Abstract
The Pms2 gene has been implicated in hereditary colon cancer and is one of several mammalian homologs of the Escherichia coli mutL DNA mismatch repair gene. To determine the effect of Pms2 inactivation on genomic integrity in vivo, hybrid transgenic mice were constructed that carry targeted disruptions at the Pms2 loci along with a chromosomally integrated mutation reporter gene. In the absence of any mutagenic treatment, mice nullizygous for Pms2 showed a 100-fold elevation in mutation frequency in all tissues examined compared with both wild-type and heterozygous litter mates. The mutation pattern in the nullizygotes was notable for frequent 1-bp deletions and insertions within mononucleotide repeat sequences, consistent with an essential role for PMS2 in the repair of replication slippage errors. Further, the results demonstrate that high rates of mutagenesis in multiple tissues are compatible with normal development and life and are not necessarily associated with accelerated aging. Also, the finding of genetic instability in all tissues tested contrasts with the limited tissue distribution of cancers in the animals, raising important questions regarding the role of mutagenesis in carcinogenesis.
Keywords: transgenic mice, supF, genetic instability, cancer
Hereditary nonpolyposis colorectal cancer (HNPCC) has been linked to germ-line mutations in the human homologs of Escherichia coli and yeast DNA mismatch repair genes (1–3). Recent work has revealed that mammalian cells contain multiple homologs of the E. coli MutS and MutL proteins, including MSH2, MSH3/DUG1/MRP1, and GTBP/MSH6 (1, 2, 4–6) and MLH1, PMS2, and PMS1 (1, 2, 7–9), respectively. Biochemical studies with mammalian or yeast cell extracts or partially purified proteins have shown that the MutS homologs can associate in heterodimers in at least two combinations (MSH2 with MSH3 and MSH2 with GTBP/MSH6), each with different activities and mismatch substrate specificities, suggesting both redundancy and divergence of function (1, 10, 11). Studies with the MutL homologs suggest that MLH1 and PMS2 form a functional heterodimer (12–14); other possible MutL complexes have yet to be characterized.
In addition, evidence is emerging that the mismatch repair gene homologs may participate in various other cellular functions, such as transcription-coupled repair (15), recombination (16), and even cell cycle regulation (17). Consequently, a deficiency in one of these homologs may disrupt genome stability in multiple ways.
Previous attempts to evaluate the specific in vivo role of each of the mammalian mismatch repair homologs have been based for the most part on comparisons of changes at microsatellite loci. However, these assays report only expansion or contraction of simple repeated sequences, representing just one specific subset of all possible genetic changes. Forward mutation assays, in contrast, are capable of detecting a wide range of mutations. Such assays have been carried out with the selectable HPRT locus in colon cancer-derived cell lines, some of which are deficient in DNA mismatch repair. These studies have revealed some significant differences among the mutation patterns in different lines (18, 19). However, such tumor cell lines are likely to contain multiple genetic abnormalities besides mismatch repair gene defects, and some of these might affect DNA metabolism, complicating interpretation of the experiments.
Hence, the full impact of Pms2 inactivation on genomic integrity in vivo has not yet been elucidated. Herein we report the construction and analysis of hybrid transgenic mice carrying targeted disruptions at the Pms2 loci along with a chromosomally integrated mutation reporter gene contained within a recoverable λ phage shuttle vector. Data are presented showing that mice nullizygous for Pms2 have high levels of spontaneous mutagenesis in multiple tissues, even those not associated with an increased risk of cancer. In addition, Pms2 inactivation is shown to predispose to deletions and insertions, with a significantly lesser impact on the occurrence of base substitutions.
MATERIALS AND METHODS
Transgenic Mice.
Construction of the Pms2-deficient mice (using D3 embryonic stem cells of 129/Sv mouse origin) has been described (20). The 3340 supFG1 mice were produced as described for the 1139 mice (21) except that the supFG1 gene was substituted for supF in the λ vector. Both were derived from the C57BL/6 mouse background. Construction of the supFG1 gene has been described (22). The Pms2 genotype of the mice was determined by the presence or absence of the targeted insertion by PCR amplification of a region in the Pms2 gene as described (20). The presence of the supFG1 or supF gene in the mice was also confirmed by PCR amplification, as described (21) .
λ Shuttle Vector Rescue and Analysis.
High molecular weight DNA was prepared from selected mouse tissues as described (21). λ vector rescue from the mouse DNA was carried out with λ in vitro packaging extracts (21, 23, 24). Packaging extracts were made as described (23), except that a new E. coli lysogen, NM759 [E. coli K12 recA56 Δ(mcrA) e14° Δ(mrr-hsd-mcr) (λimm434 cIts b2 red3 Dam15 Sam7)/λ] was used instead of BHB2690 for the preparation of the sonicate extract (24). This lysogen produces extracts that are deficient in methyl-directed restriction activity that would otherwise degrade DNA methylated in the mammalian pattern and reduce the yield of rescued phage (24, 25). The mouse DNA was incubated in the λ in vitro packaging extracts at a concentration of 0.05 μg/μl for 2 h at 37°C. The packaged phage were diluted in 10 mM Tris·HCl, pH 8.0/5 mM MgCl2, adsorbed to PG901 [E. coli C1a lacZ125 (am)], and plated in 0.6% top agar on LB plates in the presence of 5-bromo-4-chloro-3-indolyl β-d-galactoside (1.6 mg/ml) and isopropyl β-d-thiogalactoside (1.3 mg/ml), as described (23). Phage with functional supF genes suppress the nonsense mutation in the host bacteria β-galactosidase gene, allowing synthesis of active enzyme capable of metabolizing 5-bromo-4-chloro-3-indolyl β-d-galactoside, thereby yielding blue plaques. Phage with inactivating supF mutations produce colorless plaques.
Mutation Sequencing.
After plaque purification, PCR amplification of the supF gene sequences was carried out with the GeneAmp kit (Perkin–Elmer/Cetus). Sequence analysis of the PCR products was performed as described (26).
RESULTS
Construction of Hybrid Transgenic Mice.
To elucidate in a well-defined system the in vivo role of the MutL homologs, mice deficient for Pms2 were constructed by gene targeting in embryonic stem cells (20). Initial studies of the Pms2 nullizygotes revealed increased microsatellite mutation, sterility in males, and the development of lymphomas and sarcomas within the first year of age (20). To study genetic instability in multiple tissues from these mice, we designed an in vivo forward mutation assay. The Pms2-deficient mice were bred with transgenic mice carrying the supF tRNA suppressor gene as a mutation reporter gene within a chromosomally integrated recoverable λ phage shuttle vector (21, 23). By using λ in vitro packaging extracts, the vector DNA can be identified, excised, and packaged from within the mouse DNA into viable λ particles for analysis in bacteria of supF mutations that occurred in the animals (23, 24). These supF mice therefore provide an in vivo mutation reporter system that can detect base substitutions, as well as deletions and insertions. Consequently, this system for reporting mutations is more sensitive and complete than the standard PCR-based assay of microsatellite stability.
To enhance the sensitivity of the assay even further, we used two independent lines of supF mice, one (1139) carrying the standard supF gene (21) and another (3340) with a supF gene modified to contain extended simple sequence repeats (supFG1) (22). Male supF-positive mice were bred with Pms2 heterozygous females to construct hybrid transgenic mice. The F1 mice were crossed to produce F2 mice that were wild type, heterozygous, or nullizygous at the Pms2 loci and that also carried the supF transgene.
Analysis of Spontaneous Mutagenesis in Vivo.
Three independent sets of 3340/Pms2 male litter mates were identified for analysis at 12 weeks of age. The mice were maintained under standard animal husbandry conditions in the same cage. There was no known exposure to any mutagenic or genotoxic agent. DNA was prepared from several different tissues of the mice and used for vector rescue (via in vitro packaging) and reporter gene analysis (Table 1).
Table 1.
Spontaneous mutagenesis of the supFG1 transgene in Pms2-deficient 3340 mice
| Mouse/tissue | Mutation frequency (×10−5) | No. mutants/ total no. |
|---|---|---|
| Wild type | ||
| Mouse F2-17 | ||
| Skin | ≤2 | 0/72,990 |
| Liver | 4 | 6/170,260 |
| Mouse F3-9 | ||
| Skin | ≤2 | 0/72,240 |
| Colon | 2 | 1/45,620 |
| Total | 2 | 7/361,110 |
| Heterozygous | ||
| Mouse F2-18 | ||
| Skin | 5 | 5/92,730 |
| Liver | 4 | 5/131,500 |
| Spleen | ≤2 | 0/75,630 |
| Colon | ≤2 | 0/78,620 |
| Mouse F2-4 | ||
| Skin | 1 | 1/87,690 |
| Mouse F3-13 | ||
| Skin | 2 | 1/51,600 |
| Total | 2 | 12/517,760 |
| Nullisygous | ||
| Mouse F2-19 | ||
| Skin | 237 | 302/127,700 |
| Liver | 132 | 55/41,700 |
| Spleen | 226 | 33/14,620 |
| Colon | 286 | 201/70,360 |
| Brain | 270 | 53/19,610 |
| Lung | 206 | 35/16,980 |
| Mouse F2-29 | ||
| Skin | 177 | 27/15,270 |
| Spleen | 182 | 27/14,850 |
| Colon | 187 | 37/19,780 |
| Mouse F3-11 | ||
| Skin | 173 | 58/33,590 |
| Liver | 118 | 54/45,650 |
| Total | 210 | 882/420,110 |
The frequency of mutations in the wild-type mice was in the range of 1 to 2 × 10−5, consistent with previous observations of baseline mutation frequencies in such transgenic animal systems (21, 25, 27). However, the nullizygote samples consistently showed 100-fold or more elevations in mutation frequencies in all tissues and mice tested (Table 1). Significant tissue to tissue differences were not detected, with the largest difference being 2-fold (between liver and colon in F2–19). However, colon in F2–29 showed a slightly lower frequency than in F2–19, and there were similar slight fluctuations in the data from the skin samples from the three nullizygous mice. Analysis of the heterozygotes revealed mutation frequencies similar to those found in the wild-type animals.
In the 1139/Pms2 hybrids, the nullizygotes showed only a 5-fold overall increase in mutation frequency (Table 2). However, the 1139 mice were previously found to have an unusually high frequency of spontaneous deletions due to locus-specific effects at the transgene integration site (21). When only supF point mutations are considered, the nullizygotes again show a substantial elevation in mutation frequency of approximately 25-fold relative to the wild-type mice (Table 2).
Table 2.
Spontaneous mutagenesis of the supF transgene in Pms2-deficient 1139 mice
| Mouse/tissue | Mutation frequency (×10−5) | No. mutants/total no. | % point mutations | % deletions | Frequency of point mutations (×10−5) |
|---|---|---|---|---|---|
| Wild type | |||||
| Skin | 39 | 199/511,950 | |||
| Liver | 29 | 47/199,010 | |||
| Spleen | 35 | 82/232,580 | |||
| Total | 35 | 328/943,540 | 6 | 94 | 2 |
| Nullizygous | |||||
| Skin | 117 | 163/138,910 | |||
| Liver | 139 | 165/118,860 | |||
| Spleen | 154 | 151/98,360 | |||
| Total | 135 | 479/356,120 | 42 | 58 | 57 |
Mutation Patterns.
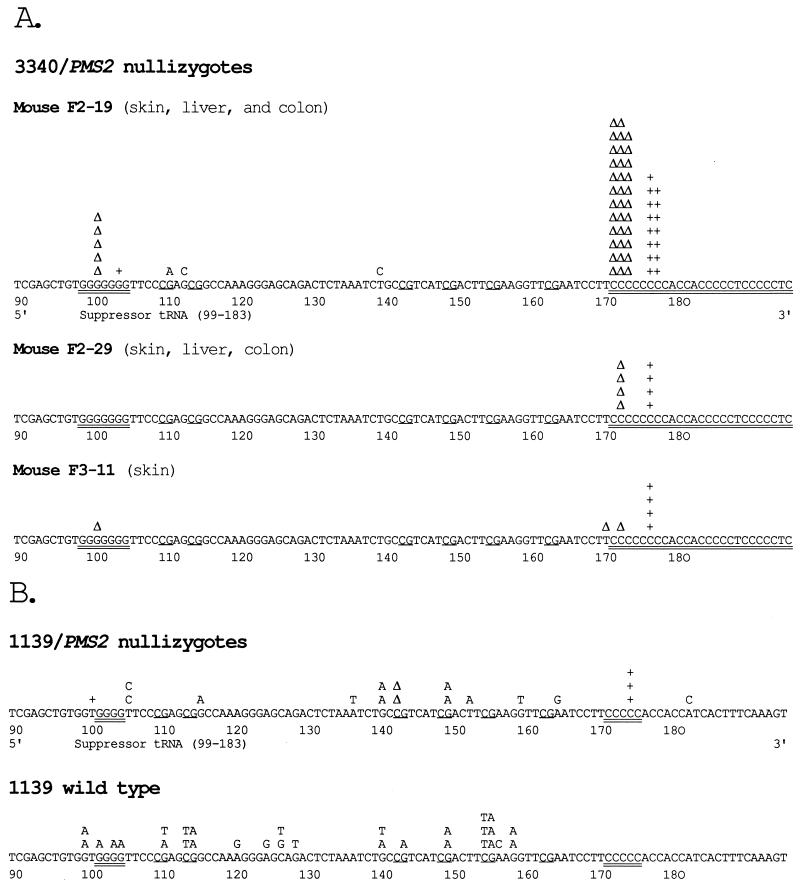
The mutations arising in the nullizygous mice were analyzed at the sequence level (Fig. 1 and Table 3). Almost all of the mutations in the 3340/Pms2 nullizygotes (71 out of 74) were single-base pair deletions or insertions within the G·C base pair repeats at positions 99–105 and 172–179. The eight G·C base pair run at positions 172–179 is a particular hot spot. Note that this stretch constitutes a subset of a longer G·C-rich region that extends beyond the end of the tRNA gene at position 183. This pattern was seen in multiple tissues in three different nullizygous mice from three different litters (Fig. 1A).
Figure 1.
Sequences of reporter gene mutations in Pms2 nullizygous mice. (A) Mutations in the supFG1 gene in 3340/Pms2 nullizygotes. Mutations from three different mice from independent litters are shown, with the tissues of origin indicated. Base substitutions are listed above the original sequence, and single-base pair deletions or insertions are indicated by Δ or +, respectively, above the corresponding site. Mononucleotide repeat sequences are highlighted by double underlining. CpG sequences, potential sites of cytosine methylation, are noted by a single underline. (B) Mutations in the supF gene in 1139/Pms2 nullizygotes and in wild-type 1139 mice, as indicated.
Table 3.
Mutations in Pms2 nullizygous mice
| Mutation | 3340/PMS2 nullizygotes, no. | 1139/PMS2 nullizygotes, no. | 1139 wild-type mice, no. |
|---|---|---|---|
| C·G → T·A | 1 | 5 | 23 |
| T·A → C·G | 1 | 3 | 2 |
| C·G → A·T | 0 | 1 | 3 |
| C·G → G·C | 1 | 0 | 1 |
| T·A → A·T | 0 | 2 | 0 |
| T·A → G·C | 0 | 1 | 1 |
| C·G → T·A at CpG | 1 | 2 | 14 |
| +1 insertion | 24 | 4 | 0 |
| −1 deletion | 47 | 2 | 0 |
| Total | 74 | 18 | 30 |
In the 1139 mice, single-base pair insertions and deletions were less prominent (6 of 18 mutations) but were still more frequent than in the wild-type mice (0 of 30 mutations) (Fig. 1B and Table 3). The difference in mutation pattern between the 3340 and the 1139 mice can be attributed in part to the differences in the reporter genes. SupFG1 in 3340 mice has two runs of 7 and 8 G·C base pairs, respectively. The longest repeat sequence in the supF gene in the 1139 mice, however, is a 5 G·C base pair stretch at positions 171–176. We propose that this shorter region is less susceptible to slippage replication errors (28) and is, therefore, less dependent on Pms2 function for stability.
The base substitutions in the nullizygotes were mostly transitions (Table 3). Because there were so few base substitutions in the 3340/Pms2 nullizygotes (only 3 of 74 mutations), the main comparison to be made is between the 1139/Pms2 nullizygotes and the wild-type mice. Overall, the spectra of base substitutions are similar except that the proportion of C → T transitions at CpG sites in the nullizygotes is only 11%, compared with 47% in the wild type. Because CpG sites are subject to cytosine methylation in mammalian cells, transitions at these positions are thought to arise from the propensity of 5-methylcytosine to deaminate to thymidine. Although such mutations do occur in the Pms2 nullizygotes, they are under-represented relative to those in the wild-type animals, suggesting that Pms2 is not essential for repair of these lesions. This is consistent with reports of an alternate repair pathway involving a glycosylase specific for the G·U and G·T mismatches that are produced by deamination (29, 30). The paucity of methylcytosine-related transitions in the nullizygotes is not due to undermethylation of the transgene sequences. The supF transgenes in both the 3340 and 1139 mice are each heavily methylated, as determined by comparison of susceptibility of the mouse genomic DNA to HpaII and MspI digestion (data not shown).
DISCUSSION
We have detected high somatic mutation frequencies in Pms2 nullizygous mice of up to 100-fold above the wild-type background. Mice heterozygous for the Pms2 disruption did not show increased spontaneous mutagenesis, suggesting that a single functional allele provides near normal repair activity and that Pms2-related genetic instability may, therefore, be recessive. However, we cannot rule out the possibility that the heterozygotes do have a low level of genetic instability, since slight increases in mutation frequency (such as 2-fold or less) are difficult to reliably detect in this assay, even with the analysis of large numbers of animals and phage. In light of the apparent participation of mismatch repair factors in aspects of DNA repair other than mismatch correction (15, 31), differences between the heterozygotes and the wild-type mice may eventually be revealed by studies of induced mutagenesis.
The heterozygous mice mimic the situation in most patients with the HNPCC syndrome, who are also heterozygous at one of the mismatch repair loci in their somatic cells (1, 3). Their tumors, however, are characterized by loss or inactivation of the second allele, leading to genomic instability. Such loss of heterozygosity events should also be possible in mice, thus predisposing the affected cell lineages to supF mutations. However, the low mutation frequencies in the heterozygotes suggest that such events are rare.
The spectrum of mutations in the nullizygotes is consistent with a primary role for Pms2 in the correction of slippage errors occurring during replication of repeated sequences (28). By comparison of the results in the supF and supFG1 reporter genes, we can conclude that seven or more mononucleotides are particularly unstable in vivo. This is consistent with the frequent finding of frameshift mutations within a 10 A·T base-pair run in the transforming growth factor β receptor gene in a series of colon cancer cell lines (32, 33). Such long mononucleotide runs are also particularly common within promoters and introns. For example, the c-myc promoter contains several extended runs of G·C base pairs (34), and the binding site for the Sp1 transcription factor is highly G+C-rich (35). We would predict that these regulatory regions would be especially unstable in the setting of Pms2 deficiency, leading to accumulating abnormalities in gene expression and thereby contributing to aberrant cellular function and eventually carcinogenesis.
The reduced frequency of C → T transitions at CpG sites in the Pms2 nullizygotes is in contrast to the results of Andrew and colleagues (S. E. Andrew, A. H. Reitmar, J. Fox, L. Hsiao, A. Francis, M. McKinnon, T. W. Mak, and F. Jirik, personal communication), who have found a higher frequency of such mutations in mice deficient in the MutS homolog Msh2. Since they used a different mutation reporter gene (lacI), the discrepancy could be attributed to the known propensity for such mutations in the lacI transgene (27). Also, the different loci of integration of the reporter genes may influence the mutation pattern, as has occasionally been seen in transgenic constructs (21, 36). It is also possible, however, that Msh2 has a role in the repair of the G·T mismatches that result from deamination of methylcytosine.
In addition, the mutation frequency in the Msh2-deficient mice studied by Andrew et al. (personal communication) is somewhat less than that seen in the Pms2 nullizygotes. Some of this difference may again be explained by differences in the reporter genes. Nonetheless, the comparsion indicates that Pms2 deficiency results in a degree of genetic instability at least as severe as that caused by lack of Msh2. Hence, the observation that Msh2 mutations are more common in HNPPC patients than are Pms2 mutations (2, 8) cannot be attributed to a greater impact of Msh2 mutations on genetic instability. Possibly, it could relate to differences in the mutation spectra, although these differences need to be confirmed in other target genes. More likely, Msh2 mutations may be more prevalent because of other factors, such as an increased susceptibility of the Msh2 locus itself to mutation or to loss of heterozygosity.
The nullizygous mice showed increased mutation in all tissues tested, including skin, liver, spleen, colon, brain, and lung. However, these animals appear to be predisposed to only lymphomas and sarcomas (20). This disparity shows that mutagenesis is just one element in carcinogenesis, highlighting the complexity of cancer etiology. Additional factors could include tissue damage and aberrant mitogenic stimuli. However, the particular factors that influence the tissue distribution of neoplasms in the mice remain to be determined. In HNPCC patients, the range of tumors is also limited; but it is different from that seen in the mice, with epithelial cancers of the colon most common. One explanation for this difference could be that most of the HNPCC patients are genotypically heterozygotes, and inactivation of the second mismatch repair gene allele is a critical step in the progression to neoplasm (1, 3, 37). Tissue-specific factors that vary among species, such as differential exposure to diet-derived genotoxic agents in the colon, may play an important part in promoting the loss of heterozygosity.
Several unusual patients, however, have been found to lack mismatch repair activity in their nonneoplastic tissues and in their cancers (38, 39). Two of these were determined to be heterozygous at the Pms2 locus, but in each case the mutant allele was found to code for a truncated protein with apparent dominant negative activity. In terms of Pms2-related mismatch repair function, the phenotype of these patients is, therefore, similar to that of the Pms2-nullizygous mice; yet, they developed colorectal carcinomas, not lymphomas or sarcomas. This difference suggests that there may be additional species-specific factors influencing tumor distribution. Determining whether these factors are genetic in nature (such as differences in the controls on growth and differentiation) or reflect environmental and life style differences will require further study.
Nonetheless, the Pms2 nullizygous mice described here show, remarkably, that high levels of spontaneous somatic mutations are compatible with mostly normal development and life, except for early onset carcinogenesis and male infertility (20). It might be predicted that such profound genetic instability would also cause accelerated aging, but this has not yet been demonstrated in the mice. Recently, Werner syndrome, a disease of premature aging and predisposition to malignancy, has been linked to a gene encoding a putative DNA helicase (40). In this disorder, the lack of helicase activity is hypothesized to lead to the accumulation of genetic abnormalities, producing early cellular senescence. However, mutations in Werner cells include a high proportion of large deletions (41), rather than point mutations as seen in the Pms2 mice. Whether this difference in mutation pattern accounts for the differences in carcinogenesis and aging remains to be determined.
Acknowledgments
We thank E. G. Leach, T. Y. Reynolds, E. G. Gunther, G. Wang, R. Franklin, S. J. Baserga, and L. Cabral for their help; and we thank S. Andrew and F. Jirik for sharing their results with Msh2-deficient mice. This work was supported by the Charles E. Culpeper Foundation, the Leukemia Society of America, and the National Institutes of Health (Grant ES05775).
ABBREVIATION
- HNPCC
hereditary nonpolyposis colorectal cancer
References
- 1.Kolodner R. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 2.Modrich P, Lahue R. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 3.de la Chapelle A, Peltomaki P. Annu Rev Genet. 1995;29:329–348. doi: 10.1146/annurev.ge.29.120195.001553. [DOI] [PubMed] [Google Scholar]
- 4.Fishel R, Lescoe M K, Rao M R, Copeland N G, Jenkins N A, Garber J, Kane M, Kolodner R. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 5.Leach F S, Nicolaides N C, Papadopoulos N, Liu B, Jen J, et al. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 6.Papadopoulos N, Nicolaides N C, Liu B, Parsons R, Lengauer C, Palombo F, D’Arrigo A, Markowitz S, Willson J K, Kinzler K W, Jirieny J, Vogelstein B. Science. 1995;268:1915–1917. doi: 10.1126/science.7604266. [DOI] [PubMed] [Google Scholar]
- 7.Bronner C E, Baker S M, Morrison P T, Warren G, Smith L G, Lescoe M K, Kane M, Earabino C, Lipford J, Lindblom A, Tannergard P, Bollag R J, Godwin A R, Ward D C, Nordenskjold M, Fishel R, Kolodner R, Liskay R M. Nature (London) 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 8.Nicolaides N C, Papadopoulos N, Liu B, Wei Y F, Carter K C, Ruben S M, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, Adams M D, Venter J C, Dunlop M G, Hamilton S R, Petersen G M, de la Chapelle A, Vogelstein B, Kinzler K W. Nature (London) 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 9.Papadopoulos N, Nicolaides N C, Wei Y F, Ruben S M, Carter K C, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, Adams M D, Venter J C, Hamilton S R, Petersen G M, Watson P, Lynch H T, Peltomaki P, Mecklin J, de la Chapelle A, Kinzler K V, Vogelstein B. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 10.Palombo F, Gallinari P, Iaccarino I, Lettieri T, Hughes M, D’Arrigo A, Truong O, Hsuan J J, Jiricny J. Science. 1995;268:1912–1914. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- 11.Drummond J T, Li G M, Longley M J, Modrich P. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 12.Prolla T A, Pang Q, Alani E, Kolodner R D, Liskay R M. Science. 1994;265:1091–1093. doi: 10.1126/science.8066446. [DOI] [PubMed] [Google Scholar]
- 13.Prolla T A, Christie D M, Liskay R M. Mol Cell Biol. 1994;14:407–415. doi: 10.1128/mcb.14.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li G M, Modrich P. Proc Natl Acad Sci USA. 1995;92:1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mellon I, Rajpal D K, Koi M, Boland C R, Champe G N. Science. 1996;272:557–560. doi: 10.1126/science.272.5261.557. [DOI] [PubMed] [Google Scholar]
- 16.Worth L, Jr, Clark S, Radman M, Modrich P. Proc Natl Acad Sci USA. 1994;91:3238–3241. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hawn M T, Umar A, Carethers J M, Marra G, Kunkel T A, Boland C R, Koi M. Cancer Res. 1995;55:3721–3725. [PubMed] [Google Scholar]
- 18.Bhattacharyya N P, Skandalis A, Ganesh A, Groden J, Meuth M. Proc Natl Acad Sci USA. 1994;91:6319–6323. doi: 10.1073/pnas.91.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eshleman J R, Lang E Z, Bowerfind G K, Parsons R, Vogelstein B, Willson J K V, Veigl M L, Sedwick W D, Markowitz S D. Oncogene. 1995;10:33–37. [PubMed] [Google Scholar]
- 20.Baker S M, Bronner C E, Zhang L, Plug A W, Robatzek M, Warren G, Elliott E A, Yu J, Ashley T, Arnheim N, Flavell R A, Liskay R M. Cell. 1995;82:309–319. doi: 10.1016/0092-8674(95)90318-6. [DOI] [PubMed] [Google Scholar]
- 21.Leach E G, Gunther E J, Yeasky T M, Gibson L H, Yang-Feng T L, Glazer P M. Mutagenesis. 1996;11:49–56. doi: 10.1093/mutage/11.1.49. [DOI] [PubMed] [Google Scholar]
- 22.Wang G, Levy D D, Seidman M M, Glazer P M. Mol Cell Biol. 1995;15:1759–1768. doi: 10.1128/mcb.15.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glazer P M, Sarkar S N, Summers W C. Proc Natl Acad Sci USA. 1986;83:1041–1044. doi: 10.1073/pnas.83.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gunther E J, Murray N E, Glazer P M. Nucleic Acids Res. 1993;21:3903–3904. doi: 10.1093/nar/21.16.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gossen J A, de Leeuw W J, Tan C H, Zwarthoff E C, Berends F, Lohman P H, Knook D L, Vijg J. Proc Natl Acad Sci USA. 1989;86:7971–7975. doi: 10.1073/pnas.86.20.7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Havre P A, Gunther E J, Gasparro F P, Glazer P M. Proc Natl Acad Sci USA. 1993;90:7879–7883. doi: 10.1073/pnas.90.16.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohler S W, Provost G S, Fieck A, Kretz P L, Bullock W O, Sorge J A, Putman D L, Short J M. Proc Natl Acad Sci USA. 1991;88:7958–7962. doi: 10.1073/pnas.88.18.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzah E, Inoye M. Cold Spring Harbor Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 29.Wiebauer K, Jiricny J. Proc Natl Acad Sci USA. 1990;87:5842–5845. doi: 10.1073/pnas.87.15.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neddermann P, Jiricny J. Proc Natl Acad Sci USA. 1994;91:1642–1646. doi: 10.1073/pnas.91.5.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Branch P, Aquilina G, Bignami M, Karran P. Nature (London) 1993;362:652–654. doi: 10.1038/362652a0. [DOI] [PubMed] [Google Scholar]
- 32.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L Z, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B, Brattain M, Willson J K V. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 33.Parsons R, Myeroff L L, Liu B, Willson J K, Markowitz S D, Kinzler K W, Vogelstein B. Cancer Res. 1995;55:5548–5550. [PubMed] [Google Scholar]
- 34.Postel E H, Flint S J, Kessler D J, Hogan M E. Proc Natl Acad Sci USA. 1991;88:8227–8231. doi: 10.1073/pnas.88.18.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Briggs M R, Kadonaga J T, Bell S P, Tjian R. Science. 1986;234:47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- 36.Gossen J A, de Leeuw W J, Verwest A, Lohman P H, Vijg J. Mutat Res. 1991;250:423–429. doi: 10.1016/0027-5107(91)90198-w. [DOI] [PubMed] [Google Scholar]
- 37.Modrich P. Science. 1994;266:1959–1960. doi: 10.1126/science.7801122. [DOI] [PubMed] [Google Scholar]
- 38.Parsons R, Li G M, Longley M, Modrich P, Liu B, Berk T, Hamilton S R, Kinzler K W, Vogelstein B. Science. 1995;268:738–740. doi: 10.1126/science.7632227. [DOI] [PubMed] [Google Scholar]
- 39.Hamilton S R, Liu B, Parsons R E, Papadopoulos N, Jen J, Powell S M, Krush A J, Wood P A, Taqi F, Booker S V, Petersen G M, Offerhaus G J A, Tersmette A C, Giardiello F M, Vogelstein B, Kinzler K W. N Engl J Med. 1995;332:839–847. doi: 10.1056/NEJM199503303321302. [DOI] [PubMed] [Google Scholar]
- 40.Yu C, Oshima J, Fu Y, Wijsman E M, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin G M, Mulligan J, Schellenberg G D. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 41.Fukuchi K, Martin G M, Monnat R J., Jr Proc Natl Acad Sci USA. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]