Abstract
The existence of reliable mtDNA reference sequences for each species is of great relevance in a variety of fields, from phylogenetic and population genetics studies to pathogenetic determination of mtDNA variants in humans or in animal models of mtDNA-linked diseases. We present compelling evidence for the existence of sequencing errors on the current mouse mtDNA reference sequence. This includes the deletion of a full codon in two genes, the substitution of one amino acid on five occasions and also the involvement of tRNA and rRNA genes. The conclusions are supported by: (i) the re-sequencing of the original cell line used by Bibb and Clayton, the LA9 cell line, (ii) the sequencing of a second L-derivative clone (L929), and (iii) the comparison with 12 other mtDNA sequences from live mice, 10 of them maternally related with the mouse from which the L cells were generated. Two of the latest sequences are reported for the first time in this study (Balb/cJ and C57BL/6J). In addition, we found that both the LA9 and L929 mtDNAs also contain private clone polymorphic variants that, at least in the case of L929, promote functional impairment of the oxidative phosphorylation system. Conse quently, the mtDNA of the strain used for the mouse genome project (C57BL/6J) is proposed as the new standard for the mouse mtDNA sequence.
INTRODUCTION
The mtDNA from the cell line LA9, a derivative clone of the L cell strain, was the first mouse mtDNA to be sequenced (1) and it has been considered as the standard reference (named hereafter as LA91981). The parent L strain was generated in the 1940s from normal subcutaneous aerolar and adipose tissue of a 100-day-old male C3H/An mouse (2). However, since its publication, a number of mouse mtDNA full sequences showing important discrepancies with the LA9 reference have been reported (Table 1). These discrepancies could be due to existing polymorphisms in the mouse populations, acquired mutations during the lifetime in culture of the LA9 cell line or sequencing errors. Since mouse models for human mtDNA-linked diseases are starting to be generated (3–5), and because this sequence is used for phylogenetic studies including comparative analysis to estimate the potential pathogenicity of human mtDNA variants, the clarification of these uncertainties is of major interest.
Table 1. Conflicting positions in the LA9 mouse mtDNA reference sequence.
| Positiona | Gene | Cell lines | Related mouse strains | Unrelated mouse strains | Proposed referenceb | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LA91981 | L929 | LA92002 | C3H/He | AKR/J | Balb/cJ | A/J | NOD/LtJ | SKH2/J | SAMR1 | SAMP1 | SAMP8 | NZB/B1NJa | Mil Pa | C57BL/6J | ||
| 16 295 | 16 301 | 16 300 | 16 301 | 16 300 | 16 300 | 16 301 | 16 300 | 16 300 | 16 299 | 16 299 | 16 299 | 16 303 | 16 303 | 16 299 | ||
| 1767e | rRNA16S | a | g | g | g | g | g | g | g | g | g | g | g | g | g | g |
| 2256 | rRNA16S | t | t | t | t | t | t | t | t | t | t | c | c | t | t | t |
| 3815e | tRNAGln | t | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 3824e | tRNAGln | – | t | t | t | t | t | t | t | t | t | t | t | t | t | t |
| 4012e | Nd2 | a | g | g | g | g | g | g | g | g | g | g | g | g | g | g |
| 4794 | Nd2 | tc | c | tc | c | c | c | c | c | c | c | c | c | c | c | c |
| 5734e | CoI | c | t | t | t | t | t | t | t | t | t | t | t | t | t | t |
| 5737e | CoI | t | c | c | c | c | c | c | c | c | c | c | c | c | c | c |
| 6063 | CoI | c | c/ad | c | c | c | c | c | c | c | c | c | c | c | c | c |
| 6238e | CoI | g | a | a | a | a | a | a | a | a | a | a | a | a | a | a |
| 6589 | CoI | t | cd | t | t | t | t | t | t | t | t | t | t | t | t | t |
| 9348 | CoIII | a | a | a | a | g | a | a | a | g | g | g | g | g | g | g |
| 9461 | Nd3 | c | c | c | c | c | c | c | c | c | c | c | c | c | c | t |
| 9554e | Nd3 | – | aaa | aaa | aaa | aaa | aaa | aaa | aaa | aaa | aaa | aaa | aaa | aaa | aaa | aaa |
| 9818e | tRNAArg | – | aa | -a | ta | -a | -a | aa | -a | -a | – | – | – | aa | aa | – |
| 9856e | tRNAArg | c | a | a | a | a | a | a | a | a | a | a | a | a | a | a |
| 10 059e | Nd4L | – | atc | atc | atc | atc | atc | atc | atc | atc | atc | atc | atc | atc | atc | atc |
| 10 841 | Nd4 | g | g | g | g | g | g | g | g | g | g | g | a | g | g | g |
| 11 175 | Nd4 | a | a | a | a | a | a | a | a | a | a | g | g | a | a | a |
| 12 042 | Nd5 | cc | t | cc | t | t | t | t | t | t | t | t | t | t | t | t |
| 13 046 | Nd5 | t | t | t | t | t | t | t | t | t | c | c | c | t | t | t |
| 13 879 | Nd6 | 6 Cs | 6/7 Csd | 6 Cs | 6 Cs | 6 Cs | 6 Cs | 6 Cs | 6 Cs | 6 Cs | 6 Cs | 6 Cs | 6 Cs | 6 Cs | 6 Cs | 6 Cs |
| 15 823e | D-loop | t | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 16 119e | D-loop | a | c | c | c | c | c | c | c | c | c | c | c | c | c | c |
| 16 240e | D-loop | a | – | – | – | – | – | – | – | – | – | – | – | a | a | – |
| Accession no. | J01420 | AJ489607 | AY339599 | AB049357 | AB042432 | AJ512208 | NA | NA | NA | AB042523 | AB042524 | AB042809 | L07095 | L07096 | AY172335 | |
The number under the name of each sequence indicates the mtDNA total length. NA, not available.
aThe table shows differences in the mtDNA sequence at the indicated loci numbered according to Bibb et al. (1), the remaining positions are identical between all the sequences with the exception of the two more divergent ones (NZB and Mil P), which showed a high number of differences not indicated. The positions at which the LA91981 sequence is likely to be wrong are followed by an e.
bThe heteroplasmic locus at position 5172 is not indicated.
cIn culture, mutations acquired by LA9.
dIn culture, mutations acquired by L929.
eHighly polymorphic locus.
Information obtained from live animals is available for the full mtDNA sequence of 10 different Mus musculus strains. These are: (i) C3H/He and six other strains belonging together to the group B (Castle’s mice) of mouse-inbred strains (6) [A/J (7), NZB/B1NJ (8), AKR/J and its derivative strains SAMR1, SAMP1 and SAMP8]; (ii) a congenic derivative from the group A (Swiss mice) NOD/LtJ inbred strain, NOD.NON-H2nb1/LtJ (7); (iii) the spontaneous mutant strain SKH2/J (7); and (iv) the Mil P mouse strain, descendant from a wild female mouse caught near Pavia, Italy (8). We report here another two mtDNA sequences of mouse-inbred strains, Balb/cJ, a group B inbred strain (6), sequenced at Zaragoza University, and C57BL/6J, a group E inbred strain (6), obtained at the Sanger Institute from the same source of DNA used for the sequencing of the mouse nuclear genome. In addition, we have performed the full sequencing of the LA9 cell line mtDNA (called hereafter LA92002) and an additional L-derived cell clone (L929) mtDNA.
MATERIALS AND METHODS
Numbering of the oligonucleotides used
For standardization we have referred the numbering of the starting and ending nucleotides of the different primers used in this research to the C57BL/6J mtDNA.
Sequence analysis
The comparison of mtDNA sequences from the different mouse strains and cell lines was performed using BLAST (9).
mtDNA sequencing
Total DNA was isolated from cells by digestion with proteinase K in buffer TE (10 mM Tris, 1 mM EDTA, pH 7.5), containing 0.5% SDS and ribonuclease A, purified by extraction with phenol–chloroform–isoamyl alcohol and precipitated with ethanol. Seventeen overlapping segments of 1000–1500 bp covering the entire mtDNA were amplified by PCR, with the appropriate oligodeoxynucleotides. The PCR products were purified using the Rapid PCR purification system of Gibco BRL, and the purified double-stranded fragments were directly sequenced by the chain termination method, using nested primers.
Polymorphysm analysis
Quantification of the CoI 6589 mutation was achieved by RFLP analysis, and quantification of the Nd6C insertion was carried out by allele-specific termination of primer extension as described elsewhere (10).
To confirm the CoI A6063C mutation, RFLP analysis was carried out. Thus, a 198 bp fragment was amplified by PCR with the following primers: (i) Fw, GCATCTGTTCTGATTCTTTGGGCACCCAGAAGTTTAaATT (positions 6023–6062, forward primer carrying a mismatch indicated by the lower case a); (ii) Rev, TCTAATCCTACTGTGAATtTGTGGTGGGCTC (positions 6190–6220, reverse primer carrying a mismatch indicated by the lower case t in order to create an internal restriction site).
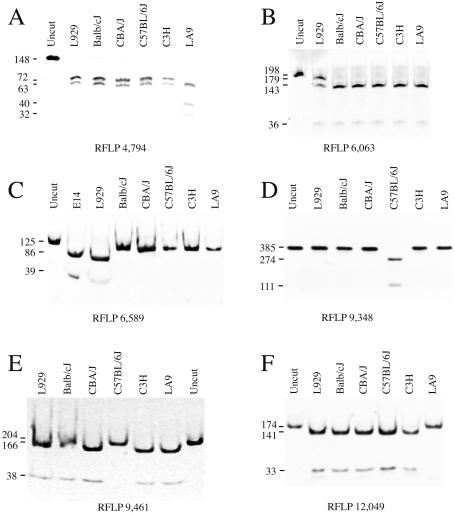
The primer-generated mutations together with the C6063 wild-type version create two recognition sites for AcsI and produce three bands of 143, 36 and 19 bp upon digestion with this enzyme. The restriction site that produces the 143 and 36 bp bands is disrupted when the mutation is present and a new band of 179 bp appears. Therefore, an internal control for full digestion with AcsI is present in the analysis. Fragments were analyzed by electophoresis in a 5% polyacrylamide gel. Notice that the 19 bp band is lost in Figure 1B.
Figure 1.
RFLP confirmation and degree of heteroplasmy of private mutations observed in LA9 (A and F) and L929 mtDNAs (B and C) and specific polymorphysms detected in C57BL/6J mtDNA (D and E). The number under each panel indicates the nucleotide position responsible for the RFLP. The amplification products and the different restriction enzymes used are described in Material and Methods.
For the CoIII A9348G polymorphism analysis, a 385 bp fragment containing the 9348 site was amplified by PCR with the following primers: (i) Fw, CGAAACCACATAAATCAAGCCC (positions 9072–9093); (ii) Rev, CTCTCTTCTGGGTTTATTCAGA (positions 9435–9456).
The G9348 version generates a recognition site for AspI while this restriction site is disrupted when the polymorphism A9348 is present. Fragments were visualized by electrophoresis in a 2% agarose gel containing 0.5 µg/ml ethidium bromide.
The Nd3 C9461T polymorphism was confirmed by RFLP analysis. Thus, a 204 bp fragment containing the 9461 site was amplified by PCR with the following primers: (i) Fw, TTCCAATTAGTAGATTCTGAATAAACCCAGAAGAGAGTgAT (positions 9420–9460, forward primer carrying a mismatch indicated by the lower case g); (ii) Rev, AAATTTTATTGAGAATGGTAGACG (positions 9600–9623).
The combination of the primer-generated mutation together with the C9461 wild-type version produces a recognition site for BclI. Thus, the presence of the T9461 polymorphism disrupts the restriction site. Fragments were analyzed by electrophoresis in a 5% polyacrylamide gel.
To check the Nd2 4794T polymorphism present in LA92002 a 148 bp fragment was amplified by PCR with the following primers: Fw, ACTCATAGCAATAATAGCTCTACTAAACCTATTCTTTaAT (positions 4753–4792, forward primer carrying a mismatch indicated by the lower case a); Rev, TAGGGTGGAAAATATTAGGTTGGG (positions 4877–4900).
This amplicon of 148 bp contains two restriction sites for SspI that produce three DNA fragments of 72, 63 and 13 bp. In addition, the combination of the PCR-generated mutation together with the T4794 version, present in the LA92002 cell line, generates a third recognition site for SspI while this restriction site is absent when the C4794 polymorphism is present. The new restriction site allows the 72 bp band to be cut giving rise to a 40 and 32 bp band. Notice that the 13 bp band is lost in Figure 1A. Fragments were visualized by electrophoresis in a 10% polyacrylamide gel.
To analyze the Nd5 T12049C polymorphism present in LA92002 (equivalent to the 12048 in C57BL/6J) a 174 bp fragment containing this site was amplified with the following primers: Fw, CTGTAGCCCTTTTTGTCACATGATCAATTATAgAA (positions 12013–12047, forward primer carrying a mismatch indicated by the lower case g); Rev, TTCCCACCCCTTCTCAGCCAATG (positions 12 164–12 186).
This PCR-generated mutation creates a recognition site for EcoRI that is disrupted in the LA92002 cell line (12049C). The restriction fragments generated were analyzed using 5% polyacrylamide gel electrophoresis.
Allele-specific termination of primer extension analysis
To determine the length of the A track at positions 5172–5182, allele-specific termination of primer extension was carried out. Thus, a 344 bp fragment containing the OL region was amplified by PCR with the following primers: (i) Fw, CAAACAAAAACTAAACCCAACC (positions 4862–4883); (ii) Rev, CTCTACTAAGACTTCTACCGCC (positions 5184–5205).
The PCR-amplified fragments were used as templates, and the 5′ end 32P-labeled OL-PE oligodeoxynucleotide was used as a primer: OL-PE, CTTCAATCTACTTCTACCGCCG (positions 5150–5171). Nucleotide concentrations were 50 µM dATP and 500 µM ddTTP.
Primer extension was also used to verify the length of a track of A residues starting at position 9821 in the tRNAArg gene. Thus, a 377–379 bp fragment containing this region was amplified by PCR with the following primers: (i) Fw, CTACTTCCACTACCATGAGC (positions 9672–9691); (ii) Rev, GTATGGAGCTTATGGAGTTGG (positions 10 028–10 048).
In this case, the 5′ end 32P-labeled Arg-PE oligodeoxynucleotide was used as primer: Arg-PE, GGATTAGAATGAACAGAGTAAATGGTAATTAGTTT (positions 9786–9820).
The primer-extended products generated were analyzed using 8% polyacrylamide/7 M urea sequencing gels, dried and exposed for autoradiography.
RESULTS
The comparison of the mtDNAs from the different mouse strains reveals that only the sequences of NZB/B1NJ and Mil P mtDNA are very divergent from the LA91981 mtDNA (identities = 16 197 and 12 gaps for NZB/B1NJ; identities = 16 202 and 12 gaps for Mil P) and from each other (identities = 16 236 and no gaps). On the contrary, the mtDNA sequence of all the other mice strains investigated turned out to be quite similar to each other and to LA91981 mtDNA (Table 1). Interestingly, in the case of the inbred strains, the predicted evolutionary relationships between these mtDNAs (data not shown), agree very well with their genealogy (6). This observation strongly suggests a recent common female ancestor for all the inbred strains investigated, except for the NZB/B1NJ, regardless if they belong to groups A (Swiss mice), B (Castle’s mice) or E (C57-related strains) (6).
As shown in Table 1, the C3H/He, Balb/cJ, A/J and NOD/LtJ inbred strains derived mtDNAs are identical in all but one locus, at the tRNAArg gene (position 9818 in the LA91981 sequence and position 9821 in the remaining inbred mice sequences). This difference is mainly due to the variation in the length of a stretch of As in the tRNAArg DHU-loop, which seems to be a highly polymorphic locus in mice (7). Besides this locus, other mouse strains differ from the former group in only one (AKR/J and SKH2/J), two (SAMR1 and C57BL/6J), four (SAMP1) or five (SAMP8) additional bases. Remarkably, the mtDNA non-coding regions (also called D-loops) of all these mouse strains are identical, while they differ from the genetically distant mouse strains NZB/B1NJ and Mil P (eight mismatches and one nucleotide gap with the former and nine mismatches and one nucleotide gap with Mil P, whereas between NZB/B1NJ and Mil P there are seven mismatches with no gaps). The observation of changes in the coding sequences between the related mouse strains, while the mtDNA non-coding region is strictly conserved, is in sharp contrast with what has been described in human mtDNA (11 and references therein).
It should be mentioned that the number of nucleotides in the related mouse inbred strains ranges from 16 299 to 16 301 bp, the differences being due to changes in the length of the stretch of As in the tRNAArg DHU-loop. In addition, in the unrelated strains NZB/B1NJ and Mil P, there are insertions of a base in the tRNACys T-loop (data not shown) and one nucleotide in the mtDNA non-coding region (D-loop) that enlarge the size of the sequence to 16 303 bp.
The comparison of the mouse inbred mtDNA sequences with LA91981 reveals differences in 15 loci: eight in open reading frames (ORFs), four in tRNA or rRNA genes, and only three in the D-loop. Remarkably, only one of the substitutions within the ORFs is silent, the remainder provoking the insertion of a full codon in the reading frame of two protein-coding genes (Nd3 and Nd4L) and the substitution of one amino acid in five loci (Table 2). The fact that LA91981 lacks two full codons, and has only eight As in the polymorphic locus at the tRNAArg DHU-loop explains the shorter size of its mtDNA (16 295 bp). The size variation in the mtDNA of the different sources makes the description of their comparison complicated. Thus, to avoid confusion we have chosen to give the actual numbering of the sequence that we are commenting on followed, when different, by the corresponding number of the LA91981 in brackets.
Table 2. Coded amino acid of the sequence variants described for mtDNAa.
| Nucleotide position | Protein | Amino acid position | Cell lines | Mouse strains | Proposed reference | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LA91981 | L929 | LA92002 | C3H/He | AKR/J | Balb/cJ | A/J | NOD/LtJ | SKH2/J | SAMR1 | SAMP1 | SAMP8 | NZB/B1NJ | Mil P | C57BL/6J | |||
| 4012e | ND2 | 33 | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu |
| 4794b | ND2 | 294 | Ile | Thr | Ile | Thr | Thr | Thr | Thr | Thr | Thr | Thr | Thr | Thr | Thr | Thr | Thr |
| 5734e | COI | 136 | Pro | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu |
| 5737e | COI | 137 | Val | Ala | Ala | Ala | Ala | Ala | Ala | Ala | Ala | Ala | Ala | Ala | Ala | Ala | Ala |
| 6063c | COI | 246 | Leu | Leu/Ile | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu | Leu |
| 6238e | COI | 304 | Cys | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr |
| 6589c | COI | 421 | Val | Ala | Val | Val | Val | Val | Val | Val | Val | Val | Val | Val | Val | Val | Val |
| 9348 | COIII | 248 | Ile | Ile | Ile | Ile | Val | Ile | Ile | Ile | Val | Val | Val | Val | Val | Val | Val |
| 9461e | ND3 | 1 | Met | Met | Met | Met | Met | Met | Met | Met | Met | Met | Met | Met | Met | Met | Met |
| 9554e | ND3 | 33 | – | Lys | Lys | Lys | Lys | Lys | Lys | Lys | Lys | Lys | Lys | Lys | Lys | Lys | Lys |
| 10 059e | ND4L | 64 | – | Ile | Ile | Ile | Ile | Ile | Ile | Ile | Ile | Ile | Ile | Ile | Ile | Ile | Ile |
| 10 841 | ND4 | 227 | Gly | Gly | Gly | Gly | Gly | Gly | Gly | Gly | Gly | Gly | Gly | Gly | Gly | Gly | Gly |
| 11 175 | ND4 | 339 | Ser | Ser | Ser | Ser | Ser | Ser | Ser | Ser | Ser | Ser | Pro | Pro | Ser | Ser | Ser |
| 12 042b | ND5 | 103 | Leu | Phe | Leu | Phe | Phe | Phe | Phe | Phe | Phe | Phe | Phe | Phe | Phe | Phe | Phe |
| 13 046 | ND5 | 437 | Phe | Phe | Phe | Phe | Phe | Phe | Phe | Phe | Phe | Phe | Phe | Phe | Phe | Phe | Phe |
| 13 879c | ND6 | 42 | Gly | Gly/frameshift | Gly | Gly | Gly | Gly | Gly | Gly | Gly | Gly | Gly | Gly | Gly | Gly | Gly |
aThe table shows differences in the mtDNA sequence at the indicated loci, the remaining positions are identical between all the sequences with the exception of the two more divergent ones (NZB and Mil P) that showed a high number of differences not indicated. The positions at which the LA91981 sequence is likely to be wrong are followed by an e.
bIn culture, mutations acquired by LA9.
cIn culture, mutations acquired by L929.
In principle, the differences between LA91981 and the inbred mice strains could suggest a different genetic origin for the LA9 mtDNA and the rest of the inbred strains described to date, as seems to be the case with the NZB/B1NJ mtDNA. This interpretation could be reinforced by the presence of differences in the mtDNA D-loop sequence. Intriguingly, however, all but one of the different loci in the D-loop region, are also conserved between the mtDNA of the distant strains NZB/B1NJ and Mil P. This raised the possibility that the differences between LA91981 and the mouse-derived sequences could be due either to acquired polymorphisms during the lifetime of the cell culture or to sequencing errors in the earlier sequencing.
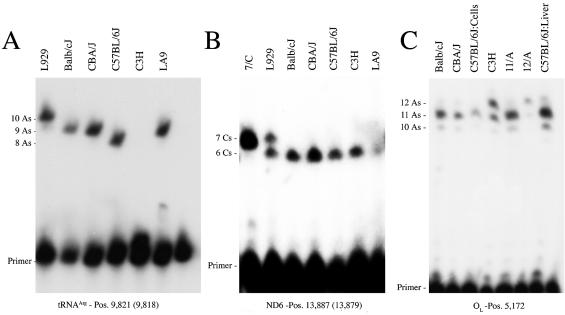
To distinguish between these two possibilities we have performed the full sequencing of the LA9 cell line mtDNA (called hereafter LA92002) and an additional L-derived cell clone (L929) mtDNA. The L929 cell clone (ATCC CCL-1) was established in 1948 by the capillary technique for single cell isolation, it being the first cell line derived from the parental L strain (12). Microsatellite analysis has allowed us to discriminate the two L-clones (LA9 and L929) from C57BL/6J, C3H/He, Balb/cJ and CBA/J, but not between them, supporting their genetic relationship (data not shown). The similarity between the mtDNAs of LA92002, L929, Balb/cJ, C57BL/6J and the rest of the inbred strains indicates that most of the problematic loci detected in the reference sequence were very likely due to sequencing errors (Table 1). However, we found at two loci the same nucleotide in LA92002 as that reported in LA91981, being discrepant with that shown in the L929 and the mouse inbred mtDNAs [positions 4794 within Nd2 and 12 049 (12 042) within Nd5]. Moreover, L929 mtDNA also showed some loci [positions 6063 and 6589 within CoI and the insertion of a C at position 13 887 (13 879) in Nd6] that differed from LA91981, LA92002 and the rest of the known mouse mtDNA sequences (Table 1). In addition, we could not resolve the discrepancy between L929 and LA91981 in the length of the stretch of As in the tRNAArg DHU-loop, since the LA92002 sequence (nine As) differs from both L929 (10 As) and LA91981 (eight As). To fully guarantee the accuracy of the obtained sequence information and to establish the heteroplasmic or homoplasmic nature of the observed variations, all the discrepant positions were confirmed by RFLP analysis (Fig. 1) or allele-specific primer extension termination (Fig. 2). We detected heteroplasmic loci in the L929 mtDNA (position 6063 in CoI), it being 18% Cwt and 82% Amt (Fig. 1B). In addition, the L929 length polymorphism due to the insertion of a C at position 13 887 (13 879) in the Nd6 gene was found to be heteroplasmic, it being present in 52% of the mtDNA molecules (Fig. 2B). The additional private polymorphisms in L929 (position 6589 within CoI) or LA9 [position 4794 in Nd2 and position 12 049 (12 042) within Nd5] were homoplasmic within the limit of resolution of the RFLP analysis (Fig. 1A, C and F). All these variations would represent clone-specific mutations acquired in culture (10,13).
Figure 2.
Allele-specific termination of primer extension assays designed to establish the size of three polymorphic mononucleotide tracks observed in the tRNAArg gene (A), the Nd6 gene (B) and the L-strand replication origin (C). Note that the extension of the C3H mtDNA sample in (A) cannot be longer than 1 nt due to its particular four Ts + nine As sequence, while the rest of the samples harbor three Ts + n As, with n = eight, nine or 10. Lines without label in (A) and (C) represent un-extended primer. Lines labeled 7/C, 11/A or 12/A in (B) or (C) represent controls for seven Cs or 11 or 12 As in the respective polymorphic track.
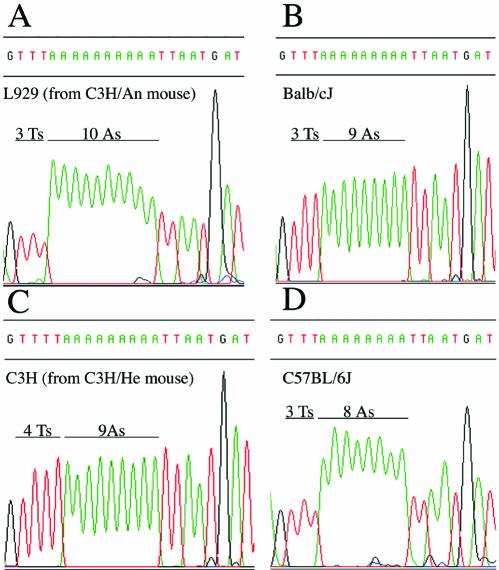
RFLP analysis confirmed that the two C57BL/6J-specific polymorphisms were also homoplasmic (Fig. 1D and E). Interestingly, we could not detect any heteroplasmic loci in the sequence of the Balb/cJ mtDNA, but we found a length track polymorphic locus within the sequence of C57BL/6J from the Sanger Institute, consisting of a stretch of either 11 or 12 As (starting position 5172) in a short non-coding segment proposed to contain the replication origin of the mtDNA L-strand (14). Allele-specific primer extension termination (Fig. 2C) demonstrated that this locus is homoplasmic in a DNA sample from a mtDNA-less cell line repopulated with either mtDNA from C57BL/6J, Balb/cJ or CBA/J mouse platelets (10,15). However, this locus is heteroplasmic for 11 and 12 As in a DNA sample isolated from C57BL/6J mouse liver and in C3H/He mouse-derived cells (Fig. 2C). Notice that the termination signal at 10 As in this figure is due to a nuclear pseudogene harboring 10 As (unpublished results). Similar heteroplasmic length variants in non-coding regions of the mtDNA from a single individual have also been documented in humans (16). Finally, the combination of sequencing analysis and allele-specific primer extension termination, allowed us to confirm the existence of all the variations proposed for the polymorphic locus at the tRNAArg DHU-loop: the length variations in the stretch of As (Figs 2A and 3), as well as the particular sequence of C3H/He-derived cells consisting of nine As plus one T (Fig. 3).
Figure 3.
Representative chromatograms that illustrate the variety of viable sequences described to date in the highly polymorphic locus at the mouse mtDNA-encoded tRNAArg gene. The source of the DNA is indicated in each panel.
It should be remarked that despite the number of confirmed errors (10), the sequencing effort performed by Bibb and co-workers in the earlier 1980s, provided a very high quality sequence with only a 0.08% error frequency.
DISCUSSION
Based on the overall data, we consider that both L929 and LA9 mtDNAs showed fixed and heteroplasmic polymorphisms acquired during their lifetime as independent clones, that were very likely not present in the mouse from which L cells were established. Worryingly enough, we have found evidence proving that the L929 clone-specific homoplasmic and heteroplasmic mutations have phenotypic consequences for the proper function of the mitochondrial respiratory chain (10,15), and this may also be the case for the LA9-specific mutations. Special mention should be made of the polymorphic locus in tRNAArg. This gene presents a stretch of eight As (LA91981, SAMR1, SAMP1, SAMP8 and C57BL/6J mtDNAs), nine As (LA92002, Balb/cJ, NOD/LtJ, SKH2/J and AKR/J), nine As plus one T (C3H/He) or 10 As (L929, A/J, Mil P and NZB/B1NJ mtDNAs). Interestingly, it has been recently reported that the length of this stretch of As can modulate the pathological expression of a nuclear locus involved in aging-related deafness in mice (7).
According to the data presented here, the reference sequence for mouse mtDNA should be reconsidered for the following reasons. (i) It contains seven substitution errors, three single nucleotide insertions and a single base and two triplet deletions. (ii) The two clone-restricted polymorphic positions in LA9 do not represent the wild-type version present in the original C3H/An mouse and they could likely be pathogenic. (iii) Since LA91981, LA92002 and L929 still diverge in the polymorphic locus at tRNAArg, a highly variable locus in mtDNAs of mouse strains, we cannot predict the more likely sequence for this locus in the original C3H/An mouse.
Therefore, we propose the sequence of C57BL/6J mtDNA as the mouse reference in order to have a true reference sequence and not a consensus one. In that way there will also be a match between the reference sequence for the nuclear and the mitochondrial genomes in mouse. Note that if these recommendations are followed, the size of the mouse mtDNA reference sequence will increase from 16 295 to 16 299 and the nucleotide numbering should be modified accordingly.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr Lluis Montoliu, Dr Allan Bradley and Dr Giuseppe Attardi for their help in establishing this collaborative effort. We also would like to thank Erika Fernández-Vizarra and Santiago Morales for their technical assistance. Our research was supported by Spanish Ministry of Education (PM-99-0082) and Instituto de Salud Carlos III (REDEMETH G03/05) grants to J.A.E., an Instituto de Salud Carlos III (REDCIEN C03/06-Grupo RC-N34-3) grant to A.P.-M., a Ramón y Cajal 2001 grant to P.F.S., a Research Resources Program for Medical Schools of the Howard Hughes Medical Institute grant to Y.B., and a Wellcome Trust grant to J.C.M. R.A.-P. and R.M.-L. are recipients of a Spanish Ministry of Education F.P.U. and, respectively, Spanish Ministry of Science and Technology F.P.I. fellowships.
DDBJ/EMBL/GenBank accession nos+ AJ489607, AY172335 and AJ512208
REFERENCES
- 1.Bibb M.J., Van Etten,R.A., Wright,C.T., Walberg,M.W. and Clayton,D.A. (1981) Sequence and gene organization of mouse mitochondrial DNA. Cell, 26, 167–180. [DOI] [PubMed] [Google Scholar]
- 2.Earle W.R., Schilling,E.L., Stark,T.H., Straus,N.P., Brown,M.F. and Shelton,E. (1943) Production of malignancy in vitro. IV. The mouse fibroblast cultures and changes seen in the living cells. J. Natl Cancer Inst., 4, 165–212. [Google Scholar]
- 3.Marchington D.R., Barlow,D. and Poulton,J. (1999) Transmitochondrial mice carrying resistance to chloramphenicol on mitochondrial DNA: developing the first mouse model of mitochondrial DNA disease. Nature Med., 5, 957–960. [DOI] [PubMed] [Google Scholar]
- 4.Inoue K., Nakada,K., Ogura,A., Isobe,K., Goto,Y., Nonaka,I. and Hayashi,J.I. (2000) Generation of mice with mitochondrial dysfunction by introducing mouse mtDNA carrying a deletion into zygotes. Nature Genet., 26, 176–181. [DOI] [PubMed] [Google Scholar]
- 5.Sligh J.E., Levy,S.E., Waymire,K.G., Allard,P., Dillehay,D.L., Nusinowitz,S., Heckenlively,J.R., MacGregor,G.R. and Wallace,D.C. (2000) Maternal germ-line transmission of mutant mtDNAs from embryonic stem cell-derived chimeric mice. Proc. Natl Acad. Sci. USA, 97, 14461–14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck J.A., Lloyd,S., Hafezparast,M., Lennon-Pierce,M., Eppig,J.T., Festing,M.F. and Fisher,E.M. (2000) Genealogies of mouse inbred strains. Nature Genet., 24, 23–25. [DOI] [PubMed] [Google Scholar]
- 7.Johnson K.R., Zheng,Q.Y., Bykhovskaya,Y., Spirina,O. and Fischel-Ghodsian,N. (2001) A nuclear-mitochondrial DNA interaction affecting hearing impairment in mice. Nature Genet., 27, 191–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loveland B., Wang,C.R., Yonekawa,H., Hermel,E. and Lindahl,K.F. (1990) Maternally transmitted histocompatibility antigen of mice: a hydrophobic peptide of a mitochondrially encoded protein. Cell, 60, 971–980. [DOI] [PubMed] [Google Scholar]
- 9.Tatusova T.A. and Madden,T.L. (1999) BLAST 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett., 174, 247–250. [DOI] [PubMed] [Google Scholar]
- 10.Acín-Pérez R., Bayona-Bafaluy,M.P., Bueno,M., Machicado,C., Fernández-Sílva,P., Pérez-Martos,A., Montoya,J., López-Pérez,M.J., Sancho,J. and Enriquez,J.A. (2003) An intragenic suppressor in the cytochrome c oxidase I gene of mouse mitochondrial DNA. Hum. Mol. Genet., 12, 329–339. [DOI] [PubMed] [Google Scholar]
- 11.Howell N., Smejkal,C.B., Mackey,D.A., Chinnery,P.F., Turnbull,D.M. and Herrnstadt,C. (2003) The pedigree rate of sequence divergence in the human mitochondrial genome: there is a difference between phylogenetic and pedigree rates Am. J. Hum. Genet., 72, 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanford K.K., Earle,W.R. and Likely,G.D. (1948) The growth in vitro of single isolated tissue cells. J. Natl Cancer Inst., 9, 229–246. [PubMed] [Google Scholar]
- 13.Herrnstadt C., Preston,G., Andrews,R., Chinnery,P., Lightowlers,R.N., Turnbull,D.M., Kubacka,I. and Howell,N. (2002) A high frequency of mtDNA polymorphisms in HeLa cell sublines. Mutat. Res., 501, 19–28. [DOI] [PubMed] [Google Scholar]
- 14.Clayton D.A. (1982) Replication of animal mitochondrial DNA. Cell, 28, 693–705. [DOI] [PubMed] [Google Scholar]
- 15.Bai Y. and Attardi,G. (1998) The mtDNA-encoded ND6 subunit of mitochondrial NADH dehydrogenase is essential for the assembly of the membrane arm and the respiratory function of the enzyme. EMBO J., 17, 4848–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trounce I., Schmiedel,J., Yen,H.C., Hosseini,S., Brown,M.D., Olson,J.J. and Wallace,D.C. (2000) Cloning of neuronal mtDNA variants in cultured cells by synaptosome fusion with mtDNA-less cells. Nucleic Acids Res., 28, 2164–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]