Abstract
We performed a yeast two-hybrid screen using p73α, which is a member of the p53 family, as bait. We found that the p53 family members were functionally associated with Daxx, which was described originally as a cytoplasmic mediator of Fas signaling, but has been identified recently as a nuclear protein that co-localizes with the promyelocytic leukemia (PML) protein and regulates transcription. Extensive yeast two-hybrid assays indicated a physical interaction between a region including the oligomerization domain (OD) of p73α (amino acids 345–380) or p53 (amino acids 319–360) and amino acids 161–311 and 667–740 (C-terminal S/P/T-rich domain) of hDaxx, which is the common binding region of Fas, ASK1 and PML. This interaction was further confirmed by in vitro GST pull-down and in vivo immunoprecipitation assays. Both Daxx and p73/p53 co-localized in nuclear dot-like structures, which are probably nuclear PML oncogenic domains (PODs) or the nuclear domain NB10. Transient co-expression of Daxx resulted in strong inhibition of p73- and p53-mediated transcriptional activation of the synthetic p53-responsive and p21WAF1 promoters. Consequently, Gal4-Daxx repressed basal transcription in a dose-dependent manner. Treatment with trichostatin A, which is an inhibitor of histone deacetylase, or PML over-expression relieved Daxx-mediated transcriptional repression of p53. The mechanism underlying PML-mediated derepression appears to be competitive binding between Daxx, p53 and PML. Taken together, these findings delineate a transcriptional regulatory network that is modulated by differential Daxx–p53–PML interactions in the nuclear PODs. Therefore, Daxx is implicated in the regulation of the cell cycle and apoptosis through transcriptional regulation of p53 and possibly its family members.
INTRODUCTION
The tumor suppressor p53 plays significant roles in growth arrest, DNA repair and apoptosis, by acting as a transcription factor to modulate the expression of various genes (1). Depending on the promoter context, the stimulus type and the cellular environment, p53 functions not only to activate gene transcription, but also to repress the expression of responsive genes (2). The repression mechanism is mediated either by direct binding to the response element or by indirect interactions with other transcriptional regulators, depending on the promoter context. It has been reported that the C-terminal oligomerization domain (OD) of p53 (3–5), histone deacetylase (HDAC) and mSin3a are involved in p53-mediated repression of transcription (6–8).
Recently, p73 and p63 were identified as members of the p53 family, and their activities have been reviewed extensively (9–12). As is the case with p53, both p73 and p63 can form oligomers, bind to canonical p53 DNA-binding sites, modulate the transcription of p53-responsive genes, and suppress growth or induce apoptosis when over-expressed in certain human tumors. However, unlike p53, the p73 and p63 isomers harbor sterile α-motif (SAM) domains, which may be involved in protein–protein interactions, at their C-termini (13). In addition, since p73 and p63 mutations are rarely found in tumors, they are unlikely to represent classical tumor suppressor genes. Therefore, the p53 family members share certain functions, but have distinct responses to extracellular signals and developmental cues at the physiological level. Thus, it is of interest to determine how the p53 family is regulated differentially at the molecular level. With this in mind, we performed a yeast two-hybrid screen, and found that hDaxx interacted with p73.
Daxx was identified initially in yeast two-hybrid screens as the protein whose C-terminal region associated with the death domain of the Fas receptor and promoted Fas-induced apoptosis (14). Later, it was reported that Daxx mediated apoptosis by linking Fas signaling to the Jun N-terminal kinase (JNK) pathway via apoptosis signal-regulating kinase 1 (ASK1) activation in the cytoplasm (15,16). Subsequent studies have suggested that, in response to Fas activation, Daxx translocates from the nucleus to the cytoplasm, where it interacts with ASK1 to promote caspase-independent cell death, thus preventing Daxx-mediated transcriptional repression in the nucleus (17,18). In contrast to the original findings (14–16), most recent studies have indicated that Daxx is present exclusively in the nucleus, where it may play a direct role in regulating transcription (19–22). Furthermore, it has been reported that Daxx is anti-apoptotic rather than pro-apoptotic (23,24). In the nucleus, Daxx is recruited to promyelocytic leukemia (PML) oncogenic domains (PODs) through interaction with the SUMO-1-modified PML (25), whereby both proteins appear to participate in a novel apoptotic pathway (26). Daxx-mediated transcriptional repression may be due to interactions with DNA methyltransferase-1 (23) and HDAC (20,27). PML appears to inhibit the repressive function of Daxx by transporting it from the condensed chromatin to PODs (20). In a similar manner, SUMO-1-modified PML can derepress Pax3 transcriptional activity, which is subject to repression by Daxx (28). Reminiscent of PML sequestration of Daxx to PODs, it has been reported recently that MSP58 translocates Daxx to the nucleolus, and thus inhibits Daxx-mediated repression of the transcriptional activity of the glucocorticoid receptor (GR) (29). Furthermore, it is of interest to note that PML also binds and targets p53 into PODs (30,31). The C-terminal sequences of PML outside of the sumoylation sites interact with the DNA-binding domain of p53. The splice variants of PML, PML-L and PML-RARα, lack this region, and thus in vitro GST pull-down assays did not detect any interactions. The PML sequestration of p53 results in the activation of p53 target genes that are involved in apoptosis and cell cycle arrest (30). This mode of PML activity depends on the abilities to form the nuclear PODs and to become sumoylated in vivo. These observations suggest that POD is a dynamic site that modulates transcriptional activities, thereby affecting cellular processes such as apoptosis (32). Therefore, we speculate that Daxx plays a regulatory role in linking the functions of PML and p53.
Daxx was identified from our yeast two-hybrid screening, which used p73 as the bait, and subjected to further investigation. We examined the sites of interaction of the p73/p53 and Daxx proteins, the biological significance of this interaction, and the role of PML in the p53–Daxx interaction. We observed in vitro and in vivo physical interactions between the ODs of the p53 family proteins and a region near the C-terminus of hDaxx. These proteins co-localized in the nuclear bodies, and Daxx inhibited the transcriptional activities of p73 and p53 in a dose-dependent manner. The co-repressor activity of Daxx was abrogated in the presence of trichostatin A (TSA), which is an inhibitor of HDAC-1. Finally, we found that Daxx-mediated transcriptional repression was relieved by competitive association of PML3 with the Daxx co-repressor. Taken together, these observations suggest that Daxx functions as a transcriptional repressor of p53 and members of the p53 family, and that this repressive effect can be modulated by PML.
MATERIALS AND METHODS
Cell lines and cell culture
All cells used in our experiments were routinely maintained in RPMI medium 1640 supplemented with 10% heat-inactivated fetal bovine serum and penicillin/streptomycin (all from Gibco-BRL, Gaithersburg, MD).
cDNA constructions
Most cDNA constructs were described previously (8). Some were made according to standard methods (33) and verified by sequencing. Human Daxx in pBluecript KS (a kind gift from Dr Ann F. Pluta, MD) (34) was PCR-amplified and subcloned into VP16 activation domain (AD)-fused pASV3 vector for mapping the interaction domain, into hemagglutinin (HA)-pcDNA3 for immunoprecipitation and transient transfection, and into pEGFP-N1 vector (Clontech, Palo Alto, CA) for co-localization assays. To obtain GST-tagged Daxx protein, a PCR fragment encoding the C-terminus of Daxx (amino acids 607–740) was amplified and subcloned into pGEX2T vector (Amersham Pharmacia, Piscataway, NJ). Expression plasmid for Gal4-fused Daxx was created by subcloning of PCR-amplified Daxx into pG4MpolyII vector (a kind gift from Dr Pierre Chambon, Strasbourg, France). Details on individual plasmid constructs are available on request.
Yeast two-hybrid system
A human liver MATCHMAKER cDNA library in the pGAD10 prey plasmid (Clontech) was screened for proteins that interact with p73 (transactivation domain deleted; amino acids 49–636) in the pBTM116 bait plasmid using the L40 yeast reporter strain as described (8). After final identification of hDaxx by sequencing and GenBank analysis, it was systematically tested for interactions with a LexA DNA-binding domain (DBD) fusion of p73α (49–636) or other members of the p53 family by β-galactosidase assays. To precisely map the p73 interaction domain, PCR fragments encoding deletion derivatives of Daxx were amplified and subcloned into VP16 AD-fused pASV3 vector and co-transformed with a LexA DBD-fused p73α (49–636) in pBTM116 vector.
GST pull-down assay
GST fusions of p53 (amino acids 319–393) and hDaxx (amino acids 607–740) were created by cloning the appropriate insert into pGEX2T (Amersham Pharmacia). Proteins were expressed in Escherichia coli and purified on glutathione–Sepharose beads (Amersham Pharmacia) by standard methods. Full-length hDaxx, p53, p73α, p73β, p63α and p63γ were in vitro translated in 50 µl of rabbit reticulocyte lysate (Promega, Madison, WI) supplemented with [35S]methionine (Amersham Pharmacia). In vitro binding assays were performed as described previously (8).
Immunoprecipitation and western blotting
After transfection with plasmid DNAs under conditions indicated in the text, p53-null Saos2 osteosarcoma cells were washed in phosphate-buffered saline (PBS), and cell lysates were prepared by adding 1 ml of ice-cold RIPA buffer (250 mM NaCl, 50 mM HEPES pH 7.0, 0.1% NP-40 and 5 mM EDTA) supplemented with protease inhibitors and pre-cleared by incubating with protein A–Sepharose beads for 1 h. The pre-cleared lysate was then incubated with a mouse anti-HA monoclonal antibody (mAb; 1:200 dilution, 12CA5; Roche Molecular Biochemicals, Mannheim, Germany), a rabbit polyclonal anti-green fluorescent protein (GFP) antibody (1:200 dilution, sc-8334; Santa Cruz Biotechnology, Santa Cruz, CA) or a rabbit polyclonal anti-Daxx antibody (1:150 dilution, sc-7152; Santa Cruz Biotechnology), and protein A–Sepharose beads overnight at 4°C. The beads were washed five times in RIPA buffer and twice with PBS, and the immune complexes were released from the beads by boiling in sample buffer for 5 min. Following 8% SDS–PAGE, immunoprecipitation products were analyzed by immunoblotting as described previously (8).
Confocal immunofluorescence microscopy
Saos2 cells seeded on glass coverslips in 6 cm plates were transfected. Two days after transfection with either HA-p73α/p53 in pcDNA3, GFP–hDaxx in pEGFP-N1 (Clontech) or both together, cells were washed with PBS, fixed with 4% paraformaldehyde in PBS for 10 min at room temperature, and permeabilized for 4 min in PBS containing 0.3% Triton X-100, 10% goat serum at 4°C. After washing, cells were blocked in PBS containing 3% BSA for 60 min at room temperature and incubated with a mouse anti-HA mAb in blocking buffer for 2 h at room temperature. Cells were then incubated with Texas red-conjugated anti-mouse IgG (1:50 dilution, N2031; Amersham Pharmacia) for 1 h at room temperature and mounted with 50 µl of VectaShield (Vector Laboratories, Burlingame, CA). Confocal microscopy was performed on a Bio-Rad microscope (Radiance 2000 MP).
Transient transfection and CAT ELISA
p53-null Saos2 cells were transiently transfected by using the Lipofectamine transfection reagent (Gibco-BRL) according to the the manufacturer’s protocol. In 6-well plates, total amounts (2.2 µg) of DNAs were kept constant by adding SV40-driven β-gal internal control plasmid and the respective test plasmids as indicated in the text. After overnight transfection, cells were washed, fed with the complete medium, and further incubated for 24 h. Cells were then collected and extracted as described previously (8). Extracts were cleared by centrifugation, and protein concentrations were determined with the Bio-Rad protein assay dye reagent. β-Galactosidase activity was determined in 96-well plates. A 30–70 µl aliquot of the clear lysates was tested for chloramphenicol acetyltransferase (CAT) concentration in the CAT enzyme-linked immunosorbent assay (ELISA) according to the manufacturer’s instructions (Roche Molecular Biochemicals). The CAT concentration of each sample was normalized with respect to β-gal activity.
RESULTS
Daxx interactions with members of the p53 family
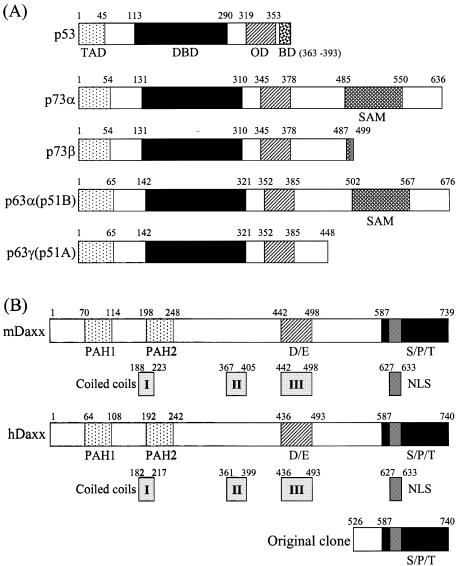
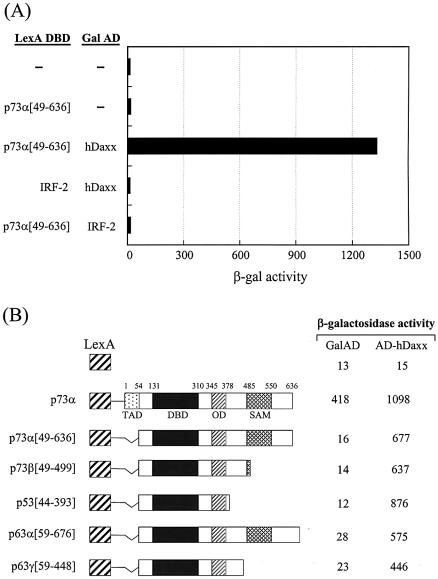
The yeast two-hybrid system was used to identify proteins that interacted with the transactivation domain (amino acids 49–636) of p73α, which is a recently identified member of the p53 family (Fig. 1A). The yeast two-hybrid screening has been described previously (8). A total of 42 unique library plasmids were obtained from the initial screen. DNA sequencing and subsequent GenBank searches indicated that four of these clones contained partial sequence of hDaxx, which comprised amino acids 526–740 (Fig. 1B). This protein interacted strongly with p73α (amino acids 49–636), but not with the unrelated IRF-2 protein in the yeast two-hybrid assays (Fig. 2A). Further yeast two-hybrid assays revealed that hDaxx interacted with other members of the p53 family, which included p73β, p53, p63α and p63γ, the transactivation domains of which were deleted for the purpose of the assay (Fig. 2B).
Figure 1.
Structural characteristics of p53 family proteins and Daxx homologs. (A) p53 family (p53, p73α, p73β, p63α and p63γ). TAD, transcription activation domain; DBD, DNA-binding domain; OD, oligomerization domain; BD, basic regulatory domain; SAM, sterile α-motif. (B) Daxx homologs (mouse and human). PAH, paired amphipathic helix; D/E, aspartate- and glutamate-rich; S/P/T, serine/proline/threonine-rich. A partial portion of hDaxx was indicated as identified by yeast two-hybrid screening. The numbers indicate amino acid positions.
Figure 2.
hDaxx interacts with the p53 family. (A) Identification of hDaxx by yeast two-hybrid screening using a human liver-derived cDNA library fused to the Gal4 activation domain (AD). (B) Interaction of the p53 family with hDaxx, analyzed by yeast two-hybrid and β-galactosidase assays using the LexA DBD-fused p53 family (in pBTM116) and the original Gal AD-fused hDaxx (in pGAD10) as described in detail in Materials and Methods.
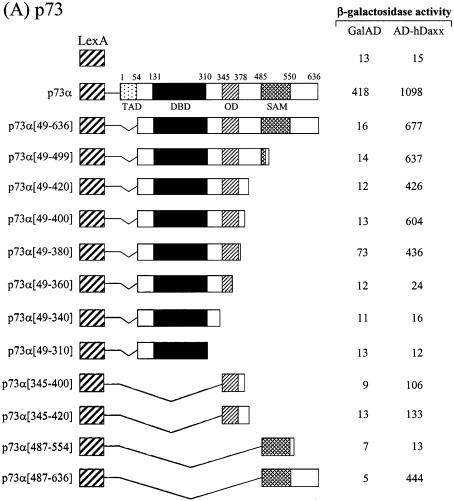
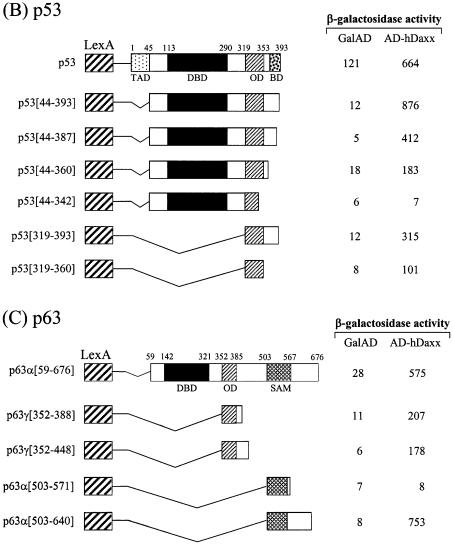
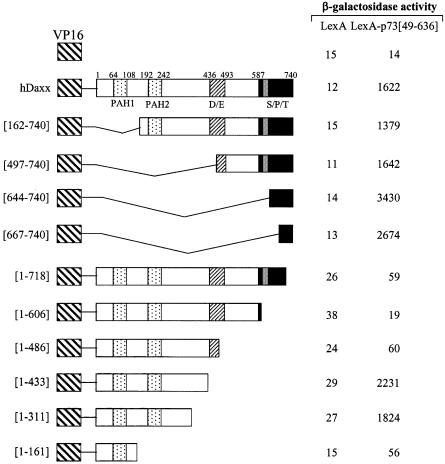
In order to map the Daxx interaction domains of the p53 family members, deletion derivatives were fused to the LexA DBD in the pBTM116 vector, and these constructs were co-transformed into the yeast strain L40, together with the original fusion of hDaxx with the Gal4 AD. Although the full-length p73α fused to the DBD showed transcriptional activity due to the presence of the transactivation domain, the interaction with Daxx was evident as shown by increased β-galactosidase activity when DBD-fused p73α was co-expressed with AD-Daxx. Assays using a series of C-terminal deletion mutants of p73α indicated that the region around the OD was required for the interaction with Daxx (Fig. 3A). Further analysis with the p73α (amino acids 49–380) and p73α (amino acids 345–400) domains revealed that the minimal interaction domain lay between amino acids 345 and 380. Interestingly, another independent interaction domain, which included the SAM domain, was located near the C-terminus. Further deletions in these regions abolished the interaction with Daxx. As was the case with p73, p53 strongly associated with Daxx. Like p73, the OD (amino acids 319–360) of p53 appeared to be at least sufficient for the interaction with Daxx. However, in the case of p53, the C-terminal regulatory region (amino acids 361–393) appeared to contribute on the increased interaction (Fig. 3B). Likewise, the interaction domain of p63 was mapped between amino acids 352 and 388, in a region that encompassed the OD (Fig. 3C). In addition, we mapped a second interaction domain near the C-terminus, in an area that included the SAM domain of p63α.
Figure 3.
Mapping of the specific region of the p53 family responsible for interaction with hDaxx. (A) p73. (B) p53. (C) p63. Yeast two-hybrid and β-galactosidase assays were performed using LexA-fused truncations of the p53 family (in pBTM116) and the original Gal AD-fused hDaxx (in pGAD10). LexA-fused mutants were almost evenly expressed in yeast as determined by western blotting analysis (data not shown).
Mapping the interaction domains of Daxx
Various deletion mutants of hDaxx were fused to the VP16 AD in the pASV3 vector, and analyzed for interactions with LexA DBD–p73α (amino acids 49–636) (Fig. 4). Neither LexA–p73α (amino acids 49–636) nor LexA alone interacted with VP16 AD. Assays with a series of N- and C-terminal deletion mutants of hDaxx revealed that the C-terminal region between amino acids 667 and 740 was required and sufficient for the interaction with p73α. Further deletions in this region abolished the interaction with p73α. However, further deletions at the N-terminus uncovered an additional interaction domain. Interestingly, the C-terminal portion of Daxx is known to interact with several proteins, such as Fas (14), ASK1 (15), HSP27 (16), Pax3 (19), ETS1 (21), Pax5 (22), PML (25) and CENP-C (34), which indicates the dynamic regulatory role of Daxx.
Figure 4.
Mapping of the hDaxx domain responsible for interaction with p73α (amino acids 49–636). Yeast two-hybrid and β-galactosidase assays were performed using LexA-fused p73α (amino acids 49–636) in pBTM116 and VP16-fused truncations of hDaxx in pASV3. VP16-fused mutants were almost evenly expressed in yeast (data not shown).
Confirmation of the p53–Daxx interaction in vitro and in mammalian cells
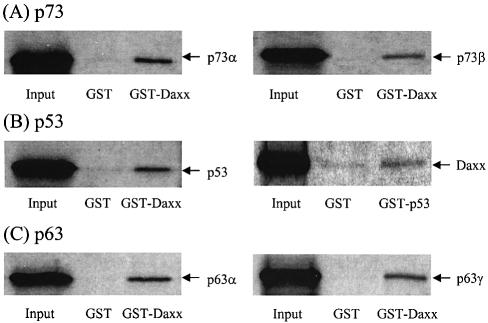
To determine whether the p53–Daxx interaction occurs in vitro, we performed GST pull-down assays in which either the GST–p53 (amino acids 319–393) or GST–Daxx (amino acids 607–740) protein was induced in E.coli, purified, and mixed with in vitro-translated [35S]methionine-labeled Daxx, p53, p73α, p73β, p63α or p63γ in the rabbit reticulocyte lysate. Binding of the p53 family members to Daxx was assessed using GST pull-down assays. Consistent with the results of the yeast two-hybrid experiments, the labeled p53 and its relatives were retained by glutathione beads that contained GST–Daxx (Fig. 5). In the reciprocal experiment, where the GST fusion was replaced with p53, the interaction between p53 and Daxx also occurred, albeit the effect was relatively weak (Fig. 5B). Overall, these data confirm that Daxx binds directly to p53 family members in vitro.
Figure 5.
GST pull-down assays. In vitro translated 35S-labeled products were incubated with GST, GST–Daxx (amino acids 607–740) or GST–p53 (amino acids 319–393) immobilized onto glutathione–Sepharose beads for 2 h. Beads were washed five times with buffer A [20 mM HEPES pH 7.5, 50 mM KCl, 2.5 mM MgCl2, 10% glycerol, 1 mM dithiothreitol (DTT), 1% NP-40 and 200 mg/ml phenylmethylsulfonyl fluoride (PMSF)]. Bound proteins were eluted with SDS sample buffer and resolved by 10% SDS–PAGE for autoradiography.
To further demonstrate that the p53 family members interacted with Daxx in mammalian cells, p53-null Saos2 cells were transfected with combinations of HA-p73α or HA-p53 and HA-Daxx or GFP–Daxx constructs. Immuno precipitation with the anti-HA antibody, and subsequent western blotting with the anti-GFP antibody revealed that both p73α and p53 interacted with Daxx in vivo under over-expressed conditions (Fig. 6A). In reciprocal experiments, in which untagged p73α or p53 and GFP–Daxx were co-transfected, both p73α and p53 were detected in the immunoprecipitates that were obtained using the anti-GFP antibody (Fig. 6B and C; left panel). These interactions were also demonstrated using the anti-Daxx antibody for immunoprecipitation, and with the anti-p73α and anti-p53 antibodies that were used to probe the blotted precipitates (Fig. 6B and C; right panel). These data indicate that p73/p53 and Daxx combine in mammalian cells to form physical complexes.
Figure 6.
In vivo interaction assay by immunoprecipitation. Transfection conditions in p53-null Saos2 cells were as follows: (A) GFP–Daxx and empty HA-pcDNA3 vector, HA-p73α or HA-p53; (B, left) p73α without tag (in pcDNA3) and empty GFP vector or GFP–Daxx; (B, right) Daxx and empty pcDNA3 or p73α; (C, left) p53 in pcDNA3 and empty GFP vector or GFP–Daxx; (C, right) Daxx and empty pcDNA3 or p53. After transfections, cells were lysed in RIPA buffer (250 mM NaCl, 50 mM HEPES pH 7.0, 0.1% NP40 and 5 mM EDTA) supplemented with protease inhibitors and pre-cleared by incubating with protein A–Sepharose for 1 h. Immuno precipitation was carried out with anti-HA, anti-Daxx or anti-GFP antibody. After resolution by 10% SDS–PAGE, western blotting was carried out using anti-GFP (A), anti-p73α (B) and anti-p53 (C) antibodies. A band in the control sample (C, right panel, Daxx alone) corresponds to the heavy chain of anti-Daxx antibody that can be detected by western blotting. The band is close to that of p53 because of running the gel for a short time.
The p53 family members and Daxx co-localize in nuclear body-like structures
We determined the subcellular co-localizations of p73α or p53 with Daxx. Thus, p53-null Saos2 cells were co-transfected transiently with plasmids that encoded HA-p73α or HA-p53 and GFP–Daxx, and analyzed by immunofluorescence and confocal laser microscopy. Consistent with previous reports, Daxx localized primarily to the nuclear spots, which probably correspond to PML bodies or PODs (25,26). Both p73α and p53 yielded diffuse nuclear red fluorescence following Texas red staining (data not shown). However, when the p53 family members and Daxx were co-expressed, the p53 family members localized to distinct spots within the nucleus (Fig. 7A and B). These findings are reminiscent of previous reports that showed that PML targets p53 into the PML nuclear bodies (30,31).
Figure 7.
Immunofluorescence assays using confocal microscopy. Transfected Saos2 cells with GFP–Daxx and HA-p73α (A) or HA-p53 (B) were replated on slides. Cells were fixed in 4% paraformaldehyde for 20 min at room temperature and were covered with blocking solution (5% goat serum in 3% bovine serum albumin in PBS) for 1 h. Cells were stained with anti-HA-Texas red-conjugated antibody for 1 h. Slides were then mounted in antifade mounting medium and analyzed by confocal microscopy.
Taken together, our in vitro and in vivo interaction data suggested that the p53 family members are associated physiologically with Daxx, which prompted us to investigate the biological significance of this interaction in vivo.
Daxx modulates the transcriptional activity of p53 family proteins
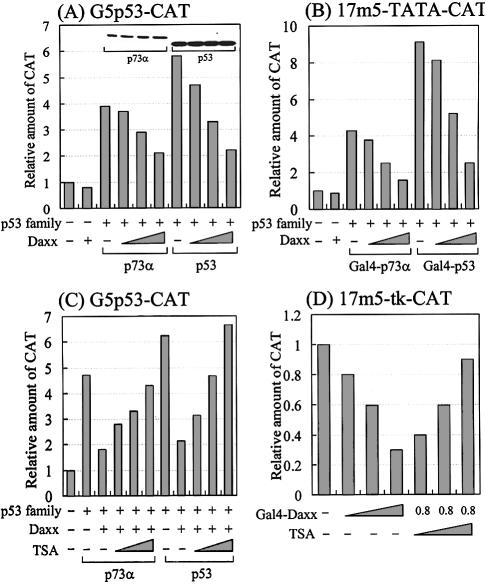
To determine the effect of Daxx on the transcriptional activities of members of the p53 family, p53-null C33A cervical carcinoma cells were transfected transiently with a fixed amount of p73α or p53 and increasing amounts of hDaxx, along with the G5p53-CAT reporter construct (the CAT reporter gene under the control of a p53-responsive element; a gift from Dr Robbins, Pittsburgh, PA). As shown in Figure 8A, Daxx inhibited the transactivation of both p73α and p53 in a dose-dependent manner. Additional transfection assays, in which the yeast Gal4-responsive 17m5-TATA-CAT reporter was used instead of G5p53-CAT, showed that Daxx also repressed the transcriptional activities of Gal4-fused p73α and p53, without affecting Gal4 alone (Fig. 8B), which suggests that Daxx functions as a co-repressor of the p53 proteins through protein–protein interactions. To rule out the possibility that apoptosis induced by the transfected Daxx might cause transcriptional downregulation, we measured the total protein concentrations produced by each of the transfections. No differences in the total protein levels might indicate that Daxx does not induce apoptosis under our experimental conditions. Furthermore, western analysis indicated no differences in the levels of p73α and p53 regardless of increased expressions of Daxx (Fig. 8A, inset), suggesting that Daxx neither induces apoptosis of transfected cells nor modulates downregulation of p73α and p53. Overall, these data suggest that Daxx represses specifically the transcriptional activity of the p53 family members, probably through specific protein–protein interactions.
Figure 8.
Effect of Daxx on the transcriptional activity of the p53 family. Saos2 cells were transfected with a total of 2.2 µg of DNA comprising the p53 family, Daxx expression vectors, 0.2 µg of p53-responsive G5p53-CAT (A and C), 17m5-TATA-CAT (B) or 17m5-tk-CAT (D) reporter plasmid, and 0.2 µg of SV40-driven β-gal internal control plasmid. The amounts of each expression vector were as follows: (A) lane 2, Daxx 0.8 µg; lanes 3–6, p73α 0.2 µg, Daxx 0, 0.2, 0.4 and 0.8 µg; lanes 7–10, p53 0.2 µg, Daxx 0, 0.2, 0.4 and 0.8 µg. (B) lane 2, Daxx 0.8 µg; lanes 3–6, Gal4-p73α 0.2 µg, Daxx 0, 0.2, 0.4 and 0.8 µg; lanes 7–10, Gal4-p53 0.2 µg, Daxx 0, 0.2, 0.4 and 0.8 µg. (C) lanes 2–6, p73α 0.2 µg, Daxx 0.8 µg, TSA 0.1, 0.2 and 0.4 nM. (D) lane 1, Gal4 alone 0.8 µg; lanes 2–4, Gal4-Daxx (amino acids 607–740) 0.2, 0.4 and 0.8 µg; lanes 5–7, Gal4-Daxx (amino acids 607–740) 0.8 µg, TSA 0.1, 0.2 and 0.4 µM. Data represent the mean of at least three independent transfections. The relative amount of CAT was determined by CAT ELISA after normalization by β-galactosidase activity. The inset in (A) shows a representative western blot of p73α and p53 with each band corresponding with the bar immediately below.
HDAC involvement in Daxx-mediated transcriptional repression was also studied. TSA, which is a well known inhibitor of HDAC, was added to cells that were transfected with p73α or p53, the Daxx expression vectors and the G5p53-CAT reporter plasmid. As shown in Figure 8C, downregulation of the reporter gene was relieved by TSA in a dose-dependent manner. We observed similar responses in transfection assays using p21WAF1-CAT, which is a natural p53-responsive reporter (data not shown). A previous report indicated that Daxx repressed basal transcription by recruiting HDAC to the promoter (20). To confirm this finding under our experimental conditions, we performed transient transfections with Gal4-Daxx expression vector and the yeast Gal4-responsive 17m5-tk-CAT reporter plasmid (Fig. 8D). Increasing amounts of Gal4-Daxx had titratable inhibitory effects on basal transcription, resulting in ∼70% repression. Furthermore, TSA reversed Gal4-Daxx-mediated transcriptional repression in a dose-dependent manner. In summary, our transfection data suggest that histone deacetylation by HDAC is involved in Daxx-mediated transcriptional repression of the p53 family members.
PML relieves Daxx-mediated transcriptional repression
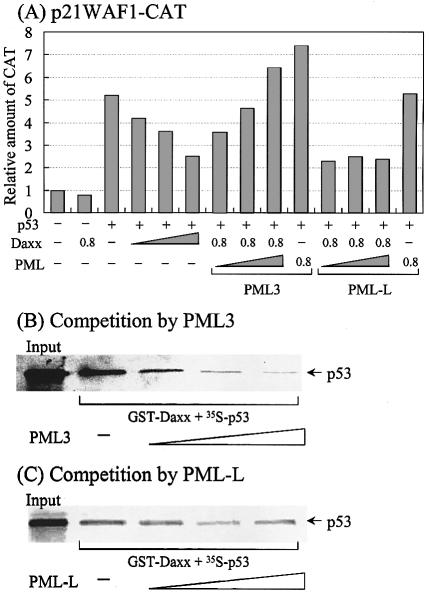
Since PML inhibits Daxx-mediated repression of basal transcription (20), and Pax3 activates transcription (28), and particularly activates p53 transcription by targeting it to the PML body (30,31), we investigated whether PML also played a role in modulating Daxx-mediated repression of p53-mediated activation. As shown in Figure 9A, the p53-responsive reporter activity of p21WAF1-CAT was gradually inhibited by increasing amounts of Daxx. This repression was relieved in a dose-dependent manner by PML3, but not by PML-L, which is another PML splice variant. This suggests that the effect is specific for PML3. When PML was expressed along with p53 in the absence of Daxx, PML3 (but not PML-L) increased the p53 activity slightly. This phenomenon may be attributed to the ability of PML3 to relieve p53 repression of endogenous Daxx. These observations corroborate the finding that PML3 (but not PML-L) interacts specifically with p53 for targeting to the PML body, thus promoting its biological functions, such as transactivation and apoptosis (31).
Figure 9.
PML3 relieves Daxx-mediated repression of p53 activated transcription possibly through the disruption of Daxx-p53 complex. (A) Transient transfection assays. Transfections were performed as described in Figure 8 except using p21WAF1-CAT as a reporter plasmid. The transfected amounts of Daxx, PML3 or PML-L were 0.2, 0.4 and 0.8 µg, respectively. The amount of p53 was 0.2 µg. Results represent the mean of three independent experiments. (B) Competition by PML3. For assays, 10 µl of in vitro translated p53 was mixed with purified GST–Daxx (amino acids 607–740) and further reacted with increasing volumes of PML3 (10, 20 and 40 µl). Afterwards, GST pull-down assays were performed as described in Figure 5. (C) Competition by PML-L. The same reactions were performed as (B) except using PML-L instead of PML3.
To gain insight into the underlying mechanism through which PML3 rescues Daxx-mediated repression of p53-activated transcription, we performed GST pull-down assays using GST–Daxx, [35S]p53 and PMLs. Both [35S]p53 and PMLs were synthesized in vitro using rabbit reticulocytes, while GST–Daxx was purified as described previously. In the absence of PMLs, Daxx was able to interact with p53 (Fig. 9B and C). However, the addition of increasing amounts of PLM3 to the reaction mixture progressively inhibited the interaction of Daxx with p53 (Fig. 9B). In contrast, PLM-L was not as effective as PML3 in disrupting the Daxx–p53 complex (Fig. 9C). These results indicate that PML3 competes specifically with Daxx for p53 binding, which is related to the previous finding that PML3, but not PML-L or RML-RARα, interacts strongly with p53 (31). Since our assays were performed in vitro under conditions that lacked the components necessary for sumoylation, it is not possible to speculate on the potential interactions between PML and Daxx, for which sumoylation of PML is required (25,26). Taken together, these results suggest that Daxx-mediated repression of p53 activation is relieved by PML3 binding to p53, which results in the observed release of p53 and the activation of p53-dependent transcription.
DISCUSSION
In this study, we identified hDaxx as a p73-interacting protein, and found that Daxx could interact with other members of the p53 family. Daxx was originally described as a cytoplasmic mediator of Fas signaling, but was recently identified as a nuclear protein that co-localizes with PML and regulates transcription. On the basis of our yeast studies, we mapped the interaction domains of the p53 family proteins and the C-terminus of Daxx. These interactions were confirmed by GST pull-down assays in vitro, and by immunoprecipitation in mammalian cells. Immunofluorescence assays using confocal microscopy revealed that both proteins co-localized in nuclear bodies, which were probably PML bodies or PODs. Various transfection assays indicated that the transcriptional activities of p73 and p53 from p73/p53-responsive promoters were inhibited in a dose-dependent manner by Daxx co-expression. The co-repressor activity of Daxx appeared to be HDAC dependent, as demonstrated by treatment with TSA, which is an inhibitor of HDAC. Furthermore, PML co-expression, specifically that of PML3, relieved Daxx-mediated repression of p53 activity, due to specific disruption of the Daxx–p53 complex by PML3. Taken together, these observations suggest that Daxx functions as a transcriptional repressor of members of the p53 family, and that the repressive effect is modulated specifically by PML3.
It has been proposed that PODs form a nuclear substructure that directs various cellular regulatory networks. PML, which is one of the major components of the nuclear PODs, is associated and co-localizes with several proteins, such as Sp100, Blm, p53, Rb, Mre11, NBS, CBP, Daxx and viral proteins (35,36). These associated proteins may direct the cellular functions of PODs, which include translation, transcription, and DNA repair processes that influence apoptosis, cell proliferation and senescence (32,37). Recently, new roles in transcriptional regulation have been reported for PODs, based on studies showing that transcriptional regulators, such as p53 (30,31), CBP (38,39), HDAC (40) and HIPK2 (8,41,42), interact with PML in PODs. In addition, the location of Daxx in the nucleus appears to determine its role in transcriptional regulation. In the absence of PML, Daxx is located mainly in the condensed chromatin, where it may associate with HDAC to repress transcription (20). However, when PML is expressed by either transfection or As2O3 induction, it targets Daxx to the PODS, which relieves the Daxx-mediated repression. This type of sequestration requires interaction between the N-terminal portion of PML, which includes the three sumoylation sites, and the C-terminal S/P/T-rich region of Daxx. Studies with PML mutants have indicated that the sumoylation of PML is essential not only for proper formation and stability of PODs (26), but also for the reversal of Daxx activity (20,28). Although PML-RARα shares the Daxx interaction domain with PML, it is not capable of reversing transcriptional repression caused by Daxx. This is probably because PML-RARα disrupts the POD structure, thereby directing Daxx to the disrupted microparticulate structures, where it retains the ability to repress transcription. It is noteworthy that PML binds p53 and targets it into PODs via the interaction between its C-terminus, which lacks sumoylation sites, and the DBD of p53 (30,31). This interaction appears to be specific for PML3, since no interactions have been noted between p53 and the splice variants PML-L and PML-RARα. However, in addition to the interactions that were determined in vitro, the sumoylation and POD localization of PML are required for p53 function in vivo. Recently, it has been reported that HIPK2, which co-localizes with PML in PODs, interacts directly with and phosphorylates p53 at Ser46. The phosphorylated p53 promotes CBP-mediated acetylation at Lys382, which results in p53 transcriptional activation and apoptosis (41,42). In this respect, our studies indicated that HIPK2 could associate functionally with HDAC, and act as a co-repressor of the p53-repressible promoters, while enhancing apoptosis and growth repression (8). Interestingly, a more recent report indicates that the HIPK2 isoform HIPK1 interacts with and phosphorylates Daxx, thereby modulating both Daxx translocation from PODs and Daxx-mediated transcriptional repression (43). Taken together, these observations suggest that PODs represent a dynamic site that modulates a network of regulatory pathways, which involve PML–HIPK–Daxx–p53–CBP interactions and protein modifications, such as the sumoylation of PML and p53, the phosphorylation of Daxx and p53, and the acetylation of p53, in response to various extracellular signals.
In terms of potential pathways in PODs, our current results describe how Daxx modulates the transcriptional activities of the p53 family. Our in vitro and in vivo binding assays revealed that Daxx associated directly with the p53 family. What are the consequences of p73/p53 interaction with Daxx and its recruitment to PODs, as described above? Our initial evidence suggests that Daxx modulates the p73/p53 transactivation activity. Co-expression of Daxx partly inhibited the transactivation function of p53 and other members of the p53 family when p53-positive CAT reporter genes were used for transfection in p53-null C33A cells. The response was HDAC dependent, as determined by treating the cells with TSA, which is an inhibitor of HDAC. The co-repressor function of Daxx was inhibited selectively by the co-expression of PML3. In corroboration of the transfection data, our GST pull-down assays indicated that PML3, but not PML-L, disrupted the Daxx–p53 complex, presumably by binding preferentially to p53, thereby freeing p53 from the repression complex and restoring its transcriptional activity. Previously, it was reported that PML sumoylation was required for proper targeting to PODs, which would relieve both Daxx-mediated repression of basal transcription (20) and Pax3-activated transcription (28). Therefore, it remains to be determined whether SUMO-1 conjugation is a prerequisite for PML to relieve Daxx-mediated repression of p53 activity, and whether targeting of PML to PODs is necessary for derepression. Nevertheless, we speculate that PML sumoylation is not required for derepression, because our GST pull-down assays were performed in vitro under conditions where the components required for sumoylation and PML targeting to PODs were lacking.
Daxx was originally identified as a cytoplasmic protein that promoted Fas-induced apoptosis (14). However, recent studies have indicated that Daxx is present exclusively in the nucleus, where it appears to play a direct role in regulating transcription (19–22). Furthermore, it has been suggested that Daxx is anti-apoptotic rather than pro-apoptotic (23,24). Therefore, it remains unclear whether Daxx is pro- or anti-apoptotic. Reflecting these complexities, our data suggest that Daxx is anti-apoptotic, since it represses the transcriptional activity of p53, which is a critical inducer of apoptosis. However, it appears that PML, which is an inducer of both p53-dependent and p53-independent apoptosis, can restore p53 activity in the presence of Daxx. In support of the pro-apoptotic role of Daxx, we found that in addition to its repressive role in p53-activated transcription, it could also enhance p53-repressible transcription of the Coll-CAT and MDR-CAT reporters (data not shown). In addition to repressing collagenase and MDR genes, p53 also regulates negatively the transcription of growth regulation genes, such as cyclin B, Bcl-2, Map4, survivin and presenilin-1 (44–48). The C-terminal OD of p53 is necessary for p53-mediated repression (3–5), and the HDACs and mSin3a also appear to be involved (6–8). These observations correlate with our finding that Daxx associates with the OD of p53, and utilizes HDAC as part of its repression mechanism. However, it remains to be determined whether Daxx expression can further repress the expression of the anti-apoptotic genes that are regulated negatively in vivo by p53. Interestingly, Daxx appears to be both pro-apoptotic and growth suppressive when overexpressed in p53-positive U2OS cells, as determined by 4′,6-diamidino-2-phenylindole (DAPI) staining, fluorescence-activated cell sorting (FACS) and colony formation analyses (data not shown). Although it is not clear whether or not the Daxx response is p53 dependent, it is possible that Daxx suppresses tumor cell growth through other members of the p53 family, such as p73 and p63. In support of this possibility, we found additional Daxx interaction domains (including the SAM domains) at the C-termini of p73 and p63. It will be interesting to determine whether such interactions are critical for Daxx function in apoptosis.
In contrast to our observations, a recent report indicated that tumorigenic mutants of p53 bind to Daxx and inhibit Daxx-dependent activation of ASK1, thereby preventing apoptosis that leads to cellular transformation (49). Under our experimental conditions, we were able to demonstrate clearly the interaction between the wild-type p53 and Daxx, which suggests a role for this interaction in the transcriptional regulation of p53. Consistent with our data, another report suggested a physical interaction between p53 and Daxx, although further investigations of the role of this complex in the regulation of transcription and apoptosis were not described (31). The discrepancies noted between our study and the other studies may be due to differences in the experimental conditions and thus need to be clarified.
Acknowledgments
ACKNOWLEDGEMENTS
We are very grateful to Dr Ann F. Pluta for hDaxx. This work was supported by grant number M1-01-KB-01-0001-02-K02-01-011-3-0 of the 21C Frontier Functional Human Genome Project from the Ministry of Science and Technology of Korea. S.-J.U. was supported by the BK21 project from the Ministry of Education and Human Resources Development.
REFERENCES
- 1.Somasundaram K. and El-Deiry,W.S. (2000) Tumor suppressor p53: regulation and function. Front. Biosci., 5, 424–437. [DOI] [PubMed] [Google Scholar]
- 2.Zhao R., Gish,K., Murphy,M., Yin,Y., Notterman,D., Hoffman,W.H., Tom,E., Mack,D.H. and Levine,A.J. (2000) Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev., 14, 981–993. [PMC free article] [PubMed] [Google Scholar]
- 3.Sang B.C., Chen,J.Y., Minna,J. and Barbosa,M.S. (1994) Distinct regions of p53 have a differential role in transcriptional activation and repression functions. Oncogene, 9, 853–859. [PubMed] [Google Scholar]
- 4.Shaulian E., Haviv,I., Shaul,Y. and Oren,M. (1995) Transcriptional repression by the C-terminal domain of p53. Oncogene, 10, 671–680. [PubMed] [Google Scholar]
- 5.Hong T.M., Chen,J.J., Peck,K., Yang,P.C. and Wu,C.W. (2001) p53 amino acids 339–346 represent the minimal p53 repression domain. J. Biol. Chem., 276, 1510–1515. [DOI] [PubMed] [Google Scholar]
- 6.Murphy M., Ahn,J., Walker,K.K., Hoffman,W.H., Evans,R.M., Levine,A.J. and George,D.L. (1999) Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev., 13, 2490–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Juan L.J., Shia,W.J., Chen,M.H., Yang,W.M., Seto,E., Lin,Y.S. and Wu,C.W. (2000) Histone deacetylases specifically down-regulate p53-dependent gene activation. J. Biol. Chem., 275, 20436–20443. [DOI] [PubMed] [Google Scholar]
- 8.Kim E.J., Park,J.S. and Um S.J. (2002) Identification and characterization of HIPK2 interacting with p73 and modulating functions of the p53 family in vivo. J. Biol. Chem., 277, 32020–32028. [DOI] [PubMed] [Google Scholar]
- 9.Kaelin W.G. Jr (1999) The p53 gene family. Oncogene, 18, 7701–7705. [DOI] [PubMed] [Google Scholar]
- 10.Lohrum M.A. and Vousden,K.H. (2000) Regulation and activation of the p53-related proteins: same family, different rules. Trends Cell Biol., 10, 197–202. [DOI] [PubMed] [Google Scholar]
- 11.Irwin M.S. and Kaelin,W.G. (2001) p53 family update: p73 and p63 develop their own identities. Cell Death Differ., 12, 337–349. [PubMed] [Google Scholar]
- 12.Yang A., Kaghad,M., Caput,D. and McKeon,F. (2002) On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet., 18, 90–95. [DOI] [PubMed] [Google Scholar]
- 13.Thanos C.D. and Bowie,J.U. (1999) p53 family members p63 and p73 are SAM domain-containing proteins. Protein Sci., 8, 1708–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X., Khosravi-Far,R., Chang,H.Y. and Baltimore,D. (1997) Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell, 89, 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang H.Y., Nishitoh,H., Yang,X., Ichijo,H. and Baltimore,D. (1998) Activation of apoptosis signal-regulating kinase 1 (ASK1) by the adapter protein Daxx. Science, 281, 1860–1863. [DOI] [PubMed] [Google Scholar]
- 16.Charette S.J., Lavoie,J.N., Lambert,H. and Landry,J. (2000) Inhibition of Daxx-mediated apoptosis by heat shock protein 27. Mol. Cell. Biol., 20, 7602–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Charette S.J., Lambert,H. and Landry,J. (2001) A kinase-independent function of Ask1 in caspase-independent cell death. J. Biol. Chem., 276, 36071–36074. [DOI] [PubMed] [Google Scholar]
- 18.Ko Y.G., Kang,Y.S., Park,H., Seol,W., Kim,J., Kim,T., Park,H.S., Choi,E.J. and Kim,S. (2001) Apoptosis signal-regulating kinase 1 controls the proapoptotic function of death-associated protein (Daxx) in the cytoplasm. J. Biol. Chem., 276, 39103–39106. [DOI] [PubMed] [Google Scholar]
- 19.Hollenbach A.D., Sublett,J.E., McPherson,C.J. and Grosveld,G. (1999) The Pax3-FKHR oncoprotein is unresponsive to the Pax3-associated repressor hDaxx. EMBO J., 18, 3702–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H., Leo,C., Zhu,J., Wu,X., O’Neil,J., Park,E.J. and Chen,J.D. (2000) Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol. Cell. Biol., 20, 1784–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li R., Pei,H., Watson,D.K. and Papas,T.S. (2000) EAP1/Daxx interacts with ETS1 and represses transcriptional activation of ETS1 target genes. Oncogene, 19, 745–753. [DOI] [PubMed] [Google Scholar]
- 22.Emelyanov A.V., Kovac,C.R., Sepulveda,M.A. and Birshtein,B.K. (2002) The interaction of Pax5 (BSAP) with Daxx can result in transcriptional activation in B cells. J. Biol. Chem., 277, 11156–11164. [DOI] [PubMed] [Google Scholar]
- 23.Michaelson J.S., Bader,D., Kuo,F., Kozak,C. and Leder,P. (1999) Loss of Daxx, a promiscuously interacting protein, results in extensive apoptosis in early mouse development. Genes Dev., 13, 1918–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michaelson J.S. and Leder,P. (2003) RNAi reveals anti-apoptotic and transcriptionally repressive activities of DAXX. J. Cell Sci., 116, 345–352. [DOI] [PubMed] [Google Scholar]
- 25.Ishov A.M., Sotnikov,A.G., Negorev,D., Vladimirova,O.V., Neff,N., Kamitani,T., Yeh,E.T., Strauss,J.F.,III and Maul,G.G. (1999) PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol., 147, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong S., Salomoni,P., Ronchetti,S., Guo,A., Ruggero,D. and Pandolfi,P.P. (2000) Promyelocytic leukemia protein (PML) and Daxx participate in a novel nuclear pathway for apoptosis. J. Exp. Med., 191, 631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hollenbach A.D., McPherson,C.J., Mientjes,E.J., Iyengar,R. and Grosveld,G. (2002) Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J. Cell Sci., 115, 3319–3330. [DOI] [PubMed] [Google Scholar]
- 28.Lehembre F., Muller,S., Pandolfi,P.P. and Dejean,A. (2001) Regulation of Pax3 transcriptional activity by SUMO-1-modified PML. Oncogene, 20, 1–9. [DOI] [PubMed] [Google Scholar]
- 29.Lin D.Y. and Shih,H.M. (2002) Essential role of the 58-kDa microspherule protein in the modulation of Daxx-dependent transcriptional repression as revealed by nucleolar sequestration. J. Biol. Chem., 277, 25446–25456. [DOI] [PubMed] [Google Scholar]
- 30.Guo A., Salomoni,P., Luo,J., Shih,A., Zhong,S., Gu,W. and Pandolfi,P.P. (2000) The function of PML in p53-dependent apoptosis. Nature Cell Biol., 2, 730–736. [DOI] [PubMed] [Google Scholar]
- 31.Fogal V., Gostissa,M., Sandy,P., Zacchi,P., Sternsdorf,T., Jensen,K., Pandolfi,P.P., Will,H., Schneider,C. and Del Sal G. (2000) Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J., 19, 6185–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salomoni P. and Pandolfi,P.P. (2002) The role of PML in tumor suppression. Cell, 108, 165–170. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 34.Pluta A.F., Earnshaw,W.C. and Goldberg,I.G. (1998) Interphase-specific association of intrinsic centromere protein CENP-C with HDaxx, a death domain-binding protein implicated in Fas-mediated cell death. J. Cell Sci., 111, 2029–2041. [DOI] [PubMed] [Google Scholar]
- 35.Pearson M. and Pelicci,P.G. (2001) PML interaction with p53 and its role in apoptosis and replicative senescence. Oncogene, 20, 7250–7256. [DOI] [PubMed] [Google Scholar]
- 36.Everett R.D. (2001) DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene, 20, 7266–7273. [DOI] [PubMed] [Google Scholar]
- 37.Borden K.L. (2002) Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol., 22, 5259–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LaMorte V.J., Dyck,J.A., Ochs,R.L. and Evans,R.M. (1998) Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc. Natl Acad. Sci. USA, 95, 4991–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Mikecz A., Zhang,S., Montminy,M., Tan,E.M. and Hemmerich,P. (2000) CREB-binding protein (CBP)/p300 and RNA polymerase II colocalize in transcriptionally active domains in the nucleus. J. Cell Biol., 150, 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu W.S., Vallian,S., Seto,E., Yang,W.M., Edmondson,D., Roth,S. and Chang,K.S. (2001) The growth suppressor PML represses transcription by functionally and physically interacting with histone deacetylases. Mol. Cell. Biol., 21, 2259–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hofmann T.G., Moller,A., Sirma,H., Zentgraf,H., Taya,Y., Droge,W., Will,H. and Schmitz,M.L. (2002) Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nature Cell Biol., 4, 1–10. [DOI] [PubMed] [Google Scholar]
- 42.D'Orazi G., Cecchinelli,B., Bruno,T., Manni,I., Higashimoto,Y., Saito,S., Gostissa,M., Coen,S., Marchetti,A., Del Sal,G., Piaggio,G., Fanciulli,M., Appella,E. and Soddu,S. (2002) Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser46 and mediates apoptosis. Nature Cell Biol., 4, 11–19. [DOI] [PubMed] [Google Scholar]
- 43.Ecsedy J.A., Michaelson,J.S. and Leder,P. (2003) Homeodomain-interacting protein kinase 1 modulates daxx localization, phosphorylation and transcriptional activity. Mol. Cell. Biol., 23, 950–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krause K., Wasner,M., Reinhard,W., Haugwitz,U., Dohna,C.L., Mossner,J. and Engeland,K. (2000) The tumour suppressor protein p53 can repress transcription of cyclin B. Nucleic Acids Res., 28, 4410–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyashita T., Harigai,M., Hanada,M. and Reed,J.C. (1994) Identification of a p53-dependent negative response element in the bcl-2 gene. Cancer Res., 54, 3131–3135. [PubMed] [Google Scholar]
- 46.Murphy M., Hinman,A. and Levine,A.J. (1996) Wild-type p53 negatively regulates the expression of a microtubule-associated protein. Genes Dev., 10, 2971–2980. [DOI] [PubMed] [Google Scholar]
- 47.Hoffman W.H., Biade,S., Zilfou,J.T., Chen,J. and Murphy,M. (2002) Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J. Biol. Chem., 277, 3247–3257. [DOI] [PubMed] [Google Scholar]
- 48.Roperch J.P., Alvaro,V., Prieur,S., Tuynder,M., Nemani,M., Lethrosne,F., Piouffre,L., Gendron,M.C., Israeli,D., Dausset,J., Oren,M., Amson,R. and Telerman,A. (1998) Inhibition of presenilin 1 expression is promoted by p53 and p21WAF1 and results in apoptosis and tumor suppression. Nature Med., 4, 835–838. [DOI] [PubMed] [Google Scholar]
- 49.Ohiro Y., Usheva,A., Kobayashi,S., Duffy,S.L., Nantz,R., Gius,D. and Horikoshi,N. (2003) Inhibition of stress-inducible kinase pathways by tumorigenic mutant p53. Mol. Cell. Biol., 23, 322–334. [DOI] [PMC free article] [PubMed] [Google Scholar]