,D. (1998) AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res., 26, 4413–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aasland R., Gibson,T.J. and Stewart,A.F. (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci., 20, 56–59. [DOI] [PubMed] [Google Scholar]
- 39.Hsieh J.J., Ernst,P., Erdjument-Bromage,H., Tempst,P. and Korsmeyer,S.J. (2003) Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol. Cell. Biol., 23, 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Owen D.J., Ornaghi,P., Yang,J.C., Lowe,N., Evans,P.R., Ballario,P., Neuhaus,D., Filetici,P. and Travers,A.A. (2000) The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase Gcn5p. EMBO J., 19, 6141–6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gruenbaum Y., Naveh-Many,T., Cedar,H. and Razin,A. (1981) Sequence specificity of methylation in higher plant DNA. Nature, 292, 860–862. [DOI] [PubMed] [Google Scholar]
- 42.Yang L., Xia,L., Wu,D.Y., Wang,H., Chansky,H.A., Schubach,W.H., Hickstein,D.D. and Zhang,Y. (2002) Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene, 21, 148–152. [DOI] [PubMed] [Google Scholar]
- 43.Yarden R.I. and Brody,L.C. (1999) BRCA1 interacts with components of the histone deacetylase complex. Proc. Natl Acad. Sci. USA, 96, 4983–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gendall A.R., Levy,Y.Y., Wilson,A. and Dean,C. (2001) The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell, 107, 525–535. [DOI] [PubMed] [Google Scholar]
- 45.Finnegan E.J., Genger,R.K., Kovac,K., Peacock,W.J. and Dennis,E.S. (1998) DNA methylation and the promotion of flowering by vernalization. Proc. Natl Acad. Sci. USA, 95, 5824–5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Genger R.K., Peacock,W.J., Dennis,E.S. and Finnegan,E.J. (2003) Opposing effects of reduced DNA methylation on flowering time in Arabidopsis thaliana. Planta, 216, 461–466. [DOI] [PubMed] [Google Scholar]
- 47.Soppe W.J., Jacobsen,S.E., Alonso-Blanco,C., Jackson,J.P., Kakutani,T., Koornneef,M. and Peeters,A.J. (2000) The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol. Cell, 6, 791–802. [DOI] [PubMed] [Google Scholar]
- 48.Tian L. and Chen,Z.J. (2001) Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl Acad. Sci. USA, 98, 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lechner T., Lusser,A., Pipal,A., Brosch,G., Loidl,A., Goralik-Schramel,M., Sendra,R., Wegener,S., Walton,J.D. and Loidl,P. (2000) RPD3-type histone deacetylases in maize embryos. Biochemistry, 39, 1683–1692. [DOI] [PubMed] [Google Scholar]
- 50.Clarke J.H., Tack,D., Findlay,K., Van Montagu,M. and Van Lijsebettens,M. (1999) The SERRATE locus controls the formation of the early juvenile leaves and phase length in Arabidopsis. Plant J., 20, 493–501. [DOI] [PubMed] [Google Scholar]
- 51.Fagard M., Boutet,S., Morel,J.B., Bellini,C. and Vaucheret,H. (2000) AGO1, QDE-2 and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi and RNA interference in animals. Proc. Natl Acad. Sci. USA, 97, 11650–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaya H., Shibahara,K.I., Taoka,K.I., Iwabuchi,M., Stillman,B. and Araki,T. (2001) FASCIATA genes for chromatin assembly factor-1 in arabidopsis maintain the cellular organization of apical meristems. Cell, 104, 131–142. [DOI] [PubMed] [Google Scholar]
- 53.Prigge M.J. and Wagner,D.R. (2001) The Arabidopsis serrate gene encodes a zinc-finger protein required for normal shoot development. Plant Cell, 13, 1263–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]