Abstract
We report the isolation and characterization for the first time of a cross-link lesion between two adjacent cytosines from the 2-methyl-1,4-naphthoquinone (menadione)-sensitized 365-nm irradiation of d(CpC). Electrospray ionization mass spectrometry (ESI-MS), tandem MS and 1H NMR results indicate that the cross-link occurs between the C5 carbon atom of one cytosine and the N4 nitrogen atom of the other cytosine. Furthermore, we synthesized d(CpC) with a 15N being incorporated on the amino group of either of the two cytosines. We then irradiated the two 15N-labeled dinucleoside monophosphates, isolated the cross-link products and characterized them by MS and multi-stage tandem MS. The latter results established unambiguously that the N4 nitrogen atom of the 3′-nucleobase is involved in the covalent bond formation between the two cytosines. This, in combination with two-dimensional nuclear Overhauser effect spectroscopy (NOESY) results, demonstrates that the cross-link arises from the formation of a covalent bond between the C5 carbon atom of the 5′ cytosine and the N4 nitrogen atom of the 3′ cytosine. We also show that the solution pH has a significant effect on the formation of the cross-link lesion, which supports that the deprotonation at the exocyclic amino group of cytosine cation radical is essential for the formation of the cross-link lesion.
INTRODUCTION
Human cells are constantly assaulted by reactive oxygen species (ROS) produced by both endogenous and exogenous pathways, and the resulting damage to DNA may have implications in a number of pathological conditions including cancer and aging (1–5). The types of mutations generated by oxidative damage of DNA depend on the oxidative reagent used and the experimental system employed. However, C→T transition and G→T transversion are the most commonly observed genetic changes (6–8).
In addition to single base substitutions, CC→TT tandem double mutations have been shown to occur as a result of DNA damage from ROS generated by metal ions or activated human leukemia cells (9–12). Since UV light is the only other source that can generate the CC→TT tandem double mutation (13,14), Loeb and co-workers (9–12) argued that the tandem double mutation can be used as a marker for oxidative DNA damage in tissues that are not exposed to UV irradiation.
Because the same mutation is produced by both UV irradiation and ROS, Loeb and coworkers (10) suggested that a primary candidate lesion that could induce the tandem double mutation might be a cross-link that forms between two adjacent cytosines. Although several ROS-induced cross-link lesions between adjacent thymine and adenine or guanine have been identified and structurally characterized (15–21), no cross-link lesion between two adjacent cytosines has been identified. GC-MS results, however, indicated that several cross-link lesions between two cytosines form upon γ irradiation of cytosine, 2′-deoxycytidine, and 2′-deoxycytidine 5′-monophosphate in N2O-saturated aqueous solution (22). It should be noted that these cross-link lesions were produced under anaerobic condition, which is different from the condition we used in this paper.
Quinones such as 2-methyl-1,4-naphthoquinone (MQ), or menadione, play important roles in aerobic respiration and photosynthesis and their function is closely associated with their redox potentials, which enable them to participate in electron transport within the cell membrane (23,24). It has been found that purine and pyrimidine bases can be efficiently consumed by UVA irradiation with MQ as a photosensitizer (25–33). ESR spin-trapping studies have demonstrated the formation of a number of free radicals under those irradiation conditions, and the radicals are similar to the species produced by hydroxyl radical reactions (34). Moreover, the products from the photosensitized reaction of some nucleobases and nucleosides closely resemble those produced by hydroxyl radical (25,30). MQ-sensitized photoirradiation of 2′-deoxycytidine can give rise to, among other products, 2′-deoxyuridine, which was suggested to be formed from an N4-centered radical arising from the deprotonation of the radical cation of 2′-deoxycytidine (35). In this context, the N4 nitrogen-centered radical on cytosine can also be induced by treatment with hypochlorite (36,37) or sulfate radical anion (38,39).
Here we report the identification and structure characterization of a new cross-link lesion from the MQ-sensitized 365-nm irradiation of dinucleoside monophosphate d(CpC), and we will show that the cross-link lesion is likely induced from an N4 nitrogen-centered radical of cytosine in d(CpC).
MATERIALS AND METHODS
Materials
All chemicals unless otherwise specified were from Sigma-Aldrich (St Louis, MO). The 2′-deoxycytidine (dC)-CE phosphoramidite and 5′-O-[(4,4′-dimethoxytriphenyl)methyl]-4-triazolouridine 3′-O-[2-cyanoethyl (N,N-diisopropylamino)phosphoramidite] were purchased from Glen Research Inc. (Sterling, VA). Silica gel, TLC plates and all the solvents were obtained from EM Science (Gibbstown, NJ).
Most 1H-NMR experiments were performed on a Varian Inova 300 instrument (Palo Alto, CA) unless otherwise specified. The residual proton signal of the solvent serves as an internal reference. Two-dimensional NOESY (nuclear Overhauser effect spectroscopy) experiments were performed on a Varian Inova 500 instrument and data were acquired at 298 K with 600 and 700 ms mixing times for d(CpC) and d(C∧pC), respectively. NOESY spectra were obtained from 2 × 256 × 2048 data matrices with 48 scans per t1 value. After two-dimensional Fourier transformation, the spectra were obtained as 2k × 2k spectra data matrices, and were phase and baseline corrected in both dimensions.
Preparation of 4-(1,2,4-triazol-1-yl)-3′,5′-O-diacetyl 2′-deoxyuridine (1)
The title compound was prepared by using Reese reagent (40). Triethylamine (3.24 ml, 0.023 mol) was added drop wise to a stirred ice-water-bath-cooled solution, which contained 1,2,4-triazole (1.68 g, 24.3 mmol), phosphoryl chloride (0.5 ml, 5.2 mmol) and acetonitrile (14 ml). To the resulting solution was added a solution of 3′,5′-O-diacetyl-2′-deoxyuridine (1.2 g, 3.8 mmol) in acetonitrile (8.5 ml) and the reaction mixture was stirred at room temperature for 90 min. Triethylamine (2.25 ml, 16 mmol) and water (0.58 ml, 32.5 mmol) were then added and, after 10 min, the solvent was removed in vacuo. The residue was partitioned between chloroform (25 ml) and saturated aqueous solution of NaHCO3 (20 ml). The aqueous layer was further extracted twice with chloroform (25 ml). The combined organic layers were dried (MgSO4) and the residue crystallized from ethanol to give 4-(1,2,4-triazol-1-yl)-3′,5′-O-diacetyl-2′-deoxyuridine (1.2 g; yield, 87%). 1H NMR (DMSO-d6): δ 9.47 (s, 1H, triazolyl-H), 8.48 (d, JH6-H5 = 8.2 Hz, 1H, H6), 8.44 (s, 1H, triazolyl-H), 7.07 (d, J = 8.2 Hz, 1H, H5), 6.17 (dd, J = 6.6, 6.7 Hz, 1H, H1′), 5.25 (m, 1H, H3′), ′4.41 (m, 1H, H4′ 4.30 (m, 2H, H5′ and H5”), 2.70–2.38 (m, 2H, H2′ and H2”), 2.11 (s, 3H, CH3-CO), 2.05 (s, 3H, CH3-CO).
Preparation of [15N4]-2′-deoxycytidine (2)
We followed a similar procedure as reported by Sowers and coworkers (41). Compound 1 (1.2 g, 3.3 mmol) was suspended in 1,4-dioxane (10 ml), to which ammonium-15N hydroxide (99 atom % 15N, 6 N in H2O, 2 ml, 12 mmol) was added. The solution was stirred at room temperature for 6 h, and the solvents were removed in vacuo. The resulting residue was dissolved in methanolic ammonia (50% saturated at 0°C, 16.5 ml). After 16 h, the products were evaporated to dryness. Crystallization of the residue from aqueous ethanol gave the desired [15N4]-2′-deoxycytidine (0.65 g; yield, 85%). 1H NMR (DMSO-d6): δ 7.8 (d, JH6-H5 = 7.2 Hz, 1H, H6), 7.13 (d, J15N-H = 87.7 Hz, 2H, 15N-H), 6.17 (dd, J = 6.2, 5.6 Hz, 1H, H1′), 5.73 (d, JH6-H5 = 7.2 Hz, 1H, H5), 5.20 (d, JOH-H3′ = 4.1 Hz, 1H, 3′OH), 4.97 (t, J = 5.6 Hz, 1H, 5′-OH), 4.21 (m, 1H, H3′), 3.78 (m, 1H, H4′), 3.35 (m, 2H, H5′and H5”, 2.16–1.19 (m, 2H, H2′and H2”).
Preparation of N4-benzoyl-2′-deoxycytidine-15N4 (3)
We used the transient protection procedure reported by Ti and coworkers (42). Compound 2 (0.6 g, 2.6 mmol) was dried three times by evaporation with dry pyridine and suspended again in dry pyridine (25 ml), to which was added trimethylchlorosilane (2.24 ml, 17.5 mmol). After the solution was stirred for 15 min, benzoyl chloride (1.66 ml, 17.5 mmol) was added and the reaction was maintained at room temperature for 2 h. The mixture was then cooled in an ice bath, to which 29% aqueous ammonia (3 ml) was added, and the mixture was stirred at room temperature for 15 min. The reaction mixture was then evaporated to near dryness and the residue was dissolved in water (50 ml). The solution was then washed with EtOAc (50 ml). Crystallization began immediately after separation of the layers. After the solution was cooled, filtration gave 3 (0.8 g; yield, 92%). 1H NMR (DMSO-d6): δ 8.42 (d, JH6-H5 = 7.8 Hz, 1H, H5), 8.03 (d, J = 7.7 Hz, 1H, Ar-H of benzoyl), 7.80 (d, JH6-H5 = 7.8 Hz, 1H, H6), 7.67–7.32 (m, 5H, Ar-H of benzoyl and 15N-H), 6.16 (dd, J = 6.2, 5.6 Hz, 1H, H1′), 5.29 (d, JOH-H3′ = 4.1 Hz, 1H, 3′-OH), 5.09 (t, JOH-H5′ = 5.13 Hz, 1H, 5′-OH), 4.26 (m, 1H, H3′), 3.90 (m, 1H, H4′), 3.64 (m, 2H, H5′ and H5”), 2.38–2.30 (m, 1H, H2′), 2.12–2.04 (m, 1H, H2”). HRMS (ESI-FTICR): calcd 331.1060 [M – H]–, found 331.1056.
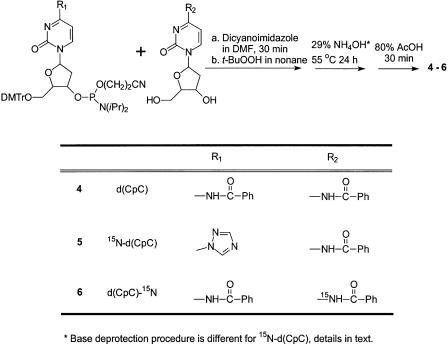
Preparation of d(CpC) (4, Scheme 1)
We followed a procedure developed in Professor John-Stephen A. Taylor’s laboratory in Washington University in St Louis (J.-S. A. Taylor, personal communication).
To an anhydrous DMF (2 ml) solution of N4-benzoyl-2′-deoxycytidine (125 mg, 0.45 mmol) and 4,5-dicyanoimidazole (106 mg, 0.90 mmol) was drop wise added 1 ml DMF solution of dC-CE phosphoramidite (250 mg, 0.30 mmol) over 30 min while stirring at room temperature. To the reaction mixture was then added 0.3-ml t-BuOOH (5–6 M solution in nonane). The reaction mixture turned yellowish from being dark yellow. After the reaction proceeded for 10 min at room temperature, TLC showed completion of the reaction. The reaction mixture was extracted with EtOAc (20 ml) and 5% NaHCO3 solution (20 ml). The organic layer was dried over anhydrous Na2SO4, concentrated, loaded onto a silica gel column and eluted with MeOH/EtOAc (1/9, v/v). The desired fractions were pooled and solvent removed in vacuo.
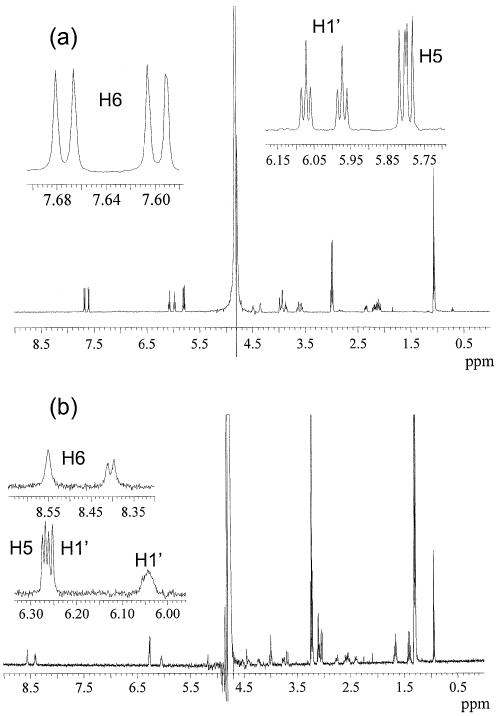
The product was suspended in 29% aqueous ammonia (5 ml) at 55°C for 24 h. The solution was dried again, and 80% acetic acid (5 ml) was added to the residue. The color of the solution changed from light yellow to red. After 30 min, the solution was concentrated and extracted with ether. Drying of the aqueous layer gave 50 mg of 4 (yield, 30%). 1H NMR (500 MHz, D2O): see Figure 6a. HRMS (ESI-FTICR): calcd 515.1292 [M – H]–, found 515.1279.
Figure 6.
1H NMR (500 MHz, D2O) of d(CpC) (a) and d(C∧pC) (b).
Preparation of 15N4-d(CpC) (5, Scheme 1)
The procedure was similar to the synthesis of d(CpC). The product was suspended in 1,4-dioxane (2 ml), to which ammonium-15N hydroxide (99 atom % 15N, 6 N in H2O, 0.4 ml, 2.4 mmol) was added. The solution was stirred at room temperature for 6 h, and the solvents were removed in vacuo. The resulting residue was dissolved in 29% aqueous ammonia (5 ml) and incubated at 55°C for 24 h. The DMTr group was removed by treatment with 80% acetic acid as described above. HRMS (ESI-FTICR): calcd 516.1262 [M – H]–, found 516.1247.
Preparation of d(CpC)-15N4 (6, Scheme 1)
The procedure was similar to the synthesis of d(CpC) except that N4-benzoyl-2′-deoxycytidine-15N4 was used instead of the N4-benzoyl-2′-deoxycytidine (Scheme 1). HRMS (ESI-FTICR): calcd 516.1262 [M – H]–, found 516.1248.
Scheme 1. Preparation of dinucleoside monophosphates d(CpC), 15N-d(CpC) and d(CpC)-15N.
Photosensitized reactions
Dinucleoside monophosphate d(CpC) (300 nmol) was dissolved in 10-ml aqueous solution that was saturated with MQ (∼0.3 mM as determined by UV absorbance at 350 nm) (43) or at other concentrations as specified in the text. The solution was then transferred to a 10.2-cm I.D. Petri dish, irradiated on ice for 30 min or other periods of time as specified. The solution was exposed to air during irradiation and two 15-W Spectroline light tubes emitting at 365 nm were used (Spectronics Corporation, Westbury, NY). The distance between the light tubes and the sample solution was ∼3 cm. The resulting solution was dried by using a Savant Speed-vac (Savant Instruments Inc., Holbrook, NY) and the dried residue was redissolved in water and subjected to HPLC analysis.
For pH-dependent studies, the irradiation was done under identical conditions except that the d(CpC) was dissolved in 10 mM phosphate buffer. Irradiation of d(TpC) and d(UpC) was carried out under the same conditions as that for d(CpC); the irradiation time for the former two, however, is 15 min. For the irradiation of duplex oligodeoxynucleotide (ODN) d(CCGGCCGGCCGGCCGG), the ODN was first annealed in a solution containing 50 mM phosphate buffer (pH 6.8) and 50 mM NaCl and dispersed in a solution with 10 mM phosphate (pH 6.8) and 10 mM NaCl for the irradiation. The concentration of MQ and irradiation time were similar to those for the irradiation of d(CpC).
HPLC
The HPLC separation was performed on a Surveyor system (ThermoFinnigan, San Jose, CA) with a photodiode array detector, and an Apollo 4.6*250 mm reverse-phase C18 column (5 µm in particle size and 300 Å in pore size; Alltech Associates Inc., Deerfield, IL) was used. The flow rate was 0.8 ml/min, and a gradient of 5-min 0–12% methanol followed by a 35-min 12–30% methanol in 10 mM ammonium formate (pH 6.3) was used. The photodiode array detector was set at 260 nm for monitoring the eluents. The HPLC fractions were then dried by using the Speed-vac. For NMR analysis, the cross-link lesion was further desalted by washing with water for 5 min, and the compound was eluted from the column by using 25% methanol in water as the mobile phase.
Electrospray ionization mass spectrometry (ESI-MS) and MSn
Most MS experiments were carried out on an LCQ Deca XP ion-trap mass spectrometer (ThermoFinnigan, San Jose, CA). An equal-volume solvent mixture of acetonitrile and water was used as the carrier and electrospray solvent, and a 1 µl aliquot of 5 µM sample solution was injected in each run. The spray voltage was 4.5 and 3.0 kV for experiments in the positive- and negative-ion modes, respectively. The mass width for precursor selection in tandem mass spectrum (MS/MS) and MSn modes was 3 m/z units. Each spectrum was obtained by averaging approximately 50 scans, and the time for each scan was 0.1 s. MS3 and MS4 were acquired by directly infusing 10 µM sample solution at a flow rate of 5 µl/min and the spectra were obtained by signal averaging for 2–3 min.
Exact-mass measurements were done on an IonSpec HiResESI external ion source FTICR mass spectrometer (IonSpec Co., Lake Forest, CA) equipped with a 4.7-T unshielded superconducting magnet and an Analytica (Branford, CT) electrospray ion source. Samples were dissolved in CH3CN/H2O (1/1, v/v) at a concentration of 5 µM and infused by a syringe pump at a flow rate of 1 µl/min. Experiments were done in the negative-ion mode and the source end plate was kept at 3.0 kV. Ions were accumulated in a hexapole for 1000–1500 ms and transported through a quadrupole ion guide to the analyzer cell by standard pulse sequences. Exact-mass measurement of the molecular ions was done by using the [M – H]– and [M – Ade – H]– ions of ApA as internal standards. For exact-mass measurements in MS/MS, the [M – H]– ions of both the analyte of interest and ApA were first isolated in the ICR cell by ejection of all other species. Sustained off-resonance irradiation/collisionally activated dissociation (SORI-CAD) of the analyte ion was performed for 1000 ms by an r.f. burst with an amplitude varied from 1.2 to 2.5 V depending on the analytes and at a frequency ∼1000 Hz from the cyclotron frequency of the analyte ion. A 200-ms pulse of nitrogen was introduced at the beginning of the irradiation period (the maximum pressure read-out on the ion gauge was 3 × 10–6 Torr). Ions were activated as a result of multiple collisions with the pulsed nitrogen gas. After a delay of 2 s, the resulting fragment ions were accelerated for detection by an r.f. sweep excitation waveform (100 V p-p). The image current was amplified and digitized at an acquisition rate of 2 MHz before Fourier transform to yield a mass spectrum. Under this condition, some of the [M – H]– ions of ApA also fragment, and the [M – H]– ions of ApA (m/z 595.1414) and adenine (m/z 134.0467) fragment were used as internal standards for calibration. All the spectra were acquired in the broadband mode and, with the internal standards, the mass accuracy for all mass measurements reported in this work was within 5 p.p.m. for the monoisotopic ion peaks.
To measure the number of exchangeable protons, the cross-link lesion was incubated in D2O overnight, dried by Speed-vac and redissolved in D2O. The resulting solution was subjected directly to ESI-MS and MS/MS analyses, and acetonitrile in D2O (50/50, v/v) was used as electrospray solvent.
RESULTS
Isolation and MS characterization of a new cross-link lesion of d(CpC)
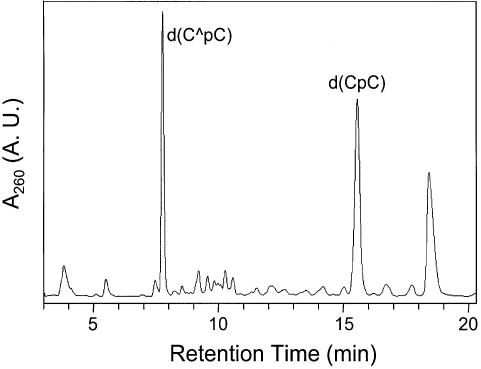
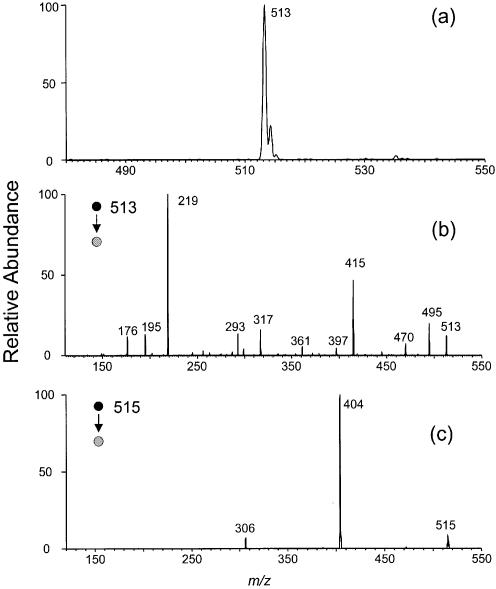
We isolated a new product from the 365-nm irradiation of d(CpC) in the presence of MQ (chromatogram for the separation of a 5-min irradiation mixture is shown in Fig. 1). Negative-ion ESI-MS of the fraction with retention time of 7.7 min shows an ion of m/z 513 (Fig. 2a). Exact-mass measurement gives m/z 513.1119 for the [M – H]– ion of the product, which indicates that the product results from d(CpC) with the elimination of two hydrogens (the calculated m/z for the deprotonated ion is 513.1135).
Figure 1.
HPLC trace for the separation of the 365-nm irradiation mixture of d(CpC) in the presence of MQ.
Figure 2.
Negative-ion ESI-MS of d(C∧pC) (a) and product-ion spectra of ESI-produced [M – H]– ions of d(C∧pC) (b) and d(CpC) (c) acquired on an ion-trap mass spectrometer.
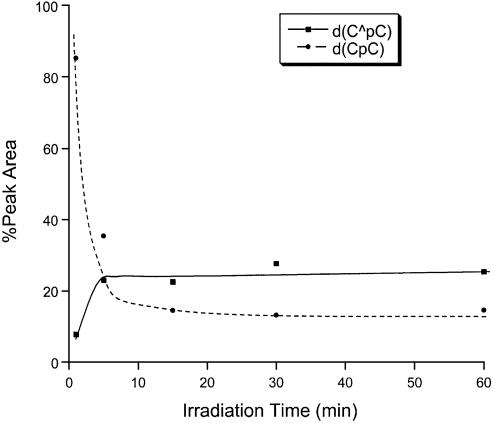
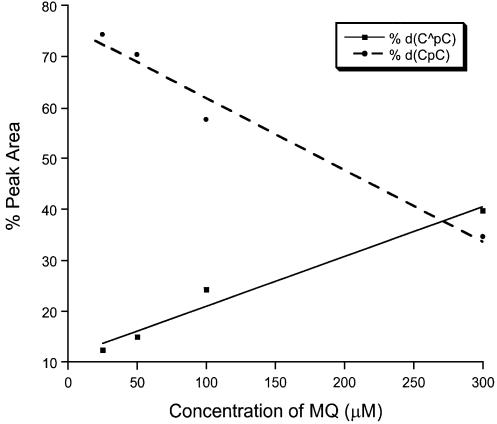
Two lines of evidence indicate that the new product is a major product formed under this irradiation condition. We varied the irradiation time while keeping the concentration of MQ and the amount of d(CpC) constant, HPLC analyses indicate that the percent yield of the new product increases at short irradiation time and it keeps almost constant after the irradiation time is >10 min (Fig. 3). Concurrent with the increase of the yield of the new product [denoted as d(C∧pC)], we observed the decrease of the starting d(CpC). That the starting material and the new product reach a plateau after a certain irradiation time may indicate that MQ is decomposed during the irradiation process. Next we irradiated the d(CpC) with different concentrations of MQ while keeping the irradiation time and the amount of d(CpC) constant. Our results again show that the percent yield of the new product increases, whereas the percentage of d(CpC) decreases, with the increasing of the concentration of MQ (Fig. 4). Furthermore, the new product is a major product formed with different concentrations of MQ.
Figure 3.
The percentages of peak areas of d(CpC) and d(C∧pC) in HPLC traces for the separation of the irradiation mixtures of d(CpC) versus irradiation time.
Figure 4.
The percentages of peak areas of d(CpC) and d(C∧pC) in HPLC traces versus the concentration of MQ.
Under typical irradiation conditions with saturated MQ and 30 min irradiation time, the yield for the formation of the new product is 9.0% as determined by HPLC analysis with the assumption that the absorption coefficient at the λmax does not change from d(CpC) to the new product. This result appears to be consistent with previous estimation that the yield for the deprotonation of the radical cation of 2′-deoxycytidine is ∼20% (35).
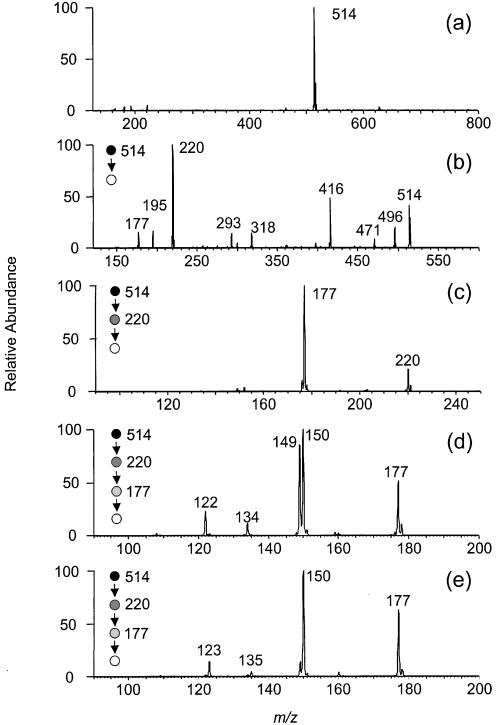
MS/MS of the ESI-produced [M – H]– ion of the product is quite different from that of the starting material (Fig. 2b and c). Whereas a neutral loss of cytosine gives the most abundant fragment ion in the latter (m/z 404), the former does not show that loss. Instead, many new fragmentation pathways open for the deprotonated ion of the new product. The ions of m/z 495 and 415 result from the loss of a H2O molecule and a 2-deoxyribose moiety, respectively. The latter ion can undergo a further loss of (HPO3 + H2O) to give an ion of m/z 317. Most strikingly, we observed an ion of m/z 219, which is the deprotonated ion of two cytosines with the loss of two hydrogens. In addition, an ion resulting from the loss of the nucleobase moiety was also produced (m/z 293), and this ion can further lose a 2-deoxyribose to yield an ion of m/z 195. The assignments of all above fragment ions have been confirmed by exact-mass measurement on an FT-ICR mass spectrometer (Table 1). The absence of the fragment ion with the loss of a single nucleobase and the presence of a unique fragment ion corresponding to two cytosines with the elimination of two hydrogens (m/z 219) demonstrate that the new product is a lesion with two cytosines being cross-linked.
Table 1. SORI-CAD MS/MS results of the [M – H]– ion of d(C∧pC).
| Calculated mass | Measured mass | Deviation (p.p.m.) | |
|---|---|---|---|
| [M – H]– | 513.1135 | 513.1154 | 3.7 |
| 513.1125 | –1.9 | ||
| 513.1156 | 4.1 | ||
| H2O loss | 495.1029 | 495.1039 | 2.0 |
| 495.1041 | 2.4 | ||
| 495.1030 | 0.2 | ||
| Deoxyribose loss | 415.0767 | 415.0767 | 0.0 |
| 415.0769 | 0.5 | ||
| 415.0763 | –1.0 | ||
| (Deoxyribose + HPO3 + H2O) loss | 317.0998 | 317.0999 | 0.3 |
| 317.1002 | 1.3 | ||
| 317.0995 | –0.9 | ||
| Base loss | 293.0426 | 293.0427 | 0.3 |
| 293.0428 | 0.7 | ||
| 293.0423 | –1.0 | ||
| [Base – H]– | 219.0630 | 219.0624 | –2.7 |
| 219.0626 | –1.8 | ||
| 219.0625 | –2.3 | ||
| [Deoxyribose + HPO3 + H2O – H]– | 195.0059 | 195.0055 | –1.8 |
| 195.0056 | –1.3 | ||
| 195.0054 | –2.3 |
Data were obtained from three independent measurements.
Measurement of the number of exchangeable protons
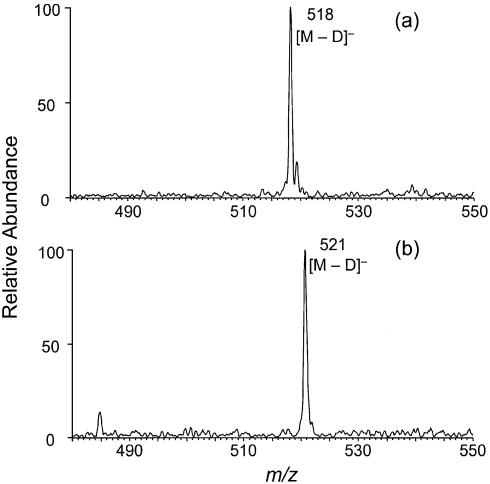
The above MS and MS/MS results show that the cross-link arises from the elimination of two hydrogens from the two cytosines. Therefore, the covalent bond that links the two cytosines may form between the C5, C6 or N4 of the two bases, which gives us nine possible structures. To determine which atoms are involved in the cross-link, we first measured the number of exchangeable protons in d(C∧pC). ESI-MS (Fig. 5a) gives m/z 518 for the [M – D]– ion of the new cross-link lesion. The result shows that five hydrogens in the cross-link lesion have been replaced with deuterons. Similar measurement with d(CpC) shows that there are six exchangeable protons in d(CpC) as expected (Fig. 5b). The loss of one active hydrogen, in combination with the above MS/MS result, demonstrates that there is one and only one amino group involved in the cross-link. Therefore, the cross-link must form via a covalent bond between the N4 nitrogen atom of one cytosine and the C5 or C6 carbon atom of the other cytosine.
Figure 5.
Negative-ion ESI-MS of d(C∧pC) (a) and d(CpC) (b) where all the active hydrogens have been replaced with deuterons. Data were acquired on an ion-trap mass spectrometer.
The above structure proposal for the new cross-link lesion is also supported by UV absorption spectroscopy measurement. UV absorption spectra showed a λmax of 289 and 271 nm for d(C∧pC) and d(CpC), respectively. The small red shift (18 nm) from the d(CpC) to d(C∧pC) is consistent with our proposed structure.
NMR characterization of the cross-link lesion
To determine whether C5 or C6 carbon atom is involved in the cross-link, we acquired the 1H-NMR of d(CpC) and d(C∧pC) (Fig. 6). Comparing with the spectrum of d(CpC), that of d(C∧pC) showed that one H5 proton resonance is absent and one of the H6 proton resonances shows as a singlet. Therefore, one H5 proton is lost upon the formation of the cross-link lesion, which demonstrates that the cross-link occurs between the C5 carbon atom of one cytosine and the N4 nitrogen atom of the other cytosine.
In addition, we carried out the 2-D NOESY experiments for both d(CpC) and d(C∧pC), which enabled us to assign all the proton resonances (Table 2). From Table 2, we note that H6 and H5 protons of the cross-link lesions shift downfield compared with the d(CpC), which might be due to the influence of the other cytosine, which is now in close proximity. Previous study with a cross-link lesion in CG, in which the C5 carbon atom of cytosine and the C8 carbon atom of the adjacent guanine is cross-linked, also showed that the H6 proton of cytosine was displaced downfield by 0.64 p.p.m. (17). In addition, the formation of the cross-link lesion affects the conformation of the 2′-deoxyriboses as manifested by the changes in chemical shifts of protons on the 2′-deoxyribose rings.
Table 2. Proton NMR chemical shift data in p.p.m. for d(CpC) (SM) and d(C∧pC) (XL) at room temperature in D2O.
| H1′ | H2′ | H2” | H3′ | H4′ | H5′ | H5” | H5 | H6 | |
|---|---|---|---|---|---|---|---|---|---|
| Cp, SM | 6.30 | 2.43 | 2.31 | 4.56 | 4.15 | 4.15 | 4.09 | 6.05 | 7.89 |
| Cp, XL | 6.25 | 2.58 | 2.54 | 4.52 | 4.43 | 3.98 | 3.67 | NA | 8.53 |
| pC, SM | 6.22 | 2.57 | 2.34 | 4.72 | 4.20 | 3.84 | 3.77 | 6.03 | 7.80 |
| PC, XL | 6.03 | 2.76 | 2.39 | 4.42 | 3.98 | 4.21 | 3.74 | 6.24 | 8.38 |
Water signal at 4.80 p.p.m. was used as reference, Cp designates the 5′ nucleoside, and pC designates the 3′ nucleoside.
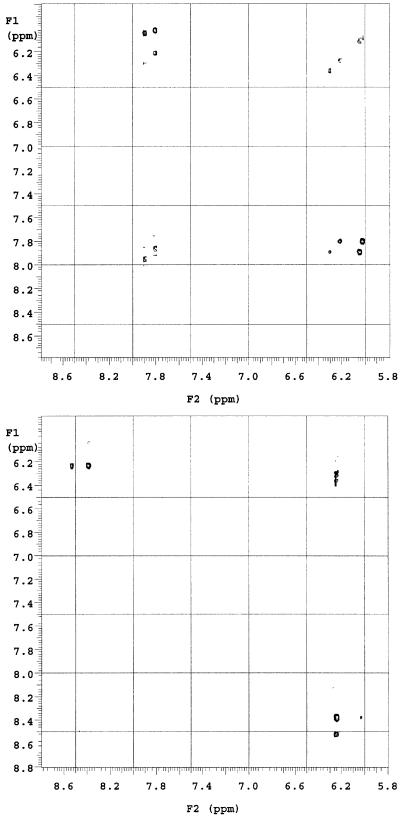
Moreover, 2-D NOESY experiments determined that the H5 hydrogen atom from the 5′ cytosine was absent in d(C∧pC). In the NOESY spectrum of d(CpC) (Fig. 7, top), we observed H6-H5 and H6-H1′ correlations for both nucleosides. In the NOESY spectrum of d(C∧pC), however, we found that the H6 proton of the 5′ cytosine correlates only with the H1′ proton of the 5′ nucleoside while the H6 proton of the 3′ cytosine correlates with both the H1′ proton and H5 proton in the 3′ nucleoside (Fig. 7, bottom).
Figure 7.
A portion of 2-D NOESY spectra of d(CpC) (top) and d(C∧pC) (bottom).
MSn of 15N-incorporated d(C∧pC)
From the above results, we determined that the cross-link lesion forms through a covalent bond between the C5 carbon atom of the 5′ cytosine and the N4 nitrogen atom of the 3′ cytosine. The involvement of the N4 nitrogen atom of the 3′ cytosine was inferred from the measurement of the number of active hydrogens. More direct evidence is, therefore, desired for supporting this conclusion. To this end, we synthesized two dinucleoside monophosphates with a 15N being incorporated on the amino group of either of the two cytosines (5,6, Scheme 1). For the brevity of discussion, we designate the dinucleoside monophosphates with 15N being incorporated on the 5′ and 3′ cytosines as 15N-d(CpC) (5) and d(CpC)-15N (6), respectively. The product-ion spectra of the [M – H]– ions of the two 15N-labeled d(CpC) are distinctive (Fig. 8a and b). The MS/MS of 15N-d(CpC) and d(CpC)-15N show a neutral loss of 15N-labeled (m/z 404) and unlabeled cytosine (m/z 405), respectively. This result is consistent with previous studies showing that collisional activation of the [M – H]– ions of dinucleoside monophosphates gives preferential loss of the 5′ base over the 3′ base (44).
Figure 8.
Product-ion spectra of the [M – H]– ions of 15N-d(CpC) (a) and d(CpC)-15N (b). Data were acquired on an ion-trap mass spectrometer.
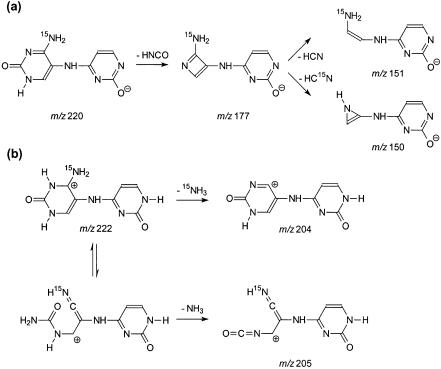
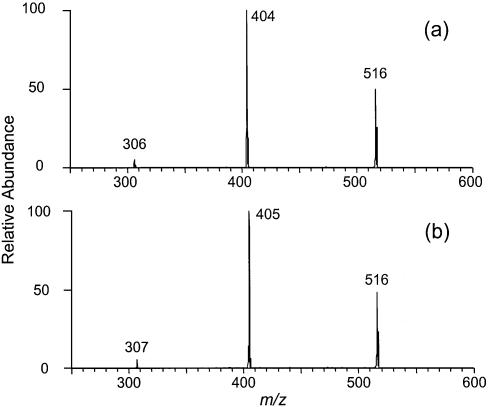
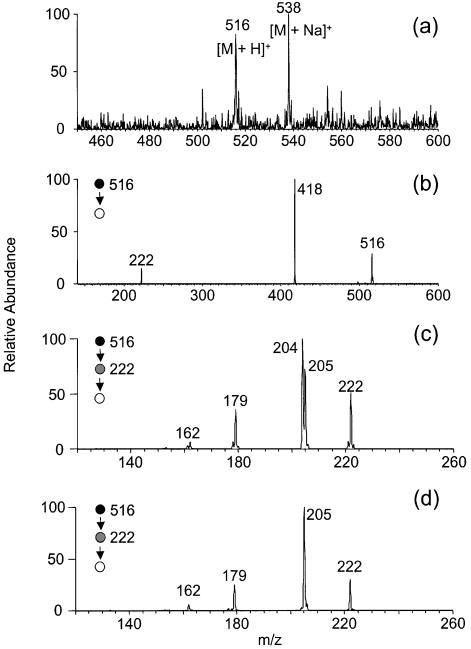
We irradiated the 15N-d(CpC) and d(CpC)-15N with 365-nm light in the presence of MQ, isolated the cross-link products by HPLC, and characterized them extensively with MS. Negative-ion ESI-MS showed an ion of m/z 514 (Fig. 9a), which is [M – H]– ion of the cross-link lesion with a 15N. Product-ion spectra of the [M – H]– ions of 15N-d(C∧pC) and d(C∧pC)-15N give identical results (the former MS/MS is shown in Fig. 9b). The MS/MS is also similar to that of the [M – H]– ion of d(C∧pC) without 15N labeling except that the ions of m/z 219, 317, 415, 495 in the latter shift to higher m/z values by 1 amu in the former, indicating that those ions bear the 15N. The results provide additional support for our assignments of fragment ions (vide supra). Similarly, MS3 of the ions of the deprotonated base moiety (m/z 220) from the 15N-d(C∧pC) and d(C∧pC)-15N gave the same results. The ion from both products can readily lose a 43-amu fragment, which is likely an HN=C=O moiety, to give an abundant ion of m/z 177 (Fig. 9c). The proposed structure for the m/z 177 ion from 15N-d(C∧pC) is shown in Scheme 2a.
Figure 9.
Negative ESI-MS of 15N-d(C∧pC) (a), Product-ion spectrum of the [M – H]– ion of 15N-d(C∧pC) (b), MS3 of the ion of m/z 220 in b (c), and MS4 (m/z 514→220→177→ for 15N-d(C∧pC) (d) and d(C∧pC)-15N (e). Data were acquired on an ion-trap mass spectrometer.
Scheme 2. Proposed structures for the fragment ions observed in Figures 9 and 10.
MS4 of the fragment ion of m/z 177, however, are different for the cross-link lesions with a 15N being labeled on 5′ or 3′ cytosines. The ion of m/z 177 from d(C∧pC)-15N can lose one and two HCN molecules to give ions of m/z 150 and 123, respectively (Fig. 9e). The m/z 177 ion from 15N-d(C∧pC), however, can lose either a HCN or HC15N molecule to yield two fragment ions, m/z 150 and 149 (the structures for those two ions were tentatively proposed as shown in Scheme 2a), with similar abundance (Fig. 9d). These two ions may further lose an HC15N and HCN, respectively, to give a common fragment ion of m/z 122 (Fig. 9e). Therefore, the 15N can be lost as a HC15N from the precursor with the 15N on the 5′ cytosine, but not from that with the 15N on the 3′ cytosine. The results demonstrate that the 3′ N4 nitrogen atom, but not the 5′ N4 nitrogen atom, is involved in the cross-link bond formation.
The cross-link lesions from the two 15N-labeled precursors were also characterized by positive-ion ESI-MS (Fig. 10a). The product-ion spectra of the [M + H]+ ions of 15N-d(C∧pC) and d(C∧pC)-15N gave identical results [Fig. 10b shows the MS/MS of the [M + H]+ ion of 15N-d(C∧pC) as an example]. MS3 of the protonated ion of the nucleobase portion (m/z 222) of 15N-d(C∧pC) and d(C∧pC)-15N, however, are different. When the 15N is on the 5′ base, the ion of m/z 222 can lose both the unlabeled (17 amu) and 15N-labeled ammonia (18 amu) to give ions of m/z 205 and 204, respectively (Fig. 10c). While the 15N is on the 3′-base, the ion of m/z 222, however, can only lose the unlabeled ammonia (17 amu) to yield an ion of m/z 205 (Fig. 10d). To better visualize the difference in ammonia losses for the two 15N-incorporated cross-link lesions, we again proposed tentatively the structures for the m/z 204 and 205 ions (Scheme 2b) observed in Figure 10c. From the proposed the fragmentation pathways, the loss of unlabeled ammonia from 15N-d(C∧pC) requires the opening of the 5′ pyrimidine ring. The 15N can be lost as an ammonia molecule when it is on the 5′ cytosine but not when it’s on the 3′ cytosine, which provides additional evidence supporting that the N4 nitrogen atom on the 3′ cytosine is covalently bonded to the C5 carbon atom of the 5′ cytosine.
Figure 10.
Positive-ion ESI-MS (a), MS/MS of the [M – H]– ion of 15N-d(C∧pC) (b), and MS3 of the protonated ions of the base portion of the cross-link from 15N-d(C∧pC) (c) and d(C∧pC)-15N (d). Data were acquired on an ion-trap mass spectrometer.
Our structural assignment is in line with the measured distances between the N4 nitrogen atom of one cytosine and the C5 carbon atom of the adjacent cytosine in B-form DNA [d(CpC) was built in software package InsightII using standard B-DNA geometry]. The distance between the N4 nitrogen atom of the 3′-cytosine and the C5 carbon atom of the 5′ cytosine is 3.94 Å, which is much shorter than the distance between N4 nitrogen atom of the 5′ cytosine and the C5 carbon atom of the 3′ cytosine (5.17 Å). The above distances suggest that the formation of a cross-link between the N4 nitrogen atom of the 3′ cytosine and the C5 carbon atom of the 5′ cytosine is more favorable than that between the N4 nitrogen atom of the 5′ cytosine and the C5 carbon atom of the 3′ cytosine. It is worth noting that we tried, but failed to isolate any cross-link lesion with a covalent bond formed between the N4 nitrogen atom of the 5′ cytosine and the C5 carbon atom of the 3′ cytosine.
In addition, we have evidence showing that the HPLC fraction eluting slightly earlier than d(C∧pC) is also a cross-link lesion, where the covalent bond appears to be between the C6 carbon atom of the 5′ cytosine and the N4 nitrogen atom of the 3′ cytosine. The detailed structure characterization is still under way, and we will report that in a separate manuscript later. The peak eluting at 18.6 min in Figure 1 gives a m/z of 221 in negative-ion ESI-MS, and it is also present in the chromatograms for the separation of irradiation mixtures of d(UpC), 15N-d(CpC) and d(CpC)-15N (data not shown). Therefore, it is likely a decomposition product of MQ, and further characterization of this compound was not pursued.
MQ-sensitized photoirradiation of d(UpC) and d(TpC)
Next we examined whether the N4-nitrogen radical of the 3′ cytosine can also attack other pyrimidine bases on the 5′ nucleoside. To this end, we irradiated d(UpC) and d(TpC), and we were able to identify a similar cross-link lesion from d(UpC) by using ESI-MS and multi-stage MS/MS (data not shown). However, we could not isolate any cross-link lesion from the irradiation of d(TpC), and the major products evolving from that irradiation have the thymine moiety being oxidized to thymine glycol (data not shown).
DISCUSSION
Formation mechanism
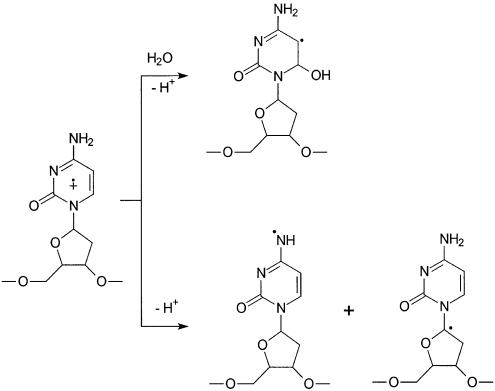
By using laser flash photolysis, Fisher and Land (45) showed that menadione-sensitized 353-nm irradiation of thymine leads to charge transfer between 3MQ and thymine to give the cation radical of the latter. Considering the triplet state energy of MQ and all four DNA bases, the authors concluded that triplet energy transfer from MQ to all DNA bases is unlikely (45). Likewise, product analysis demonstrated that radical cation of dC can form from MQ-sensitized photoirradiation (30,35). The radical cation could either deprotonate to yield a radical at N4 or C1′ site or hydrate to give 6-hydroxy-5-yl radical (Scheme 3), and the deprotonation and hydration are competitive against each other (28,35). The dC with a radical at the N4 site can deaminate to give 2′-deoxyuridine, whereas the dC with a radical at the C1′ site can undergo cleavage to form cytosine and 2-deoxy-ribono-1,4-lactone (35). The radical resulting from hydration can lead to the formation of a number of products with the C5–C6 bond in cytosine being saturated (30). In dC, the products from the hydration pathway has been shown to be much more abundant than that from the deprotonation pathway (30).
Scheme 3. Hydration and deprotonation of the cation radical of 2′-deoxycytidine.
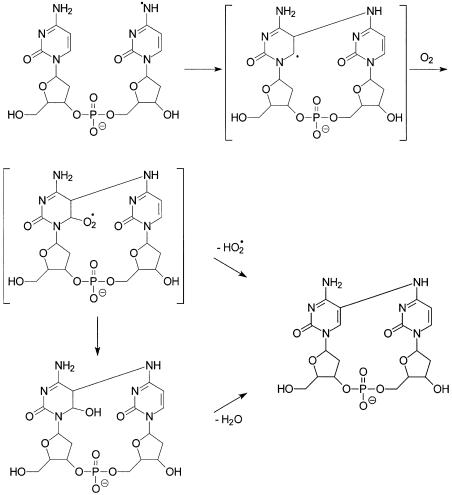
In contrast to the observation with dC, our results with different irradiation time and different concentrations of MQ indicate that the formation of the cross-link product, presumably via the N4 radical of cytosine (Scheme 4), is also a competitive process. We think that the N4 radical formed on the 3′ cytosine can attack the C5 carbon of the 5′ cytosine to give a 5,6-dihydro-6-yl radical, which can combine with molecular oxygen to give a peroxyl radical (31). In this regard, the reaction of a nitrogen-centered radical with the C5 = C6 double bond of uracil or thymine has been observed in previous studies (25,36). The peroxyl radical may lose an HO2 radical to give the cross-link lesion (Scheme 4). The driving force for the latter loss is the regeneration of an aromatic 5′ nucleobase, and the HO2· elimination has been observed in systems with an OH or NH group at the α position to the peroxyl radical (46).
Scheme 4. Proposed mechanism for the formation of d(C∧pC).
Alternatively, the secondary peroxyl radical may transform to give a 6-hydroxy-5,6-dihydro derivative through a tetroxide intermediate (46), and the latter derivative is known to undergo facile dehydration (28), which also gives the cross-link lesion (Scheme 4). Similar as 2′-deoxycytidine glycol, the 6-hydroxy-5,6-dihydrocytosine derivative is expected to undergo not only dehydration, but also deamination (47). Therefore, in addition to the cross-link lesion we identified in this paper, we expect to see the deaminated analog of the cross-link lesion. However, we were unable to find the deamination product. Therefore, the reaction through the 6-hydroxy-5,6-dihydrocytosine derivative appears to be less likely than the pathway through the direct elimination of the HO2 radical.
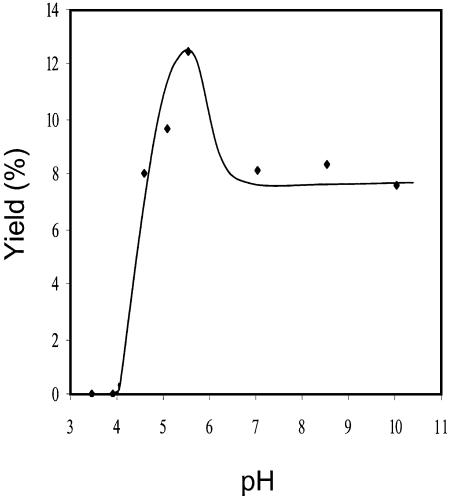
In support of the importance of the deprotonation of the cytosine radical cation in adduct formation, we found that the pH of the irradiation solution has a drastic effect on the formation of the cross-link lesion. We did the photoirradiation in 10 mM phosphate buffer with a pH ranging from 3.4 to 10.0, and strikingly, the cross-link adduct was undetectable while the pH is lower than 4 (Fig. 11), which may indicate that the low pH prevents the deprotonation of the cytosine cation radical. To our knowledge, the pKa of the radical cation of cytosine has not been measured. Based on the above product analysis at different pH, we may infer that the pKa of the radical cation of cytosine in d(CpC) is between 4 and 5. In addition, the yield increases with the increasing of the solution pH while the pH is less than 5.6. While the pH of the solution is between 7 and 10, however, the yield for the formation of the cross-link lesion is more or less constant but is lower than that at pH 5.6. The reason why the yield is lower at a higher pH is not clear at the moment.
Figure 11.
The yields for the formation of d(C∧pC) versus the pH of the solution used for the irradiation [yields were determined by using peak areas in HPLC traces and assuming that the absorption coefficients at λmax are the same for d(CpC) and d(C∧pC)].
Biological implications
One-electron oxidation of DNA has been demonstrated to result in mainly guanine damage because, among the four DNA bases, guanine is the easiest to be oxidized (48,49). Furthermore, the cation radical formed at other sites in duplex DNA may transfer to a guanine base via hole migration (50–52). Therefore, the cross-link lesion that we identified above may not be a biologically significant process in one-electron oxidation of duplex DNA per se because not much N4-centered radical of cytosine is expected to form in duplex DNA under such oxidation condition. In this respect, we did the photoirradiation of a self-complementary 16mer duplex ODN d(CCGGCCGGCCGGCCGG) under similar conditions. After digesting the irradiation mixture with nuclease P1 and alkaline phosphatase, we analyzed the products by LC-MS/MS with the ion-trap mass spectrometer. Our preliminary results indicate that no cross-link lesion formed from this duplex ODN (data not shown).
Recent EPR studies, however, demonstrate that the N4-centered radical of cytosine can also form in hypochlorite-induced damage of DNA and RNA (36,37). More importantly, the propensity for hypochlorite-induced radical formation in both nucleosides and polynucleotides are cytosine > adenine = guanine > uracil = thymine (36,37). EPR experiments also suggest that the pyrimidine-derived, nitrogen-centered radical may add to other nucleobases to give dimers with a radical on carbon atoms (36,37). HOCl is known to be produced in stimulated neutrophils (53–55). The novel cross-link lesion that we identified in this study may have implications in DNA damage induced from oxidants generated by neutrophils. In addition, it has been proposed that the π-cation of N1-substituted cytosine derivatives, which formed from photoionization or by exposure to direct effects of γ irradiation (56,57), can deprotonate at 1′ position or at the amino group. Furthermore, whether the formation of the N4-centered radical in cytosine is significant in metal ion/H2O2-mediated oxidation is unknown. We are currently investigating the formation of the cross-link lesion under those oxidation conditions.
In conclusion, a new oxidative cross-link lesion was isolated from 365-nm irradiation of dinucleoside monophosphate d(CpC) in the presence of MQ as a sensitizer. Proton NMR and exhaustive mass spectrometric characterizations show that the two adjacent cytosines are cross-linked between the C5 carbon atom of the 5′-base and the N4 nitrogen atom of the 3′-base.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Mr Yi Meng and Dr Dan Borchardt for help with the NMR experiment, and Dr Yingjie Ren and Professor John-Stephen A. Taylor for providing us with the procedure for synthesizing dinucleoside monophosphates. Funding by the NIH (grant CA 96909) is gratefully acknowledged.
REFERENCES
- 1.Ames B.N. and Gold,L.S. (1991) Endogenous mutagens and the causes of aging and cancer. Mutat. Res., 250, 3–16. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- 3.Feig D.I., Reid,T.M. and Loeb,L.A. (1994) Reactive oxygen species in tumorigenesis. Cancer Res., 54, 1890s–1894s. [PubMed] [Google Scholar]
- 4.Lindahl T. (1999) DNA lesions generated in vivo by reactive oxygen species, their accumulation and repair. NATO ASI Ser. A, 302, 251–257. [Google Scholar]
- 5.Marnett L.J. (2000) Oxyradicals and DNA damage. Carcinogenesis, 21, 361–370. [DOI] [PubMed] [Google Scholar]
- 6.Schaaper R.M. and Dunn,R.L. (1991) Spontaneous mutation in the Escherichia coli lacI gene. Genetics, 129, 317–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tkeshelashvili L.K., McBride,T., Spence,K. and Loeb,L.A. (1991) Mutation spectrum of copper-induced DNA damage. J. Biol. Chem., 266, 6401–6406. [PubMed] [Google Scholar]
- 8.McBride T.J., Preston,B.D. and Loeb,L.A. (1991) Mutagenic spectrum resulting from DNA damage by oxygen radicals. Biochemistry, 30, 207–213. [DOI] [PubMed] [Google Scholar]
- 9.Reid T.M. and Loeb,L.A. (1992) Mutagenic specificity of oxygen radicals produced by human leukemia cells. Cancer Res., 52, 1082–1086. [PubMed] [Google Scholar]
- 10.Reid T.M. and Loeb,L.A. (1993) Tandem double CC→TT mutations are produced by reactive oxygen species. Proc. Natl Acad. Sci. USA, 90, 3904–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newcomb T.G., Allen,K.J., Tkeshelashvili,L. and Loeb,L.A. (1999) Detection of tandem CC→TT mutations induced by oxygen radicals using mutation-specific PCR. Mutat. Res., 427, 21–30. [DOI] [PubMed] [Google Scholar]
- 12.Tkeshelashvili L.K., Reid,T.M., McBride,T.J. and Loeb,L.A. (1993) Nickel induces a signature mutation for oxygen free radical damage. Cancer Res., 53, 4172–4174. [PubMed] [Google Scholar]
- 13.Brash D.E., Rudolph,J.A., Simon,J.A., Lin,A., McKenna,G.J., Baden,H.P., Halperin,A.J. and Ponten,J. (1991) A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc. Natl Acad. Sci. USA, 88, 10124–10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakazawa H., English,D., Randell,P.L., Nakazawa,K., Martel,N., Armstrong,B.K. and Yamasaki,H. (1994) UV and skin cancer: specific p53 gene mutation in normal skin as a biologically relevant exposure measurement. Proc. Natl Acad. Sci. USA, 91, 360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Box H.C., Budzinski,E.E., Dawidzik,J.D., Wallace,J.C., Evans,M.S. and Gobey,J.S. (1996) Radiation-induced formation of a cross-link between base moieties of deoxyguanosine and thymidine in deoxygenated solutions of d(CpGpTpA). Radiat. Res., 145, 641–643. [PubMed] [Google Scholar]
- 16.Box H.C., Budzinski,E.E., Dawidzik,J.B., Gobey,J.S. and Freund,H.G. (1997) Free radical-induced tandem base damage in DNA oligomers. Free Radic. Biol. Med., 23, 1021–1030. [DOI] [PubMed] [Google Scholar]
- 17.Box H.C., Budzinski,E.E., Dawidzik,J.B., Wallace,J.C. and Iijima,H. (1998) Tandem lesions and other products in X-irradiated DNA oligomers. Radiat. Res., 149, 433–439. [PubMed] [Google Scholar]
- 18.Box H.C., Dawidzik,J.B. and Budzinski,E.E. (2001) Free radical-induced double lesions in DNA. Free Radic. Biol. Med., 31, 856–868. [DOI] [PubMed] [Google Scholar]
- 19.Delatour T., Douki,T., Gasparutto,D., Brochier,M.-C. and Cadet,J. (1998) A novel vicinal lesion obtained from the oxidative photosensitization of TpdG: characterization and mechanistic aspects. Chem. Res. Toxicol., 11, 1005–1013. [DOI] [PubMed] [Google Scholar]
- 20.Romieu A., Bellon,S., Gasparutto,D. and Cadet,J. (2000) Synthesis and UV photolysis of oligodeoxynucleotides that contain 5-(phenylthiomethyl)-2′-deoxyuridine: a specific photolabile precursor of 5-(2′-deoxyuridilyl)methyl radical. Org. Lett., 2, 1085–1088. [DOI] [PubMed] [Google Scholar]
- 21.Bellon S., Ravanat,J.L., Gasparutto,D. and Cadet,J. (2002) Cross-linked thymine-purine base tandem lesions: synthesis, characterization and measurement in gamma-irradiated isolated DNA. Chem. Res. Toxicol., 15, 598–606. [DOI] [PubMed] [Google Scholar]
- 22.Dizdaroglu M. and Simic,M.G. (1984) Radiation-induced crosslinking of cytosine. Radiat. Res., 100, 41–46. [PubMed] [Google Scholar]
- 23.Amesz J. (1973) The function of plastoquinone in photosynthetic electron transport. Biochim. Biophys. Acta, 301, 35–51. [DOI] [PubMed] [Google Scholar]
- 24.Klohn P.-C. and Neumann,H.-G. (1997) Impairment of respiration and oxidative phosphorylation by redox cyclers 2-nitrosofluorene and menadione. Chem.-Biol. Interact., 106, 15–28. [DOI] [PubMed] [Google Scholar]
- 25.Wagner J.R., Cadet,J. and Fisher,G.J. (1984) Photo-oxidation of thymine sensitized by 2-methyl-1,4-naphthoquinone: analysis of products including three novel photo-dimers. Photochem. Photobiol., 40, 589–597. [Google Scholar]
- 26.Wagner J.R., van Lier,J.E. and Johnston,L.J. (1990) Quinone sensitized electron transfer photooxidation of nucleic acids: chemistry of thymine and thymidine radical cations in aqueous solution. Photochem. Photobiol., 52, 333–343. [DOI] [PubMed] [Google Scholar]
- 27.Wagner J.R., van Lier,J.E., Decarroz,C., Berger,M. and Cadet,J. (1990) Photodynamic methods for oxy radical-induced DNA damage. Methods Enzymol., 186, 502–511. [DOI] [PubMed] [Google Scholar]
- 28.Cadet J. and Vigny,P. (1990) In Morrison,H. (ed.), Bioorganic Photochemistry. John Wiley, New York, NY, Vol. 1, pp. 1–272. [Google Scholar]
- 29.Wagner J.R., van Lier,J.E., Berger,M. and Cadet,J. (1994) Thymidine hydroperoxides: structural assignment, conformational features and thermal decomposition in water. J. Am. Chem. Soc., 116, 2235–2242. [Google Scholar]
- 30.Wagner J.R., Decarroz,C., Berger,M. and Cadet,J. (1999) Hydroxyl-radical-induced decomposition of 2′-deoxycytidine in aerated aqueous solutions. J. Am. Chem. Soc., 121, 4101–4110. [Google Scholar]
- 31.Ravanat J.L., Douki,T. and Cadet,J. (2001) Direct and indirect effects of UV radiation on DNA and its components. J. Photochem. Photobiol. B: Biol., 63, 88–102. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y., Liu,Z. and Dixon,C. (2002) Major adenine products from 2-methyl-1,4-naphthoquinone-sensitized photoirradiation at 365 nm. Biochem. Biophys. Res. Commun., 291, 1252–1257. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y. and Liu,Z. (2002) Mechanisms for the formation of major oxidation products of adenine upon 365-nm irradiation with 2-methyl-1,4-naphthoquinone as a sensitizer. J. Org. Chem., 67, 8507–8512. [DOI] [PubMed] [Google Scholar]
- 34.Krishna C.M., Decarroz,C., Wagner,J.R., Cadet,J. and Riesz,P. (1987) Menadione sensitized photooxidation of nucleic acid and protein constituents. An ESR and spin-trapping study. Photochem. Photobiol., 46, 175–182. [DOI] [PubMed] [Google Scholar]
- 35.Decarroz C., Wagner,J.R. and Cadet,J. (1987) Specific deprotonation reactions of the pyrimidine radical cation resulting from the menadione mediated photosensitization of 2′-deoxycytidine. Free Radic. Res. Commun., 2, 295–301. [DOI] [PubMed] [Google Scholar]
- 36.Hawkins C.L. and Davies,M.J. (2001) Hypochlorite-induced damage to nucleosides: formation of chloramines and nitrogen-centered radicals. Chem. Res. Toxicol., 14, 1071–1081. [DOI] [PubMed] [Google Scholar]
- 37.Hawkins C.L. and Davies,M.J. (2002) Hypochlorite-induced damage to DNA, RNA and polynucleotides: formation of chloramines and nitrogen-centered radicals. Chem. Res. Toxicol., 15, 83–92. [DOI] [PubMed] [Google Scholar]
- 38.Niehaus H. and Hildenbrand,K. (2000) Continuous-flow and spin-trapping EPR studies on the reactions of cytidine induced by the sulfate radical-anion in aqueous solution. Evidence for an intermediate radical-cation. J. Chem. Soc., Perkin Trans., 2, 947–952. [Google Scholar]
- 39.Naumov S., Hildenbrand,K. and von Sonntag,C. (2001) Tautomers of the N-centered radical generated by reaction of SO4–. with N1-substituted cytosines in aqueous solution. Calculation of isotropic hyperfine coupling constants by a density functional method. J. Chem. Soc., Perkin Trans., 2, 1648–1653. [Google Scholar]
- 40.Divakar K.J. and Reese,C.B. (1982) 4-(1,2,4-Triazol-1-yl)- and 4-(3-nitro-1,2,4-triazol-1-yl)-1-(.beta.-D-2,3,5-tri-O-acetylarabinofuranosyl)pyrimidin-2(1H)-ones. Valuable intermediates in the synthesis of derivatives of 1-(.beta.-D-arabinofuranosyl)cytosine (ara-C). J. Chem. Soc., Perkin Trans., 1, 1171–1176. [Google Scholar]
- 41.LaFrancois C.J., Fujimoto,J. and Sowers,L.C. (1998) Synthesis and characterization of isotopically enriched pyrimidine deoxynucleoside oxidation damage products. Chem. Res. Toxicol., 11, 75–83. [DOI] [PubMed] [Google Scholar]
- 42.Ti G.S., Gaffney,B.L. and Jones,R.A. (1982) Transient protection: efficient one-flask syntheses of protected deoxynucleosides. J. Am. Chem. Soc., 104, 1316–1319. [Google Scholar]
- 43.Douki T. and Cadet,J. (1999) Modification of DNA bases by photosensitized one-electron oxidation. Int. J. Radiat. Biol., 75, 571–581. [DOI] [PubMed] [Google Scholar]
- 44.Phillips D.R. and McCloskey,J.A. (1993) A comprehensive study of the low energy collision-induced dissociation of dinucleoside monophosphates. Int. J. Mass Spectrom. Ion Proc., 128, 61–82. [Google Scholar]
- 45.Fisher G.J. and Land,E.J. (1983) Photosensitization of pyrimidines by 2-methylnaphthoquinone in water: a laser flash photolysis study. Photochem. Photobiol., 37, 27–32. [DOI] [PubMed] [Google Scholar]
- 46.von Sonntag C. (1987) The Chemical Basis of Radiation Biology. Taylor and Francis, London, UK. [Google Scholar]
- 47.Tremblay S., Douki,T., Cadet,J. and Wagner,J.R. (1999) 2′-Deoxycytidine glycols, a missing link in the free radical-mediated oxidation of DNA. J. Biol. Chem., 274, 20833–20838. [DOI] [PubMed] [Google Scholar]
- 48.Seidel C.A.M., Schulz,A. and Sauer,M.H.M. (1996) Nucleobase-specific quenching of fluorescent dyes. 1. Nucleobase one-electron redox potentials and their correlation with static and dynamic quenching efficiencies. J. Phys. Chem., 100, 5541–5553. [Google Scholar]
- 49.Steenken S. and Jovanovic,S.V. (1997) How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc., 119, 617–618. [Google Scholar]
- 50.Barton J.K. (1998) DNA-mediated electron transfer: chemistry at a distance. Pure Appl. Chem., 70, 873–879. [Google Scholar]
- 51.Schuster G.B. (2000) Long-range charge transfer in DNA: transient structural distortions control the distance dependence. Acc. Chem. Res., 33, 253–260. [DOI] [PubMed] [Google Scholar]
- 52.Giese B. (2002) Long-distance electron transfer through DNA. Annu. Rev. Biochem., 71, 51–70. [DOI] [PubMed] [Google Scholar]
- 53.Harrison J.E. and Schultz,J. (1976) Studies on the chlorinating activity of myeloperoxidase. J. Biol. Chem., 251, 1371–1374. [PubMed] [Google Scholar]
- 54.Foote C.S., Goyne,T.E. and Lehrer,R.I. (1983) Assessment of chlorination by human neutrophils. Nature, 301, 715–716. [DOI] [PubMed] [Google Scholar]
- 55.Albrich J.M., McCarthy,C.A. and Hurst,J.K. (1981) Biological reactivity of hypochlorous acid: implications for microbicidal mechanisms of leukocyte myeloperoxidase. Proc. Natl Acad. Sci. USA, 78, 210–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bernhard W.A. (1981) Solid-state radiation chemistry of DNA: the bases. Adv. Radiat. Biol., 9, 199–280. [Google Scholar]
- 57.Sevilla M.D. (1977) In Pullman,B. and Goldblum,N. (eds), Excited States in Organic Chemistry and Biochemistry. Reidel, Dordrecht, The Netherlands, p. 15. [Google Scholar]