Abstract
The nucleocapsid (NC) protein NCp7 of the immunodeficiency virus type 1 is a small basic protein with two zinc finger motifs. NCp7 has key roles in virus replication and structure, which rely on its interactions with nucleic acids. Although most interactions involve RNAs, binding to the viral DNA is thought to be of importance to achieve protection of the DNA against cellular nucleases and its integration into the host genome. We investigated the interaction of NCp7 with plasmid DNA as a model system. The fluorescence probe YOYO-1 was used as the reporter. Binding of NCp7 to DNA caused DNA condensation, as inferred from the dramatic decrease in YOYO-1 fluorescence. Efficient condensation of DNA required the full length NCp7 with the zinc fingers. The fingerless peptide was less efficient in condensing DNA. Binding of both these NC peptides led to freezing of the segmental dynamics of DNA as revealed by anisotropy decay kinetics of YOYO-1. The truncated peptide NC(12–55) which retains the zinc fingers did not lead to DNA condensation despite its ability to bind and partially freeze the segmental motion of DNA. We propose that the histone-like property of NCp7 leading to DNA condensation contributes to viral DNA stability, in vivo.
INTRODUCTION
The nucleocapsid (NC) protein, NCp7, of the immunodeficiency virus type 1 (HIV-1) is a small basic protein characterized by two copies of a highly conserved retroviral-type zinc finger CCHC motif, which binds Zn2+ with high affinity (1). About 2000 copies of NCp7 coat the genomic RNA in the infectious virus (2,3) and protect the genomic RNA against degradation by RNAases (4). Moreover, NCp7, as the mature protein or as the Pr55 polyprotein precursor, plays critical roles in several steps of the viral life cycle. Notably, NCp7 specifically interacts with the Ψ sequence of the genomic RNA, enabling its selective encapsidation into assembling particles (5–7). NCp7 is also critical for the efficient and complete proviral DNA synthesis by promoting both the initiation and the two strand transfer steps during reverse transcription (8–17). This is related to the NCp7 chaperone properties that enable rearrangement of a nucleic acid molecule into the most stable conformation by catalyzing the breakage and re-formation of base pairs (18–21).
The major functions of NCp7 rely on its specific and non-specific interactions with nucleic acids. Initially, based on the strong affinity of murine leukemia virus nucleocapsid protein NCp10 for single-stranded DNA-cellulose and its weak affinity for double-stranded DNA-cellulose, retroviral NCs have been considered as single-stranded nucleic acid-binding proteins (22,23). Later, this idea was challenged by the observation of significant affinity of NCp7 for various double-stranded DNA sequences (24–26). Moreover, addition of NCp7 to plasmid DNA at a ratio of one molecule per 4 bp caused a complete protection from restriction nuclease digestion, suggesting that DNA is coated by the NC protein (24). Both the binding extent and the level of protection were found to depend upon the presence of both the zinc fingers and the flanking clusters of basic amino acids (24,27–31). From these observations and recent in vivo studies, it has been inferred that NCp7 binds to the newly synthesized viral DNA and protects it from cellular nucleases during reverse transcription and its transport to the nucleus up to the integration step (16,17,28,29).
With the aim of exploring the relationships between HIV-1 NCp7 and the proviral DNA we chose to characterize the structure and dynamics of complexes resulting from the interaction of NCp7 with a double-stranded plasmid DNA, taken as a model system. By using the bis-intercalating fluorescent dye YOYO-1 (32), we analyzed DNA condensation and dynamics, as well as the exposure of base pairs to solvent. We find that binding of NCp7 to DNA resulted in DNA condensation in a manner similar to that brought out by gene delivery vectors, freezing of the nanosecond segmental dynamics of DNA and protection of base pairs from solvent. Moreover, using three NCp7 derivatives (Fig. 1A), we explored the comparative influence of the zinc fingers and the flanking basic domains on the structural features of the complex.
Figure 1.
(A) NC peptides used in the present study. (B) Fluorescence emission spectra of DNA-bound YOYO-1 in the presence of NCp7 [NC(1–72)]. Samples had 400 nM of DNA phosphate, 8 nM of YOYO-1 (dye:phosphate = 1:50) in 100 mM NaCl, 20 mM HEPES at pH 7.4. (a) control; (b) 0.1 µM NC(1–72) and (c) 0.4µM NC(1–72). The excitation was at 470 nm.
MATERIALS AND METHODS
Materials
The plasmid DNA pCMV-luc (5.2 kb) was propagated and purified as described (35). Branched chain polyethylenimine (PEI) (25 kDa) was a gift from Professor J.-P. Behr, Illkirch, France. YOYO-1 (491/509) was obtained from Molecular Probes Inc. NCp7 and the related peptides were synthesized by solid-phase chemistry as described (36). Purity of the peptides analyzed by mass spectrometry was >98%. Peptide concentration was determined on a Cary 400 spectrophotometer, using an extinction coefficient at 280 nm of 12 700, 7000 and 5700 M–1 cm–1 for NC(1–72), NC(1–72)dd and NC(12–55), respectively. To protect the highly oxidizable Cys residues of NC(1–72) and NC(12–55), the peptides were stored lyophilized in their zinc-bound forms.
Samples
Plasmid DNA–YOYO-1 complexes were made by mixing equal volumes of solutions of DNA and YOYO-1 in 100 mM NaCl, 20 mM HEPES, pH 7.4. The mixed solution was incubated at room temperature for at least 5 h before use. This procedure ensured uniform distribution of YOYO-1, which tightly binds to DNA. Complexes of plasmid DNA–YOYO-1 with NCp7 and its derivatives were formed by adding the peptide to a solution of DNA–YOYO-1 complex. In the case of equilibrium measurements the samples were kept for at least 90 min for completion of condensation.
Spectroscopic measurements
An SLM 48000 spectrofluorimeter was used for steady-state fluorescence measurements.
Time-resolved fluorescence intensity and anisotropy measurements on DNA–YOYO-1–NCp7 complexes were performed by using the frequency doubled output of a Ti-Sapphire laser (Spectra Physics). The excitation wavelength was 480 nm and the emission was collected at 515 nm by using a single photon counting microchannel plate photomultiplier (Hamamatsu R3809U). Other details are given elsewhere (37). The resolution of the time-correlated single photon counting set-up was 25.5 ps and the width of the instrument response function was ∼40 ps.
Fluorescence intensity decays obtained at the magic angle were deconvoluted with the instrument response function and analyzed as a sum of exponentials:
I (t) = Σ αi exp (–t/τi)
where I (t) is the fluorescence intensity collected at the magic angle at time t and αi is the amplitude of the ith lifetime τi such that Σ αi = 1.
Time-resolved fluorescence anisotropy decays were analyzed by the following equations
I|| (t) = I(t) [1 + 2r(t)]/3 1
I⊥ (t) = I(t) [1 – r(t)]/3 2
r(t) = [I||(t) – I⊥ (t)]/[Ill(t) + 2I⊥(t)] = r0 Σ βi exp(–t/φi) 3
where r0 is the initial anisotropy and βi is the amplitude of the ith rotational correlation time φi such that Σ βi = 1. I|| and I⊥ are the intensities collected at emission polarizations parallel and perpendicular, respectively, to the polarization axis of the excitation beam. While analyzing the kinetics of anisotropy decay using equations 1 and 2, values of αi and τi [obtained from the analysis of I(t)] were kept fixed in order to reduce the number of floating parameters. This procedure results in better estimates of anisotropy decay parameters.
RESULTS
Condensation of DNA by NCp7
Condensation of plasmid DNA by NCp7 was monitored by following the fluorescence of the bis-intercalating fluorophore YOYO-1. Indeed, we have recently shown that YOYO-1 is a simple and very sensitive probe for monitoring the condensation of DNA by a variety of condensing agents (32). The quantum yield of YOYO-1 increases by several orders of magnitude during the transition from aqueous solution to its intercalating site in double-stranded DNA (32). Subsequently, when the double-stranded DNA undergoes the transition from extended (either supercoiled or relaxed) conformation to a condensed form catalyzed by a condensing agent, the quantum yield of bound YOYO-1 undergoes a drastic reduction (32). Furthermore, we had shown that this reduction is a result of a combination of formation H-dimers of YOYO-1 and fluorescence resonance energy transfer between these dimers and YOYO-1 monomer (32).
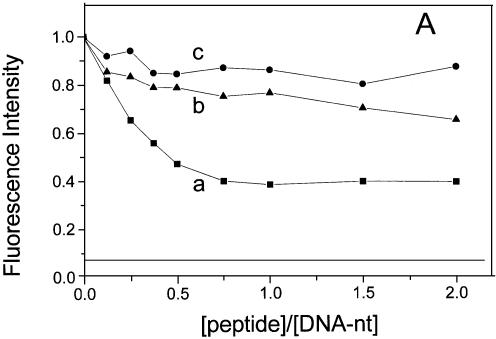
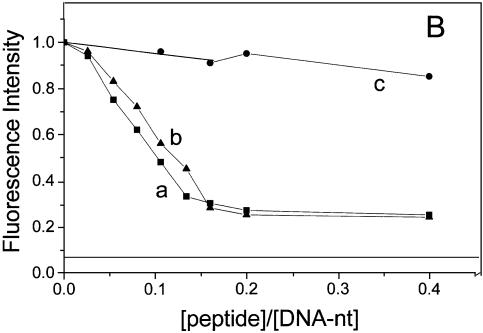
Figure 1B shows the effect of NCp7 on the fluorescence emission spectra of YOYO-1 intercalated into the DNA. In these samples the ratio of YOYO-1 to DNA phosphate was kept at 1:50 in order to visualize the condensation effect (32). In analogy to DNA-condensing agents such as PEI or cationic detergents (32), NCp7 induced a dramatic reduction in the quantum yield of YOYO-1. Figure 2 shows the titration of the fluorescence intensity with NCp7 and its derivatives at two different DNA concentrations. A progressive decrease of YOYO-1 fluorescence was observed with increasing NC(1–72) concentrations. This decrease levelled off at a plateau value that slightly depends on the oligonucleotide (nt) concentration. In contrast to NC(1–72), the NC(1–72)dd peptide, which lacks the zinc fingers, was able to condense DNA only at high concentrations of DNA. Moreover, NC(12–55), which has the zinc fingers but lacks the regions 1–11 and 56–72, was unable to condense DNA at all the concentrations used (Fig. 2A and B). Figure 2 also shows the effect of DNA condensation by PEI for comparison (horizontal line at 0.07 in Fig. 2). It can be seen that the maximum extent of fluorescence quenching by NC (65–75%) was less when compared with the >90% quenching seen with PEI. Noticeably, the observed variability in the maximum extent of fluorescence quenching by NC is probably related to the variability in the distribution of size of the nucleoprotein complexes from one experiment to another. That the NC caused a dramatic reduction in fluorescence intensity (Fig. 1B) arises from condensation of DNA and not from a direct effect of protein binding as shown by the following observation: quenching was not observed when the YOYO-1 to DNA phosphate ratio was reduced from 1:50 to 1:2000 (see for example, the mean lifetime in Table 1). These observations are similar to those seen earlier with either PEI or CTAB (32). Furthermore, direct support for condensation of DNA by the binding of NC(1–72) comes from the following observations also: (i) binding of NC to DNA leads to the formation of high molecular mass nucleoprotein complexes, which can be pelleted by centrifugation (33); and (ii) dark field electron micrographs obtained in samples having NC and DNA shows the presence of nucleoprotein complexes as dense aggregates having diameter in the range of a few hundred nanometers (34). Thus, these observations when combined with our present work indicate that YOYO-1 fluorescence is a convenient spectroscopic marker for DNA condensation.
Figure 2.
Titration of fluorescence intensity of DNA-bound YOYO-1 with NCp7 and the related peptides. (a) NC(1–72); (b) NC(1–72)dd and (c) NC(12–55). The concentration of DNA phosphate was 400 nM in (A) and 5 µM in (B). Dye:phosphate ratio was kept at 1:50. Fluorescence intensities refer to equilibrium values. The horizontal lines at 0.07 correspond to PEI–DNA complex having the ratio, of primary N of PEI to DNA phosphate, of 10.
Table 1. Parameters associated with decay of fluorescence intensity and anisotropy.
| Samplea | Fluorescence lifetimes, ns (amplitude)b | Rotational correlation times, ns (amplitude)c | ||||
|---|---|---|---|---|---|---|
| |
τ1 (α1) |
τ2 (α2) |
τmeand |
φ1e (β1) |
φ2 (β2) |
φ3 (β3) |
| 1. DNA–YOYO-1 uncondensed | 5.1 (0.50) | 2.35 (0.50) | 3.76 | <0.1 (0.32) | 0.63 (0.12) | 11.1 (0.56) |
| 2. DNA–YOYO-1 + NC(1–72) | 4.65 (0.43) | 2.05 (0.57) | 3.18 | <0.1 (0.41) | 0.98 (0.11) | >100 (0.48) |
| 3. DNA–YOYO-1+ NC(1–72)dd | 5.25 (0.69) | 1.90 (0.31) | 4.21 | <0.1 (0.38) | 1.81 (0.12) | >100 (0.50) |
| 4. DNA–YOYO-1+ NC(12–55) | 5.20 (0.42) | 2.46 (0.58) | 3.62 | <0.1 (0.40) | 0.42 (0.11) | 33.5 (0.49) |
aThe concentration of plasmid DNA was 10 µM of phosphate and that of YOYO-1 was 5 nM for all the samples. The concentration of NC peptides was 4 µM. The medium was 20 mM HEPES, 100 mM NaCl at pH 7.4.
bThe errors associated with various parameters were as follows: τ1 (±0.1), τ2 (±0.2), α1 (±0.02), α2 (±0.02); φ2 (±0.30), φ3 (±4.0), β1 (±0.03), β2 (±0.03) and β3 (±0.03). The value of reduced χ2 was in the range of 1.0–1.3 in all the cases. Also, the pattern of residuals was found to be random for all the fits.
cInitial anisotropy r0 was fixed at 0.31 ± 0.01, the value obtained for YOYO-1 in glycerol, during the analysis.
dMean lifetime τmean = Σ αi τi.
eSince the uncertainty in the value of φ1 was too large only the upper limit is given.
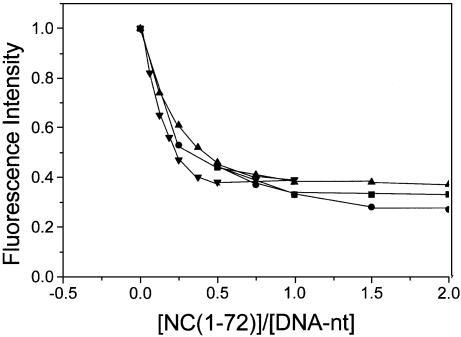
By further investigating the dependence of the extent of DNA condensation on the concentrations of NC(1–72) and DNA-nt (Fig. 3), we found that the profile of condensation largely depended on the DNA to NC molar ratio. Nearly 80% of the total decrease in fluorescence intensity occurred at [NC(1–72)]/[DNA-nt] approximately 0.2 at all the concentrations of DNA-nt (0.1–0.8 µM) used. This would suggest an apparent binding site of 2–3 bp per NCp7 molecule, which is significantly different from the binding site size of 9 bp estimated earlier (26). However, the slight deviation of the titration curves from each other (Fig. 3) could be interpreted as an indication of a deviation from stoichiometric binding. Such non-stoichiometric binding is expected to lead to the presence of unbound NC indicating that the apparent dissociation constant of NC(1–72) for plasmid DNA is in the micromolar range. However, it should be borne in mind that the observed decrease in fluorescence intensity (Figs 1–3) arose from condensation of DNA (32) subsequent to the binding of NC and not due to the binding of NC to DNA per se (see ‘Discussion’). Hence, data presented in Figures 1–3 cannot be rigorously used for the estimation of binding site size and binding constant.
Figure 3.
Titration of fluorescence intensity of DNA-bound YOYO-1 with NC(1–72) at various DNA concentrations. DNA phosphate concentrations were 0.1 µM (squares); 0.2 µM (circles); 0.4 µM (triangles) and 0.8µM (inverted triangles). Dye:phosphate ratio was 1:50.
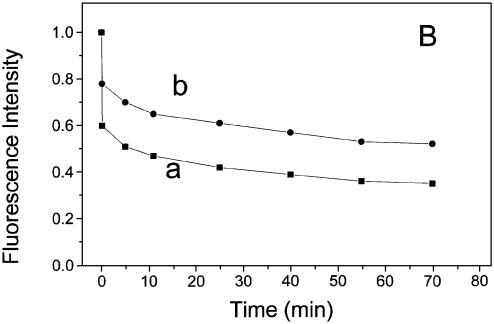
Figure 4 shows a typical time-dependence of the condensation process induced by either NC(1–72) or the fingerless peptide NC(1–72)dd. The time dependence shows that condensation is a two-step process: one that occurred within the mixing time of a few seconds and another with a half time or ∼5–20 min. The rate of the slow phase and the extent of contribution of the fast phase were higher in the case of NC(1–72) when compared with NC(1–72)dd. Furthermore, the rate of the slow phase showed only a small dependence on the concentration of DNA (compare Fig. 4A and B), suggesting that the slow phase may not represent a bimolecular process of binding of NC to the DNA. A bimolecular process would have resulted in a 5-fold decrease in the rate of the slow process when the concentration of DNA was decreased by a factor of 5. In contrast, the decrease in the apparent rate was significantly <5-fold. However, we cannot rule out, during the slow phase, a more complex process, which involves coming together of several complexes of DNA–NC in forming a large nucleoprotein structure.
Figure 4.
Time dependence of DNA condensation process monitored through YOYO-1 fluorescence intensity. At zero time DNA–YOYO-1 complex [either 5 µM (A) or 1 µM (B) of DNA phosphate] was mixed with 1 µM of either NC(1–72) (a) or NC(1–72)dd (b). Dye:phosphate ratio was kept at 1:50. Other conditions were same as in Figure 1B. Fast and slow phases of the condensation process could be seen.
Base solvent-accessibility and dynamics of NCp7–DNA complexes
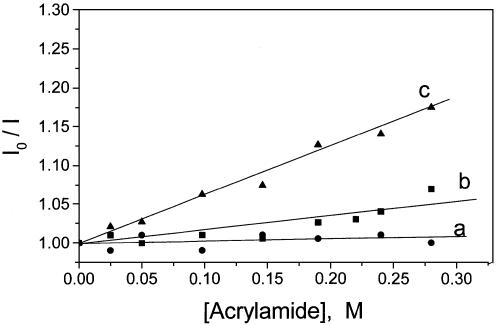
In order to check whether the bases get exposed to solvent in the condensed form of DNA, quenching of YOYO-1 fluorescence, intercalated into DNA bases by acrylamide, was monitored (Fig. 5). Solvent exposure of DNA bases could be estimated from the level of quenching of fluorescence by acrylamide (32). Although the accessibility of intercalated YOYO-1 to acrylamide was very low in both the extended and condensed forms of DNA (as shown by the very low levels of quenching), a small but reproducible increase of base exposure to solvent (as observed from the increase in slope) was observed in the presence of NCp7 (Fig. 5). In contrast to NCp7, condensation by PEI, a cross-linked polymer used in gene delivery systems, led to a much larger exposure of bases (Fig. 5) as seen earlier (32). In these experiments the ratio of YOYO-1 to oligonucleotides was kept very low (1:2000) to ensure that YOYO-1 fluorescence mainly reflected the changes in the property of individual YOYO-1 molecule rather than reflecting condensation-induced interaction among bound YOYO-1 molecules (32) seen at a high ratio such as 1:50 (Figs 1–4). Under the former condition, fluorescence lifetime (τ) did not change significantly on condensation (see later) and hence any change in slope (= KSV = τ kQ) of plots in Figure 5 reflects mainly change in accessibility (represented by the quenching constant, kQ) of YOYO-1 to acrylamide (32).
Figure 5.
Stern-Volmer plots associated with the quenching of DNA-bound YOYO-1 (10 µM DNA phosphate and 5 nM YOYO-1) by acrylamide. (Dye:phosphate ratio = 1:2000.) I0 and I are the fluorescence intensities in the absence and in the presence of acrylamide. (a) Control, slope, KSV ∼0; (b) 6 µM NCp7, KSV = 0.15 M–1; and (c) PEI at N:P = 1:5, KSV = 0.6 M–1.
Figure 6 shows the rotational dynamics of bound YOYO-1 as seen from the kinetics of decay of fluorescence anisotropy. These decay kinetics were fitted satisfactorily to a sum (equation 3) of three exponentials resulting in three correlation times (Table 1) in all the cases. The very short rotational correlation time (φ1 < 0.1 ns) arises due to the necessity of fixing the value of initial anisotropy (r0) at 0.31 ± 0.01, which was obtained for YOYO-1 in a viscous solvent. It is likely that φ1 represents the internal fast rotation of YOYO-1 and φ2 and φ3 originate from segmental dynamics of DNA backbone (32). It can be seen that the observable part of anisotropy decay kinetics is largely dominated by φ3. Hence the value of φ3 could be used in characterizing the changes in motional dynamics of DNA backbone brought about by the binding of NCp7 and its derivatives. In the uncondensed form of DNA, its backbone is highly dynamic as shown by the value of φ3 ∼11 ns similar to our earlier observations (32). Binding of NCp7 and the related peptides resulted in dramatic changes in the anisotropy decay profile reflecting changes in the motional dynamics of the DNA backbone (Fig. 6). Both NC(1–72) and the fingerless NC(1–72)dd caused significant decrease in the rate of anisotropy decay as shown by significant increase in the value of φ3 (>100 ns, this limit being the result of the limited time window offered by the fluorescence lifetime of YOYO-1) (Fig. 6 and Table 1). This could be interpreted as significant rigidification of DNA upon binding of either NC(1–72) or NC(1–72)dd. However, binding of the truncated peptide NC(12–55) retarded the decay of anisotropy (and hence DNA flexibility) to a lesser extent as shown by φ3 ∼33 ns (Fig. 6 and Table 1). However, this significant retardation effect on the rotational dynamics [φ3 ∼33 ns when compared with 11 ns in the case of NC(1–72)] is in contrast with the inability of NC(12–55) to elicit the condensation signal (Fig. 2). The ratio of YOYO-1 to DNA-nt was quite low (1:2000) in these samples for reasons mentioned above.
Figure 6.
Decay of fluorescence anisotropy of DNA-bound YOYO-1. The concentrations of DNA phosphate and YOYO-1 were 10 µM and 5 nM, respectively (dye:phosphate = 1:2000). (A) Control; (B) with 4 µM of NC(1–72); (C) with 4 µM of NC(1–72)dd; and (D) with 4 µM of NC(12–55). Parameters obtained by analysis of these traces by equation 1 are given in Table 11. Smooth lines correspond to fits according to the listed parameters.
Fluorescence intensity decay curves were satisfactorily fitted to a sum of two exponentials in all the bound forms (Table 1) similar to the behavior in uncondensed DNA (32). Furthermore, intensity decay kinetics were largely insensitive to the binding of NC peptides although some minor changes were seen (Table 1). Thus, the effect of binding NC peptides was seen mainly in anisotropy decay kinetics rather than in their fluorescence lifetimes. Concomitantly, fluorescence intensity also did not show any change on binding of NC peptides at these low levels of YOYO-1 (1:2000).
DISCUSSION
In this work we have shown by using the new DNA condensation probe YOYO-1 (32) that double-stranded DNA can be effectively condensed upon binding of HIV-1 NCp7 and its fingerless (without Zn2+) mutant. Our results (Fig. 2) point out that efficient condensation of DNA by NC requires the complete NC with the zinc fingers. The requirement for a complete NC is probably due to the fact that basic amino acids, expected to be a major driving force for condensation (through electrostatic interactions with the negatively charged DNA phosphate groups) are present fairly uniformly over the entire NC molecule (Fig. 1A). The requirement for the zinc fingers may be considered as counterintuitive especially when one notes that basic polypeptides and polycations are efficient condensing agents (32,38,39). The zinc finger-induced enhancement of condensation probably originates from specific interaction(s) between zinc finger side chain(s) and DNA bases. Stacking interaction of Trp37 side chain with bases (notably G) in DNA or RNA sequences as evidenced by NMR studies (40–42) was found to strongly contribute to the binding energy (26,43–45). Furthermore, the observation that zinc finger binding caused DNA bending (46) suggests that unique interactions are taking place between the zinc fingers and the DNA.
Moreover, the observation that NC(12–55) was unable to elicit the condensation signal (Fig. 2) shows that the basic domain of NCp7 is required for condensation. Since previous studies have shown only limited differences between the binding constants of NC(12–55) and NCp7 at 0.1 M NaCl concentration (43,44), the inability of NC(12–55) to condense DNA is probably unrelated to its binding energy. In addition, clear evidence of NC(12–55) binding to DNA came from the significant changes in the anisotropy decay parameters that followed addition of NC(12–55) to DNA (Fig. 6D and Table 1). The ability of the various NC derivatives to condense DNA correlates well with their ability to protect DNA from exonuclease digestion. While NC(1–72) was found to achieve a good protection toward nucleases (24), only a partial protection was observed with NC(1–72)dd (24) or NC mutants with imutated finger motifs (16,17,28,29) and almost no protection was afforded by NC(12–55) (Lener and Darlix, unpublished data). Moreover, in line with the hypothesis of Carteau et al. (47), NC-induced condensation of DNA may play a critical role in promoting proviral DNA integration, and notably the coupled joining of the two ends of the viral DNA at precise positions in the host cell DNA. In fact, condensation of duplex DNA by NC might facilitate association of the two proviral DNA ends and facilitate coupled integration. Tight packing of DNA has been proposed to assist end ligation by repair enzymes in radiation-resistant bacteria (48) supporting our model of NC involvement in proviral DNA integration.
Integration of the proviral DNA into the host genome may further require NC chaperone activity and notably, its nucleic acid destabilizing property. A limited destabilization of plasmid DNA by NC was evidenced by the slight increased level of accessibility of intercalated YOYO to acrylamide in NC(1–72)–DNA complexes (as shown by the increase in slope and KSV, Fig. 5) when compared with bare DNA. This destabilizing property of NCp7 on DNA is in line with previous observations with either short (25) or long duplexes (49) or hairpin loops (50–52). Both the nucleic acid condensing and destabilizing properties of NC and their consequences on the protection of the viral DNA during its nuclear import and subsequent integration may well explain why HIV-1 with mutations in the zinc fingers or in the N-terminal basic domain of NC are replication defective (15–17,28,30).
How compact is the DNA condensed by NCp7? The observation that the extent of reduction in the fluorescence intensity of YOYO-1 (Figs 2 and 3) on condensation by NCp7 is only ∼70%, which is less than the 90% reduction observed with PEI (Fig. 2) (32), suggests that the compactness of the DNA condensed by NCp7 is comparatively less. Furthermore, condensation by either NCp7 or NC(1–72)dd results in freezing of the nanosecond segmental dynamics of DNA (Fig. 6 and Table 1). The level of rigidification of the DNA backbone was higher in the cases of condensation-competent peptides NC(1–72) and NC(1–72)dd when compared with the incompetent analog NC(12–55). Further, the level of rigidification was similar to that observed with PEI (32) indicating this as one of the common features of DNA condensed by cationic polymers.
Interestingly, the condensation activity of NCp7 and the related peptides correlates also with the ability to form high molecular mass complexes with nucleic acids (53,54). For example the inability of NC(12–55) to give the condensation signal is in line with its inability to form high molecular mass complexes with polyA (53) and tRNALys (55) and various genomic RNA fragments (data not shown). Similarly, the reduced rate of DNA condensation by NC(1–72)dd [when compared with NC(1–72)] (Fig. 4) agrees with the reduced rate of formation of high molecular mass complexes (53). This suggests that the processes of condensation and formation of high molecular mass complexes are correlated with each other and probably condensation precedes formation of high molecular mass complexes. This conclusion is supported by the observation that the time course of the condensation process (half time ∼20 min, Fig. 4) is significantly shorter than the time course of high molecular mass complex formation (53), which takes several hundred minutes, although the two processes were monitored with different nucleic acids. The former process could be an intramolecular one wherein the plasmid DNA, on binding to NCp7, collapses into a globule. The inability of the truncated form NC(12–55) to either condense the DNA or form high molecular mass complexes (in spite of its ability to bind) is probably due to the absence of positively charged N- and C-terminal domains. Thus, we infer that efficient formation of compact nucleoprotein globules requires both the basic residues of the N- and C-terminal regions of NCp7 and the DNA bending ability offered by the zinc finger domain. The latter suggestion requires further investigation.
In conclusion, the DNA condensing properties and the resulting DNA rigidification evidenced in this study may confer to NCp7, a histone-like activity that is in line with its critical role of NCp7 in protecting the neosynthesized DNA from cellular nucleases and during its transport to the nucleus up to the integration step (2,16,17,24,28,29). In addition, since similar condensing properties have been inferred for model RNAs (53,54) and since efficient reverse transcription has been described within large ribonucleoprotein complexes reconstituted in vitro (4), it is likely that the condensing properties of NCp7 are relevant for additional NCp7 activities in the viral life cycle.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Mr David Lleres for the isolation and characterization of plasmid DNA, Drs Elisa Bombarda and Serena Bernacchi for zinc complexation and characterization of NC peptides and Professor N. Periasamy (TIFR, Mumbai) for providing us with the software used in the analysis of time-resolved fluorescence data. G.K. was a fellow from the Ministère de la Recherche. This work was supported by the Agence Nationale de Recherches sur le SIDA (ANRS), Sidaction, University Louis Pasteur, CNRS and the Association Francaise contre les Myopathies.
REFERENCES
- 1.Mély Y., De Rocquigny,H., Morellet,N., Roques,B.P. and Gérard,D. (1996) Zinc binding to the HIV-1 nucleocapsid protein: a thermodynamic investigation by fluorescence spectroscopy. Biochemistry, 35, 5175–5182. [DOI] [PubMed] [Google Scholar]
- 2.Darlix J.L., Lapadat-Tapolsky,M., de Rocquigny,H. and Roques,B.P. (1995) First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol., 254, 523–537. [DOI] [PubMed] [Google Scholar]
- 3.Coffin J.M. (1984) Structure of the retroviral genome. In Weiss,R., Teich,N., Varmus,H. and Coffin,J. (eds), RNA Tumor Viruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 261–368. [Google Scholar]
- 4.Tanchou V., Gabus,C., Rogemond,V. and Darlix,J.L. (1995) Formation of stable and functional HIV-1 nucleoprotein complexes in vitro. J. Mol. Biol., 252, 563–571. [DOI] [PubMed] [Google Scholar]
- 5.Aldovini A. and Young,R.A. (1990) Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol., 64, 1920–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkowitz R.D., Luban,J. and Goff,S.P. (1993) Specific binding of human immunodeficiency virus type 1 gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J. Virol., 67, 7190–7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkowitz R.D. and Goff,S.P. (1994) Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology, 202, 233–246. [DOI] [PubMed] [Google Scholar]
- 8.Barat C., Lullien,V., Schatz,O., Keith,G., Nugeyre,M.T. and Gruninger-Leitch,F. (1989) HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J., 8, 3279–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lapadat-Tapolsky M., Pernelle,C., Borie,C. and Darlix,J.L. (1995) Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res., 23, 2434–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu W., Gorelick,R.J. and Rein,A. (1994) Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol., 68, 5013–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allain B., Lapadat-Tapolsky,M., Berlioz,C. and Darlix,J.L. (1994) Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J., 13, 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo J., Henderson,L.E., Bess,J., Kane,B. and Levin,J.G. (1997) Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self priming from minus-strand strong-stop DNA. J. Virol., 71, 5178–5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J.K., Palaniappan,C., Wu,W., Fay,P.J. and Bambara,R.A. (1997) Evidence for a unique mechanism of strand transfer from the transactivation response region of HIV-1. J. Biol. Chem., 272, 16769–16777. [DOI] [PubMed] [Google Scholar]
- 14.You J.C. and McHenry,C.S. (1994) Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J. Biol. Chem., 269, 31491–31495. [PubMed] [Google Scholar]
- 15.Gorelick R.J., Gagliardi,T.D., Bosche,W.J., Wiltrout,T.A., Coren,L.V. and Chabot,D.J. (1999). Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences. Virology, 256, 92–104. [DOI] [PubMed] [Google Scholar]
- 16.Morellet N., Bouaziz,S., Petitjean,P. and Roques,B.P. (2003) NMR structure of the HIV-1 regulatory protein. J. Mol. Biol., 327, 215–227. [DOI] [PubMed] [Google Scholar]
- 17.Demene H., Dong,C.J., Ottmann,M., Rouyez,M.C., Jullian,N., Morellet,N., Mely,Y., Darlix,J.-L., Fournie-Zaluski,M.C., Saragosti,S. and Roques,B.P. (1994). 1H NMR structure and biological studies of the His23→Cys mutant nucleoprotein of HIV-1 indicate that the conformation of the first Zinc finger is critical for virus infection. Biochemistry, 33, 11707–11716. [DOI] [PubMed] [Google Scholar]
- 18.Herschlag D., Khosla,M., Tsuchihashi,Z. and Karpel,R.L. (1994) An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J., 13, 2913–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herschlag D. (1995) RNA chaperones and the RNA folding problem. J. Biol. Chem., 270, 20871–20874. [DOI] [PubMed] [Google Scholar]
- 20.Tsuchihashi Z. and Brown,P.O. (1994) DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol., 68, 5863–5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rein A., Henderson,L.E. and Levin,J.G. (1998) Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci., 23, 297–301. [DOI] [PubMed] [Google Scholar]
- 22.Davis J., Scherer,M., Tsai,W.P. and Long,C. (1976) Low-molecular-weight Rauscher leukemia virus protein with preferential binding for single-stranded RNA and DNA. J Virol., 18, 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sykora K.W. and Moelling,K. (1981) Properties of the avian viral protein p12. J. Gen. Virol., 55, 379–391. [DOI] [PubMed] [Google Scholar]
- 24.Lapadat-Tapolsky M., De Rocquigny,H., Van Gent,D., Roques,B., Plasterk,R. and Darlix,J.L. (1993) Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res., 21, 831–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urbaneja M.A., Wu,M., Casas-Finet,J.R. and Karpel,R.L. (2002) HIV-1 nucleocapsid protein as a nucleic acid chaperone: spectroscopic study of its helix-destabilizing properties, structural binding specificity and annealing activity. J. Mol. Biol., 318, 749–764. [DOI] [PubMed] [Google Scholar]
- 26.Maki A.H., Ozarowski,A., Misra,A., Urbaneja,M.A. and Casas-Finet,J.R. (2001) Phosphorescence and optically detected magnetic resonance of HIV-1 nucleocapsid protein complexes with stem–loop sequences of the genomic Psi-recognition element. Biochemistry, 40, 1403–1412. [DOI] [PubMed] [Google Scholar]
- 27.Tanchou V., Decimo,D., Pechoux,C., Lener,D., Rogemond,V., Berthoux,L., Ottmann,M. and Darlix,J.L. (1998) Role of the N-terminal zinc finger of human immunodeficiency virus type 1 nucleocapsid protein in virus structure and replication. J. Virol., 72, 4442–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buckman J.S., Bosche,W.J. and Gorelick,R.J. (2003) Human immunodeficiency virus type 1 nucleocapsid zinc fingers are required for efficient reverse transcription, initial integration process and protection of newly synthesized viral DNA. J. Virol., 77, 1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao K., Gorelick,R.J., Johnson,D.G. and Bushman,F. (2003) Cofactors for human immunodeficiency virus type 1 cDNA integration in vitro. J. Virol., 77, 1598–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berthoux L., Péchoux,C., Ottmann,M., Morel,G. and Darlix,J.-L. (1997) Mutations in the N-terminal domain of HIV-1 nucleocapsid protein affect virion core structure and proviral DNA synthesis. J. Virol., 71, 6937–6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Rocquigny H., Gabus,C., Vincent,A., Fournie-Zaluski,M.C., Roques,B. and Darlix,J.L. (1992) Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the Zinc fingers. Proc. Natl Acad. Sci. USA, 89, 6472–6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krishnamoorthy G., Duportail,G. and Mely,Y. (2002) Structure and dynamics of condensed DNA probed by 1,1′-(4,4,8,8-tetramethyl-4,8-diazaundecamethylene)bis[4-[[3-methylbenz-1,3-oxazol-2-yl]methylidine]-1,4-dihydroquinolinium] tetraiodide fluorescence. Biochemistry, 41, 15277–15287. [DOI] [PubMed] [Google Scholar]
- 33.Poljak L., Batson,S.M., Ficheux,D., Roques,B.P., Darlix,J.-L. and Kas,E. (2003) Analysis of NCp7-dependent activation of HIV-1 cDNA integration and its conservation among retroviral nucleocapsid proteins. J. Mol. Biol., 329, 411–421. [DOI] [PubMed] [Google Scholar]
- 34.Gabus C., Auxilian,S., Pechoux,C., Dormont,D., Swietnicki,W., Morillas,M., Surewicz,W., Nandi,P. and Darlix,J.-L. (2001) The prion protein has DNA strand transfer properties similar to retroviral nucleocapsid protein. J. Mol. Biol., 307, 1011–1021. [DOI] [PubMed] [Google Scholar]
- 35.Boussif O., Lezoualc’h,F., Zanata,M.A., Mergny,M.D., Scerman,D., Demeneix,B. and Behr,J.-P. (1995) A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl Acad. Sci. USA, 92, 7297–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Rocquigny H., Ficheux,D., Gabus,C., Fournie-Zaluski,M.C., Darlix,J.L. and Roques,B.P. (1991) First large scale chemical synthesis of the 72 amino acid HIV-1 nucleocapsid protein NCp7 in an active form. Biochem. Biophys. Res. Commun., 180, 1010–1018. [DOI] [PubMed] [Google Scholar]
- 37.Clamme J.-P., Bernacchi,S., Vuilleumier,C., Duportail,G. and Mely,Y. (2000) Gene transfer by cationic surfactants is essentially limited by the trapping of the surfactant/DNA complexes onto the cell membrane: a fluorescence investigation. Biochim. Biophys. Acta, 1467, 347–361. [DOI] [PubMed] [Google Scholar]
- 38.Behr J.-P. (1993) Synthetic gene-transfer vectors. Acc. Chem. Res., 26, 274–278. [DOI] [PubMed] [Google Scholar]
- 39.Tang M.X. and Szoka,F.C. (1997) The influence of polymer structure on the interactions of cationic polymers with DNA and morphology of the resulting complexes. Gene Ther., 4, 823–832. [DOI] [PubMed] [Google Scholar]
- 40.Amarasinghe G.K., De Guzman,R.N., Turner,R.B., Chancellor,K.J., Wu,R. and Summers,M.F. (2000) NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol., 301, 491–511. [DOI] [PubMed] [Google Scholar]
- 41.De Guzman R.N., Wu,Z.R., Stalling,C.C., Pappalardo,L., Borer,P.N. and Summers,M.F. (1998) Structure of the HIV-1 nucleocapsid protein bound to the SL-3 psi-like RNA recognition element. Science, 279, 384–388. [DOI] [PubMed] [Google Scholar]
- 42.Morellet N. Déméné,H. Teilleux,V., Huynh-Dinh,T., de Rocquigny,H., Fournié-Zaluski,M.C. and Roques,B.P. (1998) Structure of the complex between the HIV-1 nucleocapsid proteinNCp7 and the single-stranded pentanucleotide d(ACGCC). J. Mol. Biol., 283, 419–434. [DOI] [PubMed] [Google Scholar]
- 43.Vuilleumier C., Bombarda,E., Morellet,N., Gerard,D., Roques,B.P. and Mély,Y. (1999) Nucleic acid sequence discrimination by the HIV-1 nucleocapsid protein NCp7: a fluorescence study. Biochemistry, 38, 16816–16825. [DOI] [PubMed] [Google Scholar]
- 44.Urbaneja M.A., Kane,B.P., Johnson,D.G., Gorelick,R.J., Henderson,L.E. and Casas-Finet,J.R. (1999) Binding properties of the human immunodeficiency virus type 1 nucleocapsid protein p7 to a model RNA: elucidation of the structural determinants for function. J. Mol. Biol., 287, 59–75. [DOI] [PubMed] [Google Scholar]
- 45.Bombarda E., Ababou,A., Vuilleumier,C., Gérard,D., Roques,B.P., Piémont,E. and Mély,Y. (1999) Time-resolved fluorescence investigation of the human immunodeficiency virus type 1 nucleocapsid protein: influence of the binding of nucleic acids. Biophys. J., 76, 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imanishi M. and Sugiura,Y. (2002) Artificial DNA-bending six-zinc finger peptides with different charged linkers: distinct kinetic properties of DNA bindings. Biochemistry, 41, 1328. [DOI] [PubMed] [Google Scholar]
- 47.Carteau S., Gorelick,R.J. and Bushman,F.D. (1999) Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol., 73, 6670–6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levin-Zaidaman S., Englander,J., Shimoni,E., Sharma,A.K., Minton,K.W. and Minsky,A. (2003) Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance? Science, 299, 254–256. [DOI] [PubMed] [Google Scholar]
- 49.Williams M.C., Gorelick,R.J. and Musier-Forsyth, K (2002) Specific zinc finger architecture required for HIV-1 nucleocapsid protein’s nucleic acid chaperone function. Proc. Natl Acad. Sci. USA, 99, 8614–8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson P.E., Turner,R.B., Wu,Z.R., Hairston,L., Guo,J., Levin,J.G. and Summers,M.F. (2000) A mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription. Biochemistry, 39, 9084–9091. [DOI] [PubMed] [Google Scholar]
- 51.Bernacchi S., Stoylov,S., Piemont,E., Ficheux,D., Roques,B.P. Darlix,J.L. and Mely,Y. (2002) HIV-1 nucleocapsid protein activates transient melting of least stable parts of the secondary structure of TAR and its complimentary sequence. J. Mol. Biol., 317, 385–399. [DOI] [PubMed] [Google Scholar]
- 52.Azoulay J., Clamme,J.P., Darlix,J.L., Roques,B.P., Mély,Y. (2003) Destabilization of the HIV complementary sequence of TAR by the nucleocapsid protein through activation of conformational fluctuations. J. Mol. Biol., 326, 691–700. [DOI] [PubMed] [Google Scholar]
- 53.Stoylov S.P., Vuilleumier,C., Stoylova,E., De Rocquigny,H., Roques,B.P., Gérard,D., Mély,Y. (1997) Ordered aggregation of ribonucleic acids by the human immunodeficiency virus type 1 nucleocapsid protein. Biopolymers, 41, 301–312. [DOI] [PubMed] [Google Scholar]
- 54.Le Cam E., Coulaud,D., Delain,E., Petitjean,P., Roques,B.P., Gérard,D., Stoylova,E., Vuilleumier,C., Stoylov,S.P. and Mély,Y. (1998) Properties and growth mechanism of the ordered aggregation of a model RNA by the HIV-1 nucleocapsid protein: an electron microscopy investigation. Biopolymers, 45, 217–229. [DOI] [PubMed] [Google Scholar]
- 55.Rémy E.H., de Rocquigy,H., Petitjean,P., Muriaux,D., Teilleux,V., Paoletti,J. and Roques,B.P. (1998) The annealing of tRNALys,3 to human immunodeficiency virus type 1 primer binding site is critically dependent on the NCp7 zinc fingers structure. J. Biol. Chem., 273, 4819–4822. [DOI] [PubMed] [Google Scholar]