Abstract
Many proteins can sense the relative orientations of two sequences at distant locations in DNA: some require sites in inverted (head-to-head) orientation, others in repeat (head-to-tail) orientation. Like many restriction enzymes, the BspMI endonuclease binds two copies of its target site before cleaving DNA. Its target is an asymmetric sequence so two sites in repeat orientation differ from sites in inverted orientation. When tested against supercoiled plasmids with two sites 700 bp apart in either repeated or inverted orientations, BspMI had a higher affinity for the plasmid with repeated sites than the plasmid with inverted sites. In contrast, on linear DNA or on supercoiled DNA with sites 1605 bp apart, BspMI interacted equally with repeated or inverted sites. The ability of BspMI to detect the relative orientation of two DNA sequences thus depends on both the topology and the length of the intervening DNA. Supercoiling may restrain the juxtaposition of sites 700 bp apart to a particular alignment across the superhelical axis, but the juxtaposition of sites in linear DNA or far apart in supercoiled DNA may occur without restraint. BspMI can therefore act as a sensor of the conformational dynamics of supercoiled DNA.
INTRODUCTION
Communications between distant DNA sites play key roles in almost all genetic events (1,2): in the replication, repair and restriction of DNA; in gene expression and its regulation; in genome rearrangements by transposition and site-specific recombination (3–9). Many of these systems require sites oriented in a particular manner. Some function only when the sites are in directly repeated (head-to-tail) orientation, for example the resolvases from the Tn3-like transposons (10). Others need sites in inverted (head-to-head or tail-to-tail) orientation, for example the Type III restriction enzymes (11). Such systems can thus sense the relative orientation of two DNA sites, even when the sites are far apart along the DNA.
In some cases, the protein(s) discerns the orientation of the sites by using an energy-dependent translocation mechanism to track along the DNA from one site to the other, for example, the Type III restriction enzymes and the MutHLS repair system (4,12). In other cases, the reaction is constrained to sites in a unique orientation by the topology of the DNA: the alignment of the sites needed for the enzyme reaction may be attainable from sites in one orientation but not from sites in the opposite orientation (13). For example, the structure needed for site-specific recombination by resolvase can be assembled from directly repeated sites in a supercoiled DNA but not from inverted sites nor from sites in trans, in separate molecules of DNA (10). The topology-sensing systems require supercoiled DNA, but a role for supercoiling in determining orientation specificity has yet to be fully established. Most perturbations that remove the requirement for supercoiling also remove the requirement for the sites to be oriented in a particular manner (14,15).
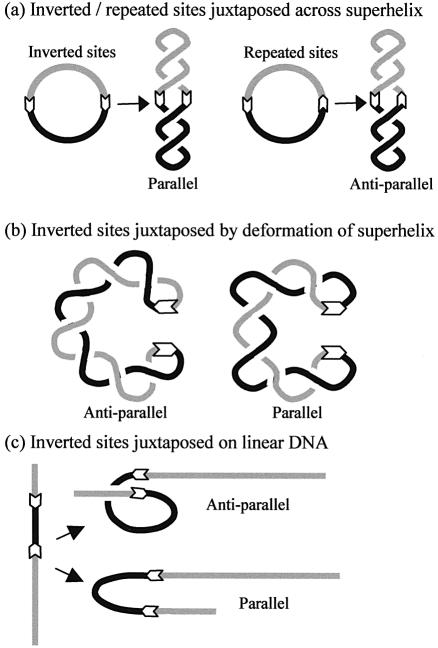
In a DNA with plectonemic supercoils (16), specific sites become juxtaposed mainly when located opposite each other across the superhelical axis, and only rarely through deformations of the superhelix (17,18). Juxtapositions across the superhelical axis automatically align directly repeated sites in a different manner from inverted sites (19,20): antiparallel and parallel, respectively (Fig. 1a). In contrast, juxtapositions by deforming the superhelix can yield both antiparallel and parallel alignments from inverted sites (Fig. 1b) and likewise from repeated sites. Moreover, supercoiled DNA of ≥4 kb is branched at typically 2 kb intervals (16). Sites ∼2 kb apart will often be in different branches and will become juxtaposed mainly by flexing the branches: both inverted and repeated sites then have equal probabilities for parallel and antiparallel encounters. The latter is also the case for juxtapositions in linear DNA (Fig. 1c). Hence, a system that recognises the alignment of two juxtaposed sites might prefer a particular orientation with closely spaced sites in supercoiled DNA, yet show no preference for that orientation with distantly spaced sites in supercoiled DNA or with sites in linear DNA. However, this test can be applied only to systems that act with similar efficiencies on supercoiled and linear DNA across varied separations.
Figure 1.
Juxtapositions of DNA sites. In all cases, white arrows mark the positions (and orientations) of two target sequences in the DNA and black and grey lines the DNA between the targets. (a) When juxtaposed across the superhelical axis, two sites in inverted orientation are aligned in parallel whereas two sites in directly repeated orientation are aligned in an antiparallel manner. (b) When sites in separate segments of the superhelix become juxtaposed by deformations of the superhelix, sites in inverted orientation can give rise to either antiparallel or parallel alignments. (c) In linear DNA, the juxtaposition of two sites in inverted orientation can give rise to either antiparallel or parallel alignments.
Many restriction enzymes interact with two copies of their recognition sequence before cutting DNA and most of these act on both supercoiled and relaxed DNA (6). The endonucleases that interact with two sites include the Type I and the Type III systems, which use translocation mechanisms to cleave DNA at sites distant from their binding sites (5). They also include many Type II enzymes that cleave DNA at fixed locations at or near their target sites. These Type II enzymes trap DNA loops by binding to two sites in cis, in the same molecule of DNA (6). However, the recognition sites for Type II enzymes are often palindromic sequences with rotational symmetry (21) and so cannot be assigned an orientation. But a group of Type II enzymes, the Type IIS systems, recognise asymmetric sequences and cleave both DNA strands at fixed positions several bases away from that sequence (22). Moreover, most of the Type IIS enzymes interact with two copies of their target sites before cleaving DNA, for example FokI, BsgI, MboII, BspMI and BfiI (23–27). These enzymes usually function better with sites in cis than with sites in trans, but it has yet to be determined for any of these enzymes if their reactions on sites in cis are affected by the relative orientation of the two sequences. If they are affected, the Type IIS enzymes could then reveal the mechanisms of how proteins sense the orientation of one DNA sequence relative to another elsewhere in the DNA.
The Type IIS enzyme studied here, BspMI, recognises the asymmetric sequence 5′-ACCTGC-3′ and cleaves top and bottom strands 4 and 8 bases downstream of this site, respectively (21). It exists in solution as a tetramer of identical subunits (26). Like the other tetrameric restriction enzymes (28,29), BspMI binds two copies of its target site and cleaves both sites concertedly. It converts a DNA with two sites directly to the final products cut at both sites, without liberating intermediates cut at one site (24). As BspMI has an asymmetric recognition sequence whose orientation can be defined, the possibility exists that its interaction with two target sites might be affected by their relative orientation. We report here that this is indeed the case and we identify some of the factors that govern this selectivity.
MATERIALS AND METHODS
Proteins
The BspMI endonuclease was purified from an over- producing strain of Escherichia coli (from R. Morgan, New England Biolabs) and its concentration (given in terms of its tetrameric form) determined as before (26). All other enzymes were purchased from commercial suppliers and used as advised by the supplier.
DNA
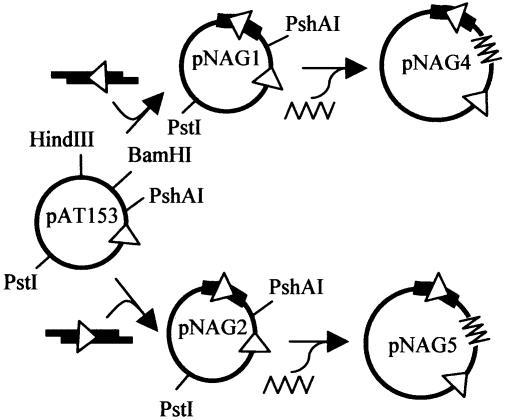
The plasmids pAT153 (30) and pNAG1 (24) were manipulated by standard procedures (31) to yield the derivatives shown in Figure 2. The duplex used to construct pNAG2 was made by annealing the oligodeoxyribonucleotide AGCTGTCATCTACCTGCCTGGACAGCATGGCC with GATCGGCCATGCTGTCCAGGCAGGTAGATGAC (from Cruachem Ltd, Glasgow, UK) at 95°C prior to slow cooling overnight. The plasmids were validated by sequencing across the sites of the insertion (University of Bristol Sequencing Service) and used to transform E.coli HB101 (31). The transformants were cultured in M9 minimal medium with 37 MBq/l [methyl-3H]thymidine (Amersham Biosciences UK Ltd) and the covalently closed form of the plasmid purified by density gradient centrifugations (28). The preparations contained mostly the supercoiled form of the monomeric plasmid, with generally <10% as either dimer or nicked open circle DNA. For reactions on linear DNA, the plasmids were cleaved with PstI, washed with phenol:chloroform and precipitated with ethanol.
Figure 2.
Plasmid substrates. The plasmid pAT153 has a single recognition site for BspMI denoted by the arrowhead (the tip points to the site of cleavage) and further restriction sites at the positions shown. To make pNAG1, pAT153 was cleaved with HindIII and BamHI and ligated to a duplex (thick line) made from two 32 base oligonucleotides: the duplex contained a double-stranded segment of 28 bp that has a recognition site for BspMI (arrowhead, orientation as indicated) and 5′ single-strand extensions of 4 bases at both ends, which match, respectively, HindIII and BamHI termini. The plasmid pNAG2 was constructed in an identical manner except that the sequence of the double-stranded segment of the duplex was reversed relative to the duplex in pNAG1. The plasmids pNAG4 and pNAG5 were constructed from pNAG1 and pNAG2, respectively, by inserting at their PshAI sites a 905 bp fragment from pBR322 (zigzag line) that lacks BspMI sites.
Reactions
Aliquots of BspMI (in BspMI dilution buffer) (24) were added to 20 vol of 3H-labelled DNA in reaction buffer (20 mM HEPES, pH 8.0, 100 mM NaCl, 1 mM dithiothreitol and 10 mM MgCl2) at 37°C. DNA concentrations were varied from 2 to 50 nM (while the reaction volume was altered from 400 to 100 µl). An aliquot of the reaction mixture was removed before adding the enzyme. Further aliquots were taken at various times after adding the enzyme (to 0.5 nM) and mixed immediately with a half volume of stop mix (26). The aliquots varied from 30 µl, from reactions with 2 nM DNA, to 4 µl, with 50 nM DNA. Twelve such samples were taken during a typical reaction. The samples were analysed by electrophoresis through agarose under conditions that separated the substrate and each of the reaction products and the concentrations of each form were determined by scintillation counting (28).
Reaction velocities were evaluated using GRAFIT (Ethricaus Software, Slough, UK) to fit the initial decrease in the concentration of the DNA substrate with time to a linear slope. The velocities (mol DNA cleaved/min) were normalised against the enzyme concentration to give turnover rates (mol DNA/mol enzyme/min). Each velocity cited here is the mean from between three and five repeats: error bars denote the standard deviations. The variations in reaction velocity with substrate concentration were fitted by GRAFIT directly to the Michaelis–Menten equation to yield Vmax and Km values.
RESULTS
Plasmids
The BspMI restriction enzyme cleaves plasmids with one copy of its recognition sequence at very low rates, though its activity against such substrates is enhanced by oligoduplexes containing the cognate sequence (26,32). Hence, as with most Type IIS enzymes, BspMI interacts with two copies of its recognition site. It probably cuts DNA with one site by means of interactions in trans, bridging sites on separate molecules of DNA. The effective concentration of one DNA site in the vicinity of another is higher for two sites in cis, in the same chain, than for sites in trans (7). Consequently, BspMI has a much lower Km for DNA with two recognition sites than DNA with one site, and it usually cleaves the two-site DNA more rapidly than the one-site DNA (24,26). The two-site DNA used previously had sites in direct repeat. Whether BspMI is affected by the orientation of its sites was not determined.
A plasmid with one site for BspMI, pAT153 (30), was manipulated to give two isogenic derivatives with two sites, pNAG1 (24) and pNAG2 (Fig. 2). The only difference between pNAG1 and pNAG2 is the orientation of a 28 bp segment of DNA that carries the second BspMI site: in pNAG1, the two sites are in direct repeat; in pNAG2, the two sites are in inverted orientation (Fig. 2). The derivatives are the same size, 3344 bp, and contain the same length of DNA between the recognition sequences for BspMI: 700 bp in one arc, 2632 bp in the other. Since restriction enzymes are often affected by sequences flanking their target sites (33), the sequences around the newly introduced BspMI sites in pNAG1 and pNAG2 were kept the same as the original site in pAT153: for 7 bp upstream and for 15 bp downstream of the site. The latter spans the sites of cleavage of both top and bottom strands, 4 and 8 bp downstream, respectively.
Reactions on supercoiled DNA
Previous work on BspMI had used the supercoiled forms of pAT153 and pNAG1, the plasmids with one and with two repeated sites for BspMI (24,26). This was extended here by using both the supercoiled and the linear forms of not only pAT153 and pNAG1 but also pNAG2, the plasmid with two sites in inverted orientation (Fig. 3). Steady-state conditions were employed, with lower concentrations of enzyme than DNA, so that only a small fraction of the DNA would be bound to the enzyme during the reaction. The DNA observed during the reaction is then mainly free rather than enzyme-bound DNA. Since these substrates yield several products, cleaved in one or both strands at one or both sites, reaction velocities were determined from the initial linear decrease in the concentration of the substrate with time, rather than from the appearance of any single product.
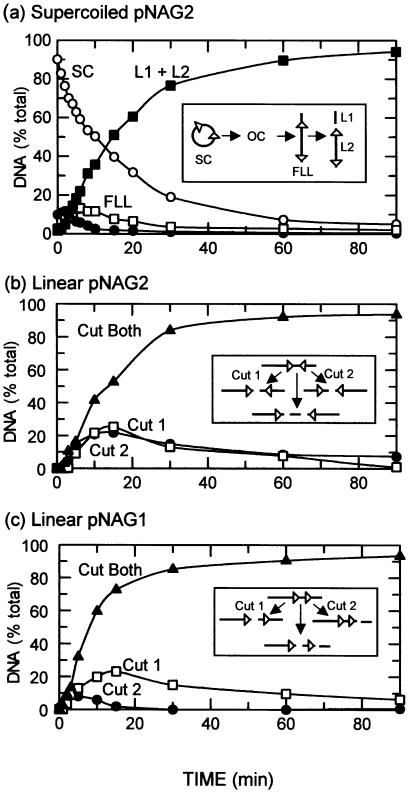
Figure 3.
Concerted reactions. The reactions contained 0.5 nM BspMI endonuclease and 5 nM DNA in reaction buffer at 37°C. Samples were withdrawn from the reactions at various times and analysed as in Materials and Methods to determine the amounts of each form of the DNA shown in the inserts. In (a), the substrate was supercoiled pNAG2, a plasmid with two BspMI sites in inverted orientation: white circles, the intact supercoiled substrate (SC); black circles, open circle DNA cut in one strand (OC); white squares, the full-length linear form cut in both strands at one site (FLL); black squares, the mean of the two linear fragments from cutting both sites in both strands (L1/L2). In (b), the substrate was PstI-linearised pNAG2: white squares, the DNA cut solely at site 1; black circles, the DNA cut solely at site 2; black triangles, the DNA cut at both sites 1 and 2. In (c), the substrate was PstI-linearised pNAG1, a DNA with two BspMI sites in repeat orientation: white squares, the DNA cut solely at site 1; black circles, the DNA cut solely at site 2; black triangles, the DNA cut at both sites 1 and 2.
BspMI cleaved the plasmid with two sites in inverted orientation in a concerted manner (Fig. 3a), in much the same way as the plasmid with repeated sites (26). The majority of the supercoiled (SC) DNA was cleaved directly to its final products, the two linear DNA fragments generated by cutting both strands at both sites (L1 and L2): only small amounts of either the open circle (OC) form of the DNA, cut in one strand at one or both sites, or the full-length linear (FLL) DNA, from cutting both strands at one site, were released from the enzyme. However, the FLL form can come from cutting the circular DNA at either one of its two BspMI sites.
Reactions on linear DNA
To see if BspMI cuts the two-site plasmid preferentially at one site over the other, linear substrates were employed that yield distinct products from cleaving one site compared to the other. For example, after cleaving pNAG2 with PstI (Fig. 3b), a BspMI reaction at site 1 (the original site from pAT153) cuts the 3344 bp substrate into fragments of 795 and 2549 bp; a BspMI reaction at site 2 (the newly introduced site) yields fragments of 1503 and 1841 bp; while cleavage of both sites yields three fragments of 795, 708 and 1841 bp. The fragments of 2549, 1503 and 708 bp thus mark, respectively, the amounts of the DNA cut only at site 1 (but not at 2), only at site 2 and at both sites. When BspMI was added to the linear form of pNAG2, the substrate with inverted sites, the majority of the DNA was cleaved concertedly to directly give the final products cut at both sites: only small amounts of the DNA were liberated after cutting at site 1 alone or site 2 alone (Fig. 3b). However, the two singly cut fragments from linear pNAG2 were formed in equal yield. BspMI thus has the same intrinsic activity at each site.
The linear form of pNAG1, the DNA with two BspMI sites in direct repeat, was also cleaved in a concerted manner: again, only small fractions of the DNA were liberated after cutting just one site (Fig. 3c). However, in contrast to the above, the two singly cut products were not formed in equal yields. Instead, much more was cleaved at site 1 than at site 2.
The difference between the inverted and the repeated orientations is probably a consequence of BspMI cutting DNA on one side of its recognition site, 4/8 bp away. DNA cleaved by BspMI thus still possesses the intact recognition sequence, either to the ‘left’ or to the ‘right’ of the point of cleavage, depending on the orientation of the site. In a linear DNA with head-to-head (inverted) sites, the two fragments from cutting at site 1 alone and the two from cutting at site 2 alone all carry one copy of the recognition sequence (insert to Fig. 3b). On the other hand, in the linear DNA with head-to-tail (repeated) sites, the two fragments from cutting at site 1 both carry one copy of the recognition sequence, but one of the two fragments from cutting at site 2 carries two copies of the sequence, while the other has none (Fig. 3c). DNA with the recognition sequence for BspMI, but not the cleavage site, can activate BspMI on a DNA with one target site (26). Hence, the fragment that has two copies of the recognition sequence may be processed rapidly to the final product cut at both sites and so fail to accumulate during the reaction, if the site next to the point of cleavage activates the cleavage of the other site. This proposal also accounts for why the amount of singly cut DNA produced during the reaction on the circular substrate is smaller than that on the linear substrate: the FLL form from cutting either site on the circular DNA still possesses two intact copies of the recognition sequence (Fig. 3a).
The BspMI endonuclease exists as a tetramer of identical subunits (26), each of which presumably contains one active site. Given either supercoiled or linear DNA with two recognition sites, in either directly repeated or inverted orientations, BspMI usually cleaves both strands at both sites within the lifetime of the DNA–protein complex (Fig. 3). Hence, regardless of the arrangement of the two sites, the tetramer must still be able to engage at the same time all four of the target phosphodiester bonds in the two-site substrate. However, the affinity of the enzyme for two sites in direct repeat may still differ from inverted sites. The concurrent binding of BspMI to two sites in a particular orientation could impose an energetically unfavourable configuration on the intervening DNA and so reduce the affinity of the enzyme for sites in that orientation.
Affinities for sites in supercoiled DNA
To assess the relative affinities of BspMI for two recognition sites in repeated or inverted orientations and for two sites in trans, the steady-state kinetics of its reactions on the supercoiled forms of pAT153, pNAG1 and pNAG2 were examined at various substrate concentrations (Fig. 4a). The first of these, pAT153, has one BspMI site and is cleaved by means of trans interactions across two molecules of the plasmid. The latter two have two BspMI sites 700 bp apart, in repeated or inverted orientations, respectively, and are cleaved mainly by cis reactions spanning sites in a single DNA. The changes in reaction velocity with DNA concentration were analysed to yield values for Vmax and Km (Table 1). [In low ionic strength buffers, the Km of BspMI for pNAG1 is too low to measure. The lowest practicable concentration of this DNA (2 nM) gives the Vmax rate (26). A higher ionic strength was used here.]
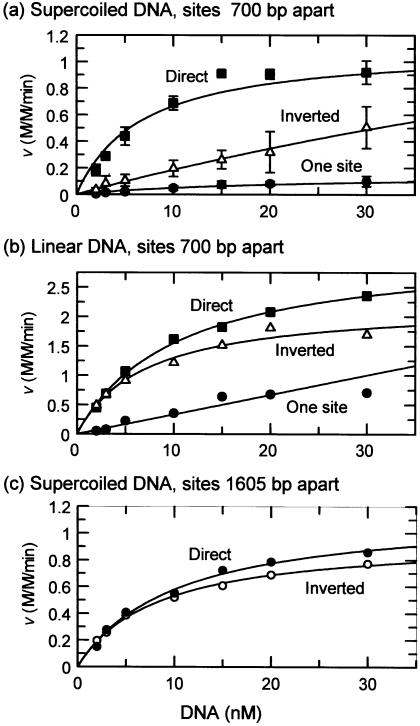
Figure 4.
Steady-state kinetics. The reactions, in reaction buffer at 37°C, contained 0.5 nM BspMI endonuclease and DNA at the concentration indicated on the x-axis. In (a), the substrates were the supercoiled forms of: pAT153, a plasmid with one BspMI site (black circles); pNAG1, a plasmid with two directly repeated BspMI sites 700 bp apart (black squares); pNAG2, a plasmid with two inverted sites 700 bp apart (white triangles). In (b), the substrates were linear DNA, from cutting the following with PstI: pAT153 (black circles); pNAG1 (black squares); pNAG2 (white triangles). In (c), the substrates were the supercoiled forms of: pNAG4, directly repeated BspMI sites 1605 bp apart (black circles); pNAG5, inverted sites 1605 bp apart (white circles). Rates of DNA cleavage, measured from the initial linear decrease in substrate concentration with time, were normalised to give the turnover rates on the y-axis. The values are the mean from between three and five independent repeats of each reaction: the error bars in (a) mark the standard deviations [in (b) and (c), error bars are not shown as those from the directly repeated sites overlap those from the inverted sites]. The lines drawn are the best fits to the Michaelis–Menten equation: the corresponding values for Vmax and Km (and the associated error margins) are given in Table 1.
Table 1. Kinetic parameters.
| Substrate | Site spacing (bp) | Site orientation | Vmax (mol DNA/min/mol enzyme) | Km (nM) |
|---|---|---|---|---|
| Supercoiled pNAG1 | 700 | Repeated | 1.1 ± 0.1 | 7 ± 2 |
| Supercoiled pNAG2 | 700 | Inverted | ∼2.5 ± 0.5 | ∼130 ± 30 |
| Linear pNAG1 | 700 | Repeated | 3.2 ± 0.1 | 8 ± 1 |
| Linear pNAG2 | 700 | Inverted | 2.2 ± 0.2 | 10 ± 1 |
| Supercoiled pNAG4 | 1605 | Repeated | 1.0 ± 0.1 | 10 ± 1 |
| Supercoiled pNAG5 | 1605 | Inverted | 1.2 ± 0.1 | 7 ± 2 |
The rates at which BspMI cleaved the plasmid with two sites in direct repeat increased with increasing DNA concentration up to a maximal rate, in the hyperbolic manner expected for a system following Michaelis–Menten kinetics (Fig. 4a). Throughout the concentration range examined, this DNA was cleaved more rapidly than the plasmid with one BspMI site. The plasmid with inverted sites was also cleaved more rapidly than the one-site plasmid, but at all concentrations tested this DNA was cleaved more slowly than the DNA with repeated sites (Fig. 4a). Moreover, instead of showing saturation kinetics, the rates of the reactions on the plasmid with inverted sites increased more or less linearly with increasing amounts of DNA throughout the accessible concentration range (even concentrations >50 nM were insufficient to approach saturation; data not shown).
Even though the v versus [S] data for the plasmid with inverted sites can be fitted to a Vmax and a Km (Table 1), the data pertain solely to reactions at substrate concentrations far below the Km, so the resultant values are ill determined. Nevertheless, the Km of BspMI for the inverted sites in pNAG2 is clearly much larger than that for the repeated sites in pNAG1, perhaps 20 times larger, while the two substrates have Vmax values that differ by less than 3-fold. Thus, on supercoiled DNA with two recognition sites 700 bp apart, BspMI forms a synaptic complex much more readily from directly repeated sites than from inverted sites, but the turnover rate of the complex with repeated sites is similar to that with inverted sites.
Affinities for sites in linear DNA
The plasmids pAT153, pNAG1 and pNAG2 were cleaved with PstI and the linear DNA used as substrates for steady-state reactions (Fig. 4b). The linear form of pNAG1, the DNA with two BspMI sites in direct repeat, had a similar Km to the same DNA in its supercoiled configuration, but a higher Vmax (Table 1). The rate-limiting step in the reaction pathway for BspMI occurs after the initial binding to the DNA but before phosphodiester hydrolysis and is most likely coupled to the formation of the synaptic complex bridging the sites (26). This may occur faster in linear DNA than in supercoiled DNA.
The DNA with one BspMI site was also cleaved more rapidly in its linear form than in its supercoiled form, by an even larger factor than that for the two-site DNA. Consequently, at all concentrations tested, the difference between the rates of cleavage of the substrates with one BspMI site and with two repeated sites was smaller with linear DNA than with supercoiled DNA. Interactions between sites in trans, on two separate molecules of DNA, occur more readily when at least one of the molecules is linear than when both are supercoiled (28). The linear DNA with one site was also cleaved more slowly than the linear DNA with two inverted sites.
The linear DNA with two sites in inverted orientation behaved, however, in a different manner from the same DNA in its supercoiled configuration. The rate of cleavage of the supercoiled plasmid with two inverted sites had increased more or less linearly with increasing DNA concentrations (Fig. 4a), but the rate of cleavage of the same DNA in its linear form followed a hyperbolic curve to a maximum at a relatively low DNA concentration (Fig. 4b). BspMI therefore has a much smaller Km for inverted sites in linear DNA than for inverted sites in supercoiled DNA (Table 1). Moreover, both the Km and Vmax for the linear DNA with inverted sites are similar to those for the linear DNA with repeated sites (Table 1). Thus, in supercoiled DNA with sites 700 bp apart, BspMI has a higher affinity for sites in repeat orientation than for sites in inverted orientation. Yet in linear DNA with sites 700 bp apart, BspMI has similar affinities for repeated and inverted sites.
Increased site separation
Further derivatives of pNAG1 and pNAG2 were constructed by inserting into the 700 bp segment between their BspMI sites a 905 bp piece of DNA that lacks the recognition site (Fig. 2). The derivatives, pNAG4 and pNAG5, have two BspMI sites flanked by identical sequences, but now separated by 1605 bp. In pNAG4, the sites are in repeat orientation, while pNAG5 has inverted sites. The increase in the length of DNA between the BspMI sites also increases the overall size of the plasmids, from 3344 bp for pNAG1/2 to 4249 bp for pNAG4/5. The larger plasmids will thus generally have branched supercoils while the smaller plasmids will often lack branches (16).
The supercoiled forms of the plasmids with increased inter-site spacings were also used as substrates for steady-state reactions (Fig. 4c). As before (Fig. 3a), BspMI cleaved these plasmids in a concerted fashion: almost all of the DNA was cleaved directly to the final products cut at both sites, largely bypassing the DNA cut at a single site (data not shown). With directly repeated sites, BspMI had similar values for both Vmax and Km on pNAG4, the plasmid with sites 1605 bp apart, as pNAG1, the plasmid with sites 700 bp apart (Table 1). In contrast, with inverted sites, BspMI had a much lower Km for sites separated by 1605 bp, in pNAG5, than for sites separated by 700 bp, in pNAG2 (Table 1). Consequently, on supercoiled DNA with sites 700 bp apart, BspMI has a greater affinity for repeated sites than inverted sites. Yet when the sites are separated by 1605 bp, its affinity for the sites in direct repeat is similar to that for the sites in inverted orientation.
DISCUSSION
It now appears that most restriction enzymes interact with two copies of their recognition sites before cleaving DNA (6). Many of these enzymes have palindromic recognition sites with the same 5′→3′ sequence in the ‘top’ and ‘bottom’ strands (21). Such sequences are unaltered by reversing the site within the DNA, so their orientation cannot be specified. The restriction enzymes that might be affected by the orientation of one site relative to another are exclusively those that recognise asymmetric sequences: for example, the Type I, the Type IIS and the Type III systems (5,22). DNA cleavage by the Type I and Type III enzymes normally follows the collision of two molecules of the enzyme that are both translocating DNA adjacent to their recognition sites (34). The Type I enzymes translocate bidirectionally, on both sides of the recognition site (35), while the Type III enzymes act unidirectionally (12). The former are thus unaffected by whether their sites are in repeat or inverse orientations (36) but the latter need sites in a particular (the inverse) orientation (11).
Outside the translocating systems, this report provides the first example of selective recognition of site orientation by a restriction enzyme. Like most Type IIS enzymes (23–27), BspMI has to interact with two copies of its recognition sequence before cleaving DNA. On supercoiled DNA with two sites 700 bp apart, it has a much higher affinity, ∼20-fold, for repeated sites over inverted sites (Table 1). However, on the linear forms of these plasmids, no significant preference was seen for repeated sites over inverted sites. Nor was any preference observed on supercoiled plasmids with 1605 bp between their BspMI sites. BspMI thus constitutes a test system to analyse how a protein recognises the orientation of one DNA sequence relative to another at a distant location in the DNA.
Many systems that recognise the orientation of two DNA sequences have an absolute requirement for sites in a particular orientation and have no activity at all on sites in the opposite orientation (1,9). BspMI differs in at least two respects from such systems. First, even in situations where BspMI prefers one orientation to the opposite orientation, it still possesses some activity against the opposite orientation. Secondly, it shows a conditional rather than a fixed preference for sites in a particular orientation, which depends on both DNA superhelicity and either the length of the intervening DNA and/or the overall size of the DNA molecule. BspMI therefore cannot employ the sort of rigorous barrier that limits, for example, resolvase to sites in direct repeat whilst excluding it from reactions with inverted sites (10).
The selective recognition of site orientation by BspMI can, however, be correlated to the scheme in Figure 1, provided that it is active only after synapsing two recognition sites in the antiparallel rather than parallel alignment. In a supercoiled DNA with sites separated by 700 bp, the preference for directly repeated sites can then be accounted for by the juxtaposition of sites across the superhelical axis generating an antiparallel alignment only from sites in direct repeat (Fig. 1a). The juxtaposition of inverted sites across the superhelical axis produces the parallel alignment. A different mode of juxtaposition, involving the deformation of the superhelical axis, is then needed to generate the active (antiparallel) complex from inverted sites (Fig. 1b).
On the other hand, distant sites in a large molecule of supercoiled DNA will usually encounter each other on collision of separate segments of the superhelix (18). Indeed, such sites will often be located in different branches of the superhelix. In these situations, the antiparallel alignment can be generated equally readily from repeated or inverted sites (Fig. 1b). This scheme can thus account for why, in 3.3 kb molecules of supercoiled DNA with two BspMI sites 0.7 kb apart, BspMI has a lower Km for repeated sites than for inverted sites, yet, in 4.3 kb supercoiled molecules with sites 1.6 kb apart, it has almost the same Km for repeated and inverted sites. Similarly, on linear DNA with sites separated by 0.7 kb, the intervening segment is considerably longer than the persistence length of DNA (37), so the antiparallel alignment will be generated just as readily from inverted sites as from repeated sites.
Why would BspMI prefer the antiparallel alignment? With another Type IIS enzyme, FokI, the binding of the dimeric form of the protein to two sites has been modelled with the sites in either the parallel or antiparallel alignment (23). Both models seem consistent with catalytic activity. However, while FokI is a monomer that associates to a dimer for its DNA cleavage reactions (38), BspMI is a tetramer of identical subunits (26). The four subunits in the BspMI tetramer are perhaps arranged in two pairs of ‘primary dimers’ (Fig. 5), as in the NgoMIV tetramer (29), but while the primary dimers of NgoMIV each contact one copy of its palindromic recognition sequence, BspMI has an asymmetric site. Hence, as in FokI (23,38), each subunit of BspMI may have a DNA-binding surface that encompasses the entire recognition sequence. If so, the binding of two copies of the recognition sequence to the two subunits within a primary dimer, say subunits 1 and 2 (Fig. 5), will give the active antiparallel alignment, while the binding to two subunits within different dimers, say subunits 1 and 4 (Fig. 5), yields the inactive parallel alignment.
Figure 5.
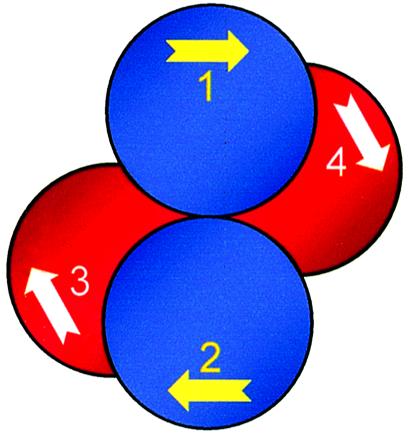
Model for BspMI. The subunits of the BspMI restriction endonuclease are illustrated as spheres, numbered 1–4. As with the NgoMIV tetramer (29), subunits 1 and 2 (in blue) are related by a two-fold axis and constitute a primary dimer, as do subunits 3 and 4 (in red). All four subunits contain a binding site for the asymmetric DNA sequence recognised by BspMI, in the direction marked by the arrow. The yellow arrows in subunits 1 and 2 indicate binding sites on the upper face of the protein and the white arrows in subunits 3 and 4 binding sites on the underside of the protein. When bound to subunits 1 and 2, the two recognition sequences are in antiparallel alignment. When bound to subunits 1 and 4, they lie parallel to each other.
Acknowledgments
ACKNOWLEDGEMENTS
We thank R. Morgan for the BspMI clone and D. Gowers, D. Scott, M. Szczelkun and A. Welsh for advice and comments. This work was supported by the Biotechnology and Biological Research Council and the Wellcome Trust.
REFERENCES
- 1.Gellert M. and Nash,H. (1987) Communication between segments of DNA during site-specific recombination. Nature, 325, 401–404. [DOI] [PubMed] [Google Scholar]
- 2.Echols H. (1990) Nucleoprotein structures initiating DNA replication, transcription and site-specific recombination. J. Biol. Chem., 265, 14697–14700. [PubMed] [Google Scholar]
- 3.Barre F.X., Soballe,B. Michel,B., Aroyo,M., Robertson,M. and Sherratt,D.J. (2001) Circles: the replication-recombination-chromosome segregation connection. Proc. Natl Acad. Sci. USA, 98, 8189–8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen D.J., Makhov,A., Grilley,M., Taylor. J., Thresher,R., Modrich,P. and Griffith,J.D. (1997) MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J., 14, 4467–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dryden D.T.F., Murray,N.E. and Rao,D.N. (2001) Nucleoside triphosphate-dependent restriction enzymes. Nucleic Acids Res., 29, 3728–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halford S.E. (2001) Hopping, jumping and looping by restriction enzymes. Biochem. Soc. Trans, 29, 363–373. [DOI] [PubMed] [Google Scholar]
- 7.Rippe K., von Hippel,P.H. and Langowski,J. (1995) Action at a distance: DNA-looping and initiation of transcription. Trends Biochem. Sci., 20, 500–506. [DOI] [PubMed] [Google Scholar]
- 8.Maniatis T. and Reed,R. (2002) An extensive network of coupling among gene expression machines. Nature, 416, 499–506. [DOI] [PubMed] [Google Scholar]
- 9.Hallet B. and Sherratt,D.J. (1997) Transposition and site-specific recombination: adapting DNA cut-and-paste mechanisms to a variety of DNA rearrangements. FEMS Microbiol. Rev., 21, 157–178. [DOI] [PubMed] [Google Scholar]
- 10.Stark W.M. and Boocock,M.A. (1995) Topological selectivity in site-specific recombination. In Sherratt,D.J. (ed.), Mobile Genetic Elements. Oxford University Press, Oxford, UK, pp. 101–129. [Google Scholar]
- 11.Meisel A., Bickle,T.A., Krüger,D.H. and Schroeder,C. (1992) Type III restriction enzymes need two inversely oriented recognition sites for DNA cleavage. Nature, 355, 467–469. [DOI] [PubMed] [Google Scholar]
- 12.Meisel A., Mackeldanz,P., Bickle,T.A., Krüger,D.H. and Schroeder,C. (1995) Type III restriction endonucleases translocate DNA in a reaction driven by recognition site-specific ATP hydrolysis. EMBO J., 14, 2958–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanaar R. and Cozzarelli,N.R. (1992) Roles of supercoiled DNA structure in DNA transactions. Curr. Opin. Struct. Biol., 2, 369–379. [Google Scholar]
- 14.Klippel A., Cloppenborg,K. and Kahmann,R. (1988) Isolation and characterization of unusual gin mutants. EMBO J., 7, 3983–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnold P.H., Blake,D.G., Grindley,N.D.F., Boocock,M.R. and Stark,W.M. (1999) Mutants of Tn3 resolvase which do not require accessory binding sites for recombination activity. EMBO J., 18, 1407–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vologodskii A.V. and Cozzarelli,N.R. (1994) Conformational and thermodynamic properties of supercoiled DNA. Annu. Rev. Biophys. Biomol. Struct., 23, 609–643. [DOI] [PubMed] [Google Scholar]
- 17.Huang J., Schlick,T. and Vologodskii,A. (2001) Dynamics of site juxtaposition in supercoiled DNA. Proc. Natl Acad. Sci. USA, 98, 968–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klenin K.V. and Langowski,J. (2001) Intrachain reactions of supercoiled DNA simulated by Brownian dynamics. Biophys. J., 81, 1924–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adzuma K. and Mizuuchi,K. (1989) Interaction of proteins located at a distance along DNA: mechanisms of target immunity in the Mu DNA strand-transfer reaction. Cell, 57, 41–47. [DOI] [PubMed] [Google Scholar]
- 20.Vologodskii A. and Cozzarelli,N.R. (1996) Effects of supercoiling on the juxtapostition and relative orientation of two DNA sites. Biophys. J., 70, 2548–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts R.J., Vincze,T., Posfai,J. and Macelis,D. (2003) REBASE: restriction enzymes and methyltransferases. Nucleic Acids Res., 31, 418–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szybalski W., Kim,S.C., Hasan,N. and Podhajska,A.J. (1991) Class-IIS restriction enzymes—a review. Gene, 100, 13–26. [DOI] [PubMed] [Google Scholar]
- 23.Vanamee E.S., Santaga,S. and Aggarwal,A.K. (2001) FokI requires two specific DNA sites for cleavage. J. Mol. Biol., 309, 69–78. [DOI] [PubMed] [Google Scholar]
- 24.Bath A.J., Milsom,S.E., Gormley,N.A. and Halford,S.E. (2002) Many type IIs restriction endonucleases interact with two recognition sites before cleaving DNA. J. Biol. Chem., 277, 4024–4033. [DOI] [PubMed] [Google Scholar]
- 25.Soundararajan M., Chang,Z., Morgan,R.D., Heslop,P. and Connolly,B.A. (2002). DNA binding and recognition by the IIs restriction endonuclease MboII. J. Biol. Chem., 277, 887–895. [DOI] [PubMed] [Google Scholar]
- 26.Gormley N.A., Hillberg,A.L. and Halford,S.E. (2002) The Type IIs restriction endonuclease BspMI is a tetramer that acts concertedly at two copies of an asymmetric DNA sequence. J. Biol. Chem., 277, 4034–4041. [DOI] [PubMed] [Google Scholar]
- 27.Lagunavicius A., Sasnauskas,G., Halford,S.E. and Siksnys,V. (2003) The metal-independent type IIs restriction enzyme BfiI is a dimer that binds two DNA sites but has only one catalytic centre. J. Mol. Biol., 326, 1051–1064. [DOI] [PubMed] [Google Scholar]
- 28.Wentzell L.M., Nobbs,T.J. and Halford,S.E. (1995) The SfiI restriction endonuclease makes a 4-strand DNA break at two copies of its recognition sequence. J. Mol. Biol., 248, 581–595. [DOI] [PubMed] [Google Scholar]
- 29.Deibert M., Grazulis,S., Sasnauskas,G., Siksnys,V. and Huber,R. (2000) Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA. Nature Struct. Biol., 7, 792–799. [DOI] [PubMed] [Google Scholar]
- 30.Twigg A.J. and Sherratt,D.J. (1980) Trans-complementable copy-number mutants of plasmid ColE1. Nature, 283, 216–218. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J. and Russsell,D.W. (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY. [Google Scholar]
- 32.Oller A.R., Vanden Broek,W., Conrad,M. and Topal,M.D. (1991) Ability of DNA and spermidine to affect the activity of restriction endonucleases from several bacterial species. Biochemistry, 30, 2543–2549. [DOI] [PubMed] [Google Scholar]
- 33.Taylor J.D. and Halford,S.E. (1992) The activity of the EcoRV restriction endonuclease is influenced by flanking DNA sequences both inside and outside the DNA-protein complex. Biochemistry, 31, 90–97. [DOI] [PubMed] [Google Scholar]
- 34.Studier F.W. and Bandyopadhyay,P.K. (1988) Model for how type I restriction enzymes select cleavage sites in DNA. Proc. Natl Acad. Sci. USA, 85, 4677–4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Firman K. and Szczelkun,M.D. (2000) Measuring motion on DNA by the type I restriction endonuclease EcoR124I using triplex displacement. EMBO J., 19, 2094–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szczelkun M.D., Dillingham,M.S., Janscak,P., Firman,K. and Halford,S.E. (1996) Repercussions of DNA tracking by the type IC restriction endonuclease EcoR124I on linear, circular and catenated substrates. EMBO J., 15, 6335–6347. [PMC free article] [PubMed] [Google Scholar]
- 37.Hagerman P.J. (1988) Flexibility of DNA. Annu. Rev. Biophys. Biophys. Chem., 17, 265–286. [DOI] [PubMed] [Google Scholar]
- 38.Bitinaite J., Wah,D.A., Aggarwal,A.K. and Schildkraut,I. (1998) FokI dimerization is required for DNA cleavage. Proc. Natl Acad. Sci. USA, 95, 10570–10575. [DOI] [PMC free article] [PubMed] [Google Scholar]