Abstract
Nucleotide substitution, insertion and deletion (indel) events are the major driving forces that have shaped genomes. Using the recently identified human ribosomal protein (RP) pseudogene sequences, we have thoroughly studied DNA mutation patterns in the human genome. We analyzed a total of 1726 processed RP pseudogene sequences, comprising more than 700 000 bases. To be sure to differentiate the sequence changes occurring in the functional genes during evolution from those occurring in pseudogenes after they were fixed in the genome, we used only pseudogene sequences originating from parts of RP genes that are identical in human and mouse. Overall, we found that nucleotide transitions are more common than transversions, by roughly a factor of two. Moreover, the substitution rates amongst the 12 possible nucleotide pairs are not homogeneous as they are affected by the type of immediately neighboring nucleotides and the overall local G+C content. Finally, our dataset is large enough that it has many indels, thus allowing for the first time statistically robust analysis of these events. Overall, we found that deletions are about three times more common than insertions (3740 versus 1291). The frequencies of both these events follow characteristic power–law behavior associated with the size of the indel. However, unexpectedly, the frequency of 3 bp deletions (in contrast to 3 bp insertions) violates this trend, being considerably higher than that of 2 bp deletions. The possible biological implications of such a 3 bp bias are discussed.
INTRODUCTION
It is important to study the patterns and frequencies of neutral mutations in the genome as these mutation events, which include nucleotide substitutions, insertions and deletions (the latter two are often collectively referred as indels), provide the molecular basis of gene and genome evolution. A very powerful approach in such study is to infer the mutation patterns by comparing the sequences of functional genes and the corresponding pseudogenes. Pseudogenes are ‘dead’ copies of genes, which were created by genomic duplication or retrotransposition (1). The latter type are often referred to as ‘processed pseudogenes’ as they are processed by the LINE1 retrotransposon machinery, i.e. reverse-transcribed from a functional mRNA transcript and integrated into the nuclear genome (2). In general, the processed pseudogenes are ‘dead-on-arrival’ because they lack promoter sequences and cannot be transcribed. Consequently, they are not under selective pressure and are free to accumulate mutations in their sequences.
Patterns of DNA mutations in human are of particular interest. They will shed light on some important evolutionary questions such as the genome stability, DNA repair and chromosome replication. From a medical perspective, most of the human inherited diseases are caused by DNA mutations (substitution or indels), thus a good understanding of the mutation process will certainly help in disease diagnostics and treatment (3,4). Previously, some investigators have studied this subject using only small number of pseudogenes (5–7); the mutation patterns in the human genome were also compared with those in the fruitfly genome (8).
As useful as pseudogenes are, there are two intrinsic problems associated with them that could potentially introduce bias in these studies. (i) When comparing the sequences between a pseudogene and a functional gene, it is often difficult to tell whether a gap in the sequence alignment was the result of an insertion event in one sequence or a deletion event in the other. In a previous study, a large number of these ambiguous indels were classified as ‘gaps’ instead of more specifically as insertions or deletions (9). (ii) Optimally, such sequence alignment and comparison should be performed between the pseudogene and the ancestral gene that the pseudogene derived from. However, because the sequence of both the functional gene and the pseudogene have evolved during evolution, it is often impossible to determine whether the difference in the sequence alignment reflects the substitutions in the pseudogenes or in the functional genes. In addition, previous to the whole-genome sequencing projects, discovery of human pseudogenes have been sporadic and only small number of sequences were available for such analysis.
Recently, we conducted a whole-genome survey of ribosomal protein (RP) processed pseudogenes (10). A total of about 2000 near complete processed pseudogenes were discovered in the human genome. Such unprecedented amount of pseudogene sequence information provided us with a good opportunity to investigate the substitution and indel patterns in great detail. RP pseudogenes also provided another unique advantage, as their sequences are well conserved among vertebrates and virtually identical among mammals (11).
In this paper, we describe an extensive study on the DNA mutation patterns as inferred from human RP pseudogenes. To separate the mutations that occurred to the functional genes during evolution from the mutations that occurred to the pseudogenes after they were fixed in the genome, we only used the pseudogene sequences that originated from the regions on the RP genes that are consensus between human and mouse. A total of 1726 processed RP pseudogenes and more than 700 000 conserved bases were analyzed in the study, which included 53 024 substitutions and over 5000 insertions or deletions. We will discuss the patterns of nucleotide mutations, neighboring biases and correlations with the genomic background. Unexpectedly, we also observed an unusual high frequency for 3 bp deletions.
MATERIALS AND METHODS
Derive consensus RP gene sequences
The protein and DNA sequences of the 79 human and mouse RP were retrieved from GenBank. For each RP, the amino acid sequences from the two species were aligned first and the DNA sequences were threaded through the amino acid alignment to obtain the DNA alignment. Coding sequences that are consensus between human and mouse were determined from the DNA alignment. In total, 89% of the coding DNA are identical (consensus) between human and mouse, 7.1% have undergone transitional substitutions, 3.6% have undergone transversions and the remaining 0.3% are deletions or insertions in one of the two species.
Remove duplicated pseudogenes
The procedures of discovering human RP pseudogenes have been described in a previous report (10). We did multiple sequence alignment for each group of pseudogenes using the program CLUSTALW (12), and obtained phylogenetic trees following neighbor-joining (NJ) method. For each RP, the phylogenetic tree was visually examined and pseudogenes that appear to have gone through recent duplications were removed. In other words, all the remaining pseudogenes were created from independent retrotransposition events. The final dataset obtained 1726 processed RP pseudogenes and more than 700 000 conserved nucleotides.
Sequence alignment between the pseudogenes and the functional genes
We ran the TFASTX program of the FASTA suit (13) to obtain the predicted amino acid sequences for each pseudogene. We then used two methods to align the pseudogene DNA sequence with the RP genes. First, we used CLUSTALW (12) to obtained the DNA alignment directly. Separately, we wrote a PERL script to do a ‘codon based’ alignment using the amino acid sequences obtained from TFASTX. The two alignments were mostly in good agreement; we further visually examined the alignments to ensure accuracy.
Simulation on deletion frequencies
We assumed that all the pseudogene sequences were already present at the beginning of the simulation and were free of indels. We further ignored all the insertions and those deletions that were longer than three nucleotides. The evolution of the pseudogenes was simulated as cycles of random deletions. We introduced three parameters in the simulation, mk, (k = 1, 2, 3), so that at every mkth cycle a deletion of length k was introduced at a random position in the sequence. We were only interested to determine the relative frequencies of the 1, 2 and 3 bp simultaneous deletion events, i.e. the ratios between m1, m2 and m3. Therefore we fixed m1 at 10, i.e. we would introduce a 1 bp deletion to the sequence at every 10th cycle.
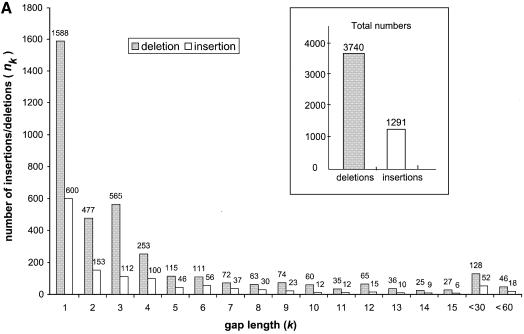
The total observed frequencies of 1, 2 and 3 bp deletions in the RP pseudogenes were 1588, 477 and 565 (Fig. 3A). From these numbers we could estimate the upper bound for the frequencies of simultaneous 2 and 3 bp deletions relative to the 1 bp deletions. We could imagine that the observed 477 2 bp deletions could be caused by either a simultaneous 2 bp deletions or accumulation of two adjacent 1 bp deletions, therefore the frequency of simultaneous 2 bp deletions must be less than or equal to 477. Similarly the frequencies of simultaneous 3 bp deletions must be less than or equal to 565. We then converted the upper bound for the deletion frequencies to the lower bound of m2 and m3, i.e. the range that we needed to survey in the simulation experiments: m2 ≥ 33, (33 = 10·1588/477), and m3 ≥ 28 (28 = 10·1588/565).
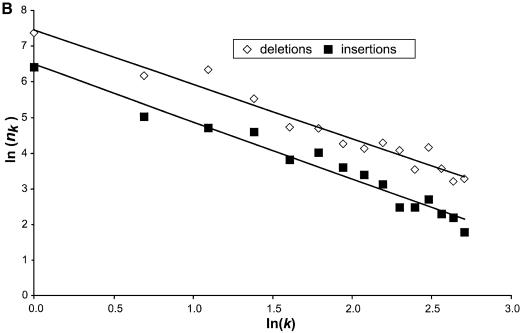
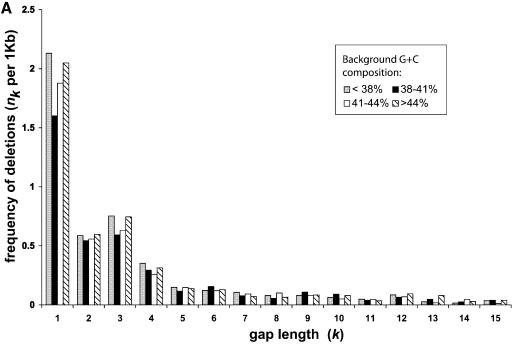
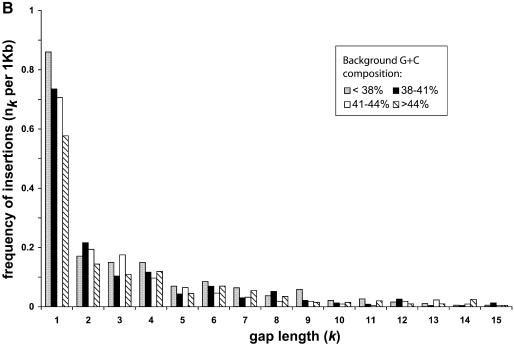
Figure 3.
(A) The length distribution of insertions and deletions in the pseudogenes. Only deletions and insertions of <60 bp are shown. The total number of insertions and deletions are shown in the inset. (B) Plots of k and nk on log scale. Deletions are shown as closed squares and insertions as open diamonds. Trend lines are fitted to the two series as well.
We started the simulation with a string of 836 105 bases, which was the total number of nucleotides in the present-day pseudogenes plus deletions and minus insertions. For all the combinations of m2 and m3 (37 ≥ m2 ≥ 33 and 35 ≥ m3 ≥ 28), we ran the simulation until the total number of 1 bp deletions reached 1588. We repeated the simulation 100 times and calculated the averaged number of 2 and 3 bp deletions and standard deviations for all the combination of m2 and m3. The pair of m2 and m3 that had the 2 and 3 bp frequencies that were most close to the observed frequencies of 477 and 565 were: m2 = 34 and m3 = 28, which produced 471 2 bp deletions and 567 3 bp deletions. Therefore, the ratio between the frequencies of 2 and 1 bp deletions was most likely to be 0.29:1 (0.29 = 10/34), and the ratio between 3 and 1 bp deletions was 0.36:1 (0.36 = 10/28).
RESULTS AND DISCUSSIONS
Transversions are more frequent than transitions
Table 1 lists some of the overall statistics of the RP pseudogene sequences that originated from the coding regions that were consensus between human and mouse. For each nucleotide type (A, G, C or T), the columns in the table give the total number of the nucleotides (‘Total, Ti’), the number of nucleotides that were missing in the pseudogenes (‘Deleted’), the number of nucleotides that remained unchanged in the pseudogenes (‘Unchanged’), the number of nucleotides that have mutated to another type (‘Substitutions, Si’), the number of nucleotides that have undergone transitional (‘Transitions’) or transversional mutations (‘Transversions’) and the ratio between the transitions and transversions (‘ratio’). Transitions refer to the substitution of a purine (A or G) by another purine or the substitution of a pyrimidine (T or C) by another pyrimidine; transversions are the substitutions of a purine by a pyrimidine or a pyrimidine by a purine. The nucleotides C and G that are part of the CpG di-nucleotides were counted separately. The bottom three rows in the table list the number of nucleotides in the pseudogenes for which the exact type in the ancestral RP genes could not be determined, i.e. these nucleotides are not conserved between mouse and human. These ambiguous positions constitute only a small fraction of the total sequences (∼7%), thus for majority of the nucleotides in the pseudogenes, we could un-ambiguously determine the consensus nucleotide in the ancestral mammalian RP genes. This demonstrated the advantages of using RP pseudogenes in this type of study. Using conserved RP gene sequences as a measuring stick avoided any ambiguity encounter by previous studies (14,15). Throughout this report, when we speak of RP pseudogenes, we strictly refer to the pseudogene sequences that were derived from the human–mouse consensus coding regions. In addition, our pseudogene dataset is much larger (at least 20 times) than the pseudogene datasets that were previously analyzed; this avoids the risk of introducing potential errors caused by small sample size.
Table 1. Summary of nucleotide substitutions in human RP pseudogenes.
| Nucleotide | Total (Ti) | Deleted | Unchanged | Substitutions (Si) | Transitions | Transversions | Ratio | |
|---|---|---|---|---|---|---|---|---|
| A | 216 954 | 4289 (2.0%) | 200 198 (92.3%) | 12 467 (5.7%) | 7564 (60.7%) | 4903 (39.3%) | 1.54 | |
| T | 143 617 | 2514 (1.8%) | 131 300 (91.4%) | 9803 (6.8%) | 6334 (64.6%) | 3469 (35.4%) | 1.83 | |
| C | ||||||||
| non-CpG | 158 686 | 2919 (1.8%) | 141 907 (89.4%) | 13 860 (8.7%) | 9533 (68.8%) | 4327 (31.2%) | 2.20 | |
| CpG | 19 913 | 517 (2.5%) | 12 979 (65.2%) | 6417 (32.2%) | 5314 (82.8%) | 1103 (17.2%) | 4.82 | |
| all | 178 599 | 3436 (1.9%) | 154 886 (86.7%) | 20 277 (11.4%) | 14 847 (73.2%) | 5430 (26.8%) | 2.73 | |
| G | ||||||||
| non-CpG | 185 117 | 3885 (2.1%) | 164 338 (88.8%) | 16 894 (9.1%) | 12 264 (72.6%) | 4630 (27.4%) | 2.65 | |
| CpG | 19 913 | 500 (2.5%) | 12 768 (64.1%) | 6645 (33.4%) | 5617 (84.5%) | 1028 (15.5%) | 5.46 | |
| all | 205 031 | 4386 (2.1%) | 177 106 (86.4%) | 23 539 (11.5%) | 17 881 (76.0%) | 5658 (24.0%) | 3.16 | |
| Total | ||||||||
| non-CpG | 704 374 | 13 607 (1.9%) | 637 743 (90.5%) | 53 024 (7.5%) | 35 695 (67.3%) | 17 329 (32.7%) | 2.06 | |
| CpG | 39 827 | 1018 (2.6%) | 25 747 (64.6%) | 13 062 (32.8%) | 10 931 (83.7%) | 2131 (16.3%) | 5.13 | |
| All | 744 201 | 14 625 (2.0%) | 663 490 (89.2%) | 66 086 (8.9%) | 46 626 (70.6%) | 19 460 (29.4%) | 2.40 | |
| A or G | 25 705 | 692 (2.7%) | 838 | |||||
| C or T | 35 826 | 714 (2.0%) | 1190 | |||||
| Non-consensus | 30.373 | 694 (2.3%) |
Except for those in the CpG di-nucleotides, on average ∼90% of the nucleotides in the RP pseudogenes remained unchanged from their original type in the genes. For those that were substituted, it was evident that for each nucleotide type, transitions were more frequent than transversions. Among the four nucleotides types, C and G had higher transition versus transversion ratios than A and T including those C and G that were not in the CpG di-nucleotides. This was largely caused by the overwhelmingly higher frequency of transitions from C to T and from G to A. Pseudogenes in general have higher G+C composition than the background genomic DNA. So once they are inserted into the genome, they tend to decay to lower G+C composition to blend into the surrounding region on the chromosome (10). Similar patterns were also observed for repetitive elements (16), collectively, this reflects a genome-wide mutational bias.
It has been known that in the human genome, most of the C residues in the CpG di-nucleotides are methylated and are frequently deaminated and mutated to T (17,18). This explained the high proportion of transitional events for C and G in the CpG di-nucleotides as shown in Table 1. As a result, the CpG di-nucleotides are greatly under-represented in the human genome.
Pattern of nucleotide substitutions
We are interested to study the substitution rates between nucleotide pairs, i.e. to determine the frequency of mutations from one type of nucleotide to another. If we denote Ti as the total number of type i nucleotides (i = A, G, C or T) in an RP gene and Ni→j as the number of times that a nucleotide is mutated from type i to type j in the corresponding pseudogene, then we can define a set of rates of substitutions as follows:
Ki→j = (Ni→j)/Ti 1
i.e. Ki→j indicates the proportion of the nucleotides of type i in the functional RP DNA that have mutated to type j in the pseudogene.
Alternatively, instead of measuring how frequently one type of nucleotide is mutated to another, we can ask the question that, given that a mutation has occurred, what is the relative frequency that it has mutated to one of the three other types? In other words, instead of normalizing Ni→j by Ti, we now normalized it by Si, i.e. the total number of mutations occurred in type i nucleotides. In equation 2, we formally define Pi→j as the ‘proportion of substitutions’. Equation 3 describes the relationship between these two different sets of statistics. Values for Ti and Si for each type of nucleotide can be found in Table 1.
It has been recognized that the nucleotide substitution rates vary in different regions of the human genome, especially among regions of different G+C composition (10,16). For each RP pseudogene, we calculated the average G+C content of the 50 000 bp flanking region and grouped them into one of four bins on the basis of their background G+C content. We divided the bins in such way that each group had roughly the same amount of DNA. Substitutions between complementary nucleotide pairs were added together and represented in a unified formula, e.g. the two complementary substitution events A→G and T→C were added and represented as A:T→G:C. Such notations are used throughout this report.
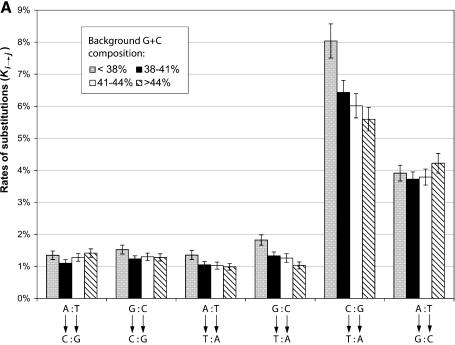
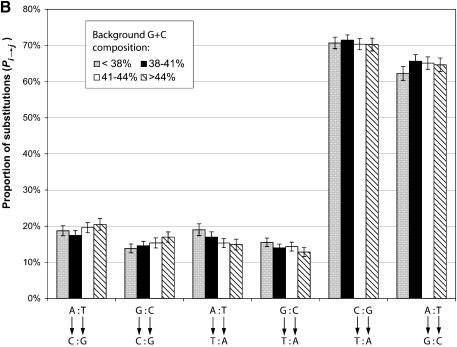
Figure 1 compares the two statistics, Ki→j and Si→j, for substitutions between the six nucleotide pairs in four different GC bins. The substitution rates Ki→j shown in Figure 1A are in good agreement with the rates previously reported, which were derived from less than a dozen human pseudogenes (8,15). As discussed in the previous section, the two transitional substitutions, which are shown on the right in the figure, were far more frequent than the transversions, shown on the left. This is true for both the un-normalized rates shown in Figure 1A and the normalized rates shown in Figure 1B. It is also obvious from Figure 1A that between the two transitional events, C:G→T:A is much more frequent than A:T→G:C regardless of the background G:C composition. This trend was more pronounced for the G+C-poor regions than the G+C-rich regions, since the local G+C composition appears to have opposite effects on the substitution rates of the two transitions. C:G→T:A substitutions are more frequent in the regions of lower G+C content and the A:T→G:C substitutions are more frequent in the regions of higher G:C content (except for the region of G:C < 38%). This can be explained by the different mutation pressure that the pseudogenes faced in the different regions of the genome as, in general, genes and pseudogenes have higher G+C composition than background genomic DNA.
Figure 1.
Substitution pattern between nucleotide pairs. Pseudogenes are grouped by their background G+C composition. (A) Substitution rates as normalized by the numbers of nucleotides of each type. Each column represents the proportion of nucleotides that have mutated to another type. Confidence intervals (95%) are also given. (B) The proportion of substitutions as normalized by the numbers of mutations that have occurred to each type. Each column represents, among total number of mutated nucleotides, the proportion of mutations from one type to another.
However, a different picture emerges in Figure 1B, especially for the two transitional events, when we compare the proportion of substitutions Si→j. Even though C:G→T:A substitutions are still more frequent than A:T→G:C, the difference between the two is much smaller. Furthermore, t-tests showed that the background G+C composition has little effect on the normalized substitution rates for these two transitions.
Neighboring effects on the substitutions
It has been proposed that nucleotide substitutions have a neighboring bias, i.e. the chance that a nucleotide is mutated and the type of nucleotide that it is mutated to are affected by the adjacent nucleotides (5,19). To study this neighboring bias, it is essential that the nucleotide in question has not changed since the time the pseudogene was inserted in the genome. Therefore we discarded those di-nucleotides in which both nucleotides have mutated in the pseudogenes. We further excluded those di-nucleotides that either were a CpG or overlapped with a CpG in the ancestral gene sequence since CpG quickly mutate to TpG in the human genome. Because of the complementary nature of the DNA, we need only to consider the neighboring effect of the 5′ nucleotide. For instance, the effect of C on T in the di-nucleotide TpC is just equivalent to the effect of G on A in the complementary di-nucleotide GpA. For each remaining di-nucleotide in the pseudogenes, we divided them into four groups according to their first (5′) nucleotide. For each group, we calculated the statistics of the substitutions for the second nucleotide in the di-nucleotides.
We defined two statistics, Kij→k and Rij→k, to characterize the neighboring effects of nucleotide i on nucleotide j in a di-nucleotide ij. First, we need to introduce the following notations: ij represents a di-nucleotide that is composed of nucleotides of type i (5′) and type j (3′); Nij→ik represents the number of times that di-nucleotide ij in the functional genes has mutated to ik in the pseudogenes. Therefore, in equation 4 we define Kij→k as the rate of a single base pair substitution from j to k in the context of a di-nucleotide ij. In other words, Kij→k represents the frequency of a dinucleotide ij becoming ik in the pseudogenes. We did not include the cases where one or more of the nucleotides in ij are deleted in the pseudogenes.
In equation 5, we also define the statistic Pij→k as the ‘proportions of substitutions’, which represents, given a mutation has occurred to nucleotide j in the original di-nucleotide ij, the chance that it has mutated to k.
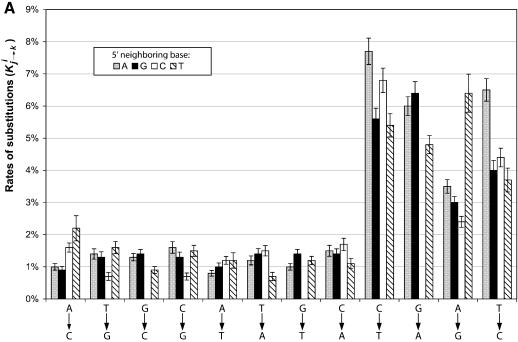
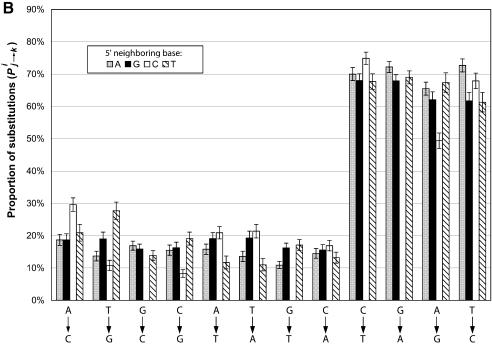
Figure 2A compares the substitution rates Kij→k for the 48 combination of i, j and k. The single nucleotide substitutions that have the same 5′ neighboring nucleotide are represented by columns of the same color. For example, the first column from the left (dark blue) indicates that ∼1% of all the di-nucleotide ApA in the RP genes has mutated to ApC in the pseudogenes. The 95% confidence intervals were also given. Similarly, Figure 2B compares the proportion of substitutions, Pij→k, for all the possible combination of i, j and k, e.g. the first column on the left indicates that, of all the mutations that occurred to the second base A in di-nucleotide ApA, 20% of them resulted in ApC. The data for substitutions G→C, G→T and G→A are not in these figures since they are substitutions from di-nucleotide CpG.
Figure 2.
Neighboring effects on the nucleotide substitution patterns. Di-nucleotides are grouped on the basis of their first (5′) nucleotide. (A) Substitution rates as normalized by the numbers of nucleotides of each type. Each column represents, given that the first nucleotide is unchanged, the chance that the second nucleotide has mutated to another type in the pseudogenes. Substitutions that have the same type of 5′ adjacent nucleotide have the same shading. (B) Proportion of substitutions as normalized by the total numbers of mutations that have occurred to the 3′ nucleotide. Each column represents, given that a mutation has occurred to the second nucleotide in the original di-nucleotide, the chance that it mutated to each one of the three other types.
It is obvious from Figure 2A that nucleotide substitutions do have significant neighboring biases. For example, the first four columns on the left in Figure 2A show that the single nucleotide substitutions A→C is more than twice as frequent in the di-nucleotides TpA than in ApA. The four transitional substitutions: C→T, G→A, A→G, T→C and the transversion T→A are also significantly affected by the 5′ neighboring base. One needs to be cautious when interpreting the rates of those substitution that result in CpG since it is likely that the majority of the resulting CpG di-nucleotides have mutated to TpG soon after the original substitution event. However, such secondary substitutions do not affect the calculated rates for other substitutions that also result in TpG such as ApG→TpG or TpA→TpG.
Similar to the analysis of single nucleotide substitutions, the variation in substitutions exhibited in Figure 2A were likely the results of two distinct but intervening factors: (i) the variation in the stability of a di-nucleotide, i.e. the likelihood that a mutation occurs to the second base in a di-nucleotide, and (ii) the variation in the directionality of the substitutions, i.e. given that a mutation has occurred, the frequency of that the nucleotide being mutated to one of the three other nucleotide types. Figure 2B shows that the proportions of substitution Pij→k clearly have a different variation pattern than the rates of substitutions Rij→k. In general, the biases exhibited in Pij→k are reduced in Figure 2B, and in some cases different trends emerged for the same type of substitutions. For example, in Figure 2B the substitution C:A→C has the largest proportion of substitutions compared with A:A→C, G:A→C and T:A→C, but in Figure 2A the rate is less than T:A→C.
We believe that the neighboring effects we have discussed above are correlated with the bias in the di-nucleotide abundances observed in the human genome, i.e. the fact that the observed di-nucleotide relative abundances significantly deviate from the expectations calculated from the frequencies of the component nucleotides (20). Such biases are mostly pronounced for di-nucleotide CpG and TpA, for which the ratios between the observed and expected abundances are 23 and 73% respectively (18). It is generally agreed that the deficiency in the CpG abundance was caused by the methylation of the cytosine bases (17), whereas the cause of the TpA deprivation in the human genome remains unclear (21). Close examination of Figure 2A shows that for all the single nucleotide substitutions from A to another type (i.e. A→G, A→C and A→T), TpA always has larger substitution rates than other di-nucleotides. Therefore the presence of T has made TpA significantly less conserved than other di-nucleotides such as ApA, GpA and CpA, which could be the cause for the under-representation of TpA in the genome.
Length distribution of insertions and deletions
Other than substitutions, genomic DNA also constantly undergoes random indels. Indels occur much less frequently than substitutions, therefore a larger amount of DNA sequences is needed to characterize them. Our pseudogene dataset is most valuable for this purpose as a larger dataset provides greater statistical significance. Figure 3A shows the length distributions of the insertions and deletions in the human RP pseudogenes. In total, 3740 deletions and 1291 insertions were observed; the ratio between the two is 2.90:1. Indels that are longer than 60 bp were not included in this analysis since it is likely that they resulted from a different mechanism. In a previous study (7), Ophir and Graur observed a similar ratio between deletions and insertions (244:84) from much fewer (93) pseudogene sequences.
The mean length of the deletions is 4.2 bp with a median at 2 bp and the mean length of insertions is 4.3 bp with median at 2 bp as well. Figure 3A illustrates that the frequency of both deletions and insertions of gap length k, nk decreases rapidly when k increases (with the exception of k = 3 for deletions). In fact, 70% of the total deletion events and 67% of the total insertion events have gap length equal or less than 3 (k ≤ 3). Furthermore, 42% of the total deletions and 46% of the total insertions are single-base indel events. This sheer dominance of short indel events is reminiscent of a power–law behavior (i.e. nk = ak–b), which exists in a wide variety of areas in biology and non-biological sciences (22). Figure 3B plots the parameters k and nk on log-scale for both insertions and deletions, and power–law trend lines were fitted with the data points. Both the insertions and the deletions can be fitted very well with a power–law trend line; the similar slopes between the two trend lines indicate that the insertions and deletions have similar parameter b in the power–law equation given above. This suggests certain similarities in the arising of these two types of indels.
Strictly speaking, the power–law relationship should be analytically defined between the gap length k and the proportion of indels of length k instead of the absolute number of indels. Following the formalism described by Gu et al. (6), in equation 6 we define fk as the probability of a random deletion or insertion to have the gap length k. In practice, we only counted k from 1 to 15.
Then the power–law relationship describing the gap length k and the proportion of nkcan be defined in 7.
fk = a × k–b 7
Regression analysis of the data shown in Figure 3B gave the following values. For deletions, a = 0.48, b = 1.51, R2 = 0.95. For insertions, a = 0.53, b = 1.60, R2 = 0.96. Obtaining exact values of a and b is important as they can be used to define gap penalty in sequence alignment programs (6). The values we obtained here provide more precise measurements than previous studies.
We have shown in the above sections that the nucleotide substitution pattern varies consistently with regard to their background G+C composition. It is intriguing to see whether the indel patterns are also affected in the same way. The RP pseudogenes were assigned into four groups according to their background G+C composition and the frequencies of indels were normalized by the amount of DNA in each group. Figure 4 shows the normalized frequency of deletions and insertions. Figure 4A illustrates that, for a deletion event, there is no clear, monotonic trend among the four groups. For k > 1, the length distributions among the four groups are practically undistinguishable. Figure 4B shows that the single base insertions are consistently more frequent in the regions of lower G+C content, but such a trend is not true for other gap lengths.
Figure 4.
The length distribution of deletions (A) and insertions (B) in the genomic regions of different G+C composition.
Higher frequency of 3 bp deletions than 2 bp deletions
It is intuitive to imagine that the indel events of longer gap length should occur less frequently than those of shorter length. While this is generally true, Figure 3A clearly shows that the frequency of 3 bp deletions (k = 3) is conspicuously higher than that of 2 and 4 bp deletions (k = 2, 4), which violates the power–law trend shown in Figure 3B. Also Figure 4A demonstrates that such higher frequency of 3 bp deletions is largely independent of the background G+C composition. We also checked to ensure that such bias was not limited to the pseudogenes derived from just a few RP genes.
As mentioned earlier, one needs to be cautious when doing sequence comparisons between the pseudogenes and the functional genes. One could argue that the higher frequency of 3 bp deletions in the pseudogenes merely reflects the existence of extra amino acids in the present-day human RP genes that were absent in the ancestral RP genes that gave rise to these pseudogenes. However, we consider this scenario very unlikely or at most that it only occurs to very few RP genes because of the following reasons. (i) Such higher frequency at k = 3 was also observed previously by other researchers (6) who manually aligned their sequences to remove potential artifacts. The ratio they observed between 3 and 2 bp deletions, 33 versus 23, was similar to that observed in this study (565 versus 477). (ii) Because we limited our sequence analysis only to the regions that are consensus between human and mouse, any bias caused by the amino acid insertions or deletions in the RP genes must have occurred before the divergence of mouse and human lineage at roughly 75 million years ago (23). However, the majority of the RP-processed pseudogenes in the human genome were created after the divergence of primates and rodents (10). (iii) Like insertions, deletions of amino acids could also occur during the evolution of RP genes; therefore one would expect the same abnormal pattern for DNA insertions in the pseudogenes, i.e. one would expect to see higher frequency for 3 bp insertions than 2 bp insertions. However, Figure 3A shows that such a phenomenon was not observed in the human genome.
Lastly, if the excess of 3 bp deletions was indeed caused by the insertion of amino acids in the functional RP genes, then one would expect to find most of these excessive deletions located between the boundaries of adjacent codons rather than in the middle of codons. Table 2 summarizes the frequency of deletions and insertions according to their positions in the codons. We define codon frame C = 1 if the indel occurs after the first base in an intact codon, C = 2 if it occurs after the second base and C = 3 after the third base, i.e. between two intact codons. If the 3 bp deletions truly follow the power–law trend, then by interpolation, we would then expect the number of 3 bp deletions to be approximately 300 and the frequency for each codon frame should be close to 100. However, Table 2 shows that the observed frequency of 3 bp deletions is significantly higher than 100 at each codon frame and is also higher than the frequency of 2 bp deletions at the same codon frame. Close examination of Table 2 shows that the 3 bp deletions actually have the highest frequency at C = 3, i.e. between adjacent codons, rather than at any of the other two frames, which seems to favor the argument that amino acid insertions caused the higher frequency of 3 bp deletions. However, we argue that such a mechanism could not fully explain the data presented in Table 2 since the 3 bp deletions also have significantly higher frequency than 2 bp deletions at the other two codon frames (C = 1, 2). It is likely that a small number of present-day RP genes do have extra amino acids that were absent in the ancestral genes, which caused the slight enrichment of 3 bp deletions at C = 3. However, such amino acid insertion/deletion events in the functional RP genes should be very minimal since the enrichment at C = 3 was not observed for 3 bp insertions. The 3 bp insertions have significantly lower frequencies than 2 bp deletions at two of three codon frames (C = 2, 3) and have a similar frequency at C = 1. In conclusion, the analysis shows that the observed excessive 3 bp deletions cannot be fully attributed to amino acid insertions in the functional RP gene sequences.
Table 2. Codon positions for insertions/deletions.
| Gap length | k = 1 | k = 2 | k = 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Codon frame | C = 1 | C = 2 | C = 3 | C = 1 | C = 2 | C = 3 | C = 1 | C = 2 | C = 3 |
| Deletions | 400 (31%) | 479 (37%) | 412 (32%) | 161 (34%) | 157 (33%) | 159 (33%) | 173 (31%) | 163 (29%) | 229 (41%) |
| Insertions | 210 (35%) | 181 (30%) | 209 (35%) | 36 (24%) | 61 (40%) | 56 (37%) | 41 (37%) | 28 (25%) | 43 (38%) |
Another possible explanation for the higher frequency of the 3 bp deletions is that this was the fortuitous combination of individual 1 and 2 bp deletions, i.e. a 1 and a 2 bp deletion happened to occur at the same position, but not necessarily at the same time, in a pseudogene, which resulted in an accumulated deletion of three bases. For this to happen, the frequency of deletions for k = 1, 2 and 3 have to be so optimal to have an accumulated distribution pattern similar to the observed pattern shown in Figure 3A. To test this hypothesis, we conducted 100 rounds of computer simulations to determine the relative frequencies of 1, 2 and 3 bp simultaneous deletions. The results showed that the most likely normalized frequencies were 1:0.29:0.36, i.e. simultaneous 3 bp deletions were 1.24 times (0.36/0.29) as likely as simultaneous 2 bp deletions. This confirmed that the simultaneous 3 bp deletions indeed have a higher frequency than 2 bp deletions.
Biological implication of the higher frequency for 3 bp deletions
It is a little surprising that the 3 bp deletions are more frequent than the 2 bp deletions in the human pseudogenes, which is reminiscent of the relative depletion of 3 bp simple repeats in the human genome (24). In order to explain such bias, it helps to discuss the mechanisms of generating these micro-deletions. It is generally agreed that DNA replication slippage (25) is the major force that causes DNA micro-deletions. This process is also responsible for generating simple sequence repeats (also known as SSRs or micro-satellites), which are prevalent in the human and other mammalian genomes (26). Similar to the 3 bp deletions, SSRs of unit length 3 (3mers) also have an unusual distribution. On average, in the intergenic regions of the genome where DNA sequences are not under selective pressure, SSRs of longer repeating units are less frequent than the SSRs of shorter repeating units. However, opposite to the general trend, it was found that SSRs of unit length 3 were significantly less frequent than the 2mers and the 4mers in the mammalian genomes (24). In the human genome, the average number of the 3mer SSRs per Mb is ∼25% of the number of the 2mer SSRs and only 34% of the 4mer SSRs; in fact the 3mer SSRs are even less frequent than the 5mer SSRs (16).
We believe that the unusual frequencies of 3mers in both simple sequence repeats and the micro-deletions are intrinsically related, as they likely reflect a mechanistic bias towards tri-nucleotides in DNA replication or/and mismatch repair processes. It was proposed that when DNA replication slippage occurs, certain DNA sequences or motifs are more likely than others to form alternative DNA conformation such as hairpin or triplex to stabilize the slipped structures (27). These stabilized DNA mismatch structures can then be reconciled by the mismatch repair system, which would consequently introduce simple sequence repeats to the chromosomes. In contrast, those mismatched DNA sequences that fail to form stable alternative structures are likely to be excised from the chromosome and therefore generate micro-deletions. It is possible that, on average, trinucleotides may have a lower capacity of forming these stable alternative conformations, therefore they may have a higher tendency of being deleted from the chromosome when mismatches occur. Such a mechanism could explain their under-representation in SSRs and over-representation in micro-deletions. However, such a conclusion based on statistical analysis is speculative in nature and experimental results are needed to fully understand the molecular basis.
CONCLUSION
The concept of ‘proportion of substitutions’ as we introduced in equation 2 is different from the ‘rate of substitutions’ as described in some of the recent reports (8,16). As we described in the text, the frequency of one nucleotide being mutated to another actually depends on two distinct aspects of nucleotide evolvement: the chance that a nucleotide is mutated and the chance that it is mutated to one particular nucleotide type. It was observed before that the substitution rates between nucleotide pairs vary greatly in regard to the background G+C composition, but we discovered that such variation was mainly caused by the difference in nucleotide stability rather than the directionality of the mutations (Fig. 1A and B). In other words, the proportion of substitutions is less dependent on the background G+C composition. We also thoroughly analyzed the neighboring-effect in the nucleotide substitutions, and studied its correlation with the biases in the di-nucleotide abundances in the human genome.
The total number of insertions and deletions that we surveyed in our study (3740 and 1291) is 15 times more than previously studied by other investigators. We not only confirmed that the deletions occurred much more frequently than insertions, but also further refined the power–law parameters that describe them. It was unexpected that we found 3 nucleotide deletions were more frequent than 2 bp deletions, for which we did thorough analysis to ensure that it was not caused by amino acid insertions in the functional genes. We hope our analysis on substitutions and indels would shed light on not only the dynamics and history of human and mammalian genomes, but also on some basic biological problems such as DNA replication, recombination and repair. The latter is demonstrated by the observed 3 bp bias in the DNA micro-deletions. We plan next to expand the indel analysis to other human repetitive elements and other genomes to confirm whether this is a universal phenomenon.
Acknowledgments
ACKNOWLEDGEMENTS
Z.Z. thanks Paul Harrison for scintillating discussions and Duncan Milburn, Yin Liu and Nat Echols for computational assistance. We also thank the two anonymous reviewers for helpful suggestions. M.G. acknowledges an NIH CEGS grant (P50HG02357-01) and the Keck Foundation for financial support.
REFERENCES
- 1.Mighell A.J., Smith,N.R., Robinson,P.A. and Markham,A.F. (2000) Vertebrate pseudogenes. FEBS Lett., 468, 109–114. [DOI] [PubMed] [Google Scholar]
- 2.Esnault C., Maestre,J. and Heidmann,T. (2000) Human line retrotransposons generate processed pseudogenes. Nature Genet., 24, 363–367. [DOI] [PubMed] [Google Scholar]
- 3.Antonarakis S.E., Krawczak,M. and Cooper,D.N. (2000) Disease-causing mutations in the human genome. Eur. J. Pediatr., 159, S173–S178. [DOI] [PubMed] [Google Scholar]
- 4.Krawczak M., Chuzhanova,N.A., Stenson,P.D., Johansen,B.N., Ball,E.V. and Cooper,D.N. (2000) Changes in primary DNA sequence complexity influence the phenotypic consequences of mutations in human gene regulatory regions. Hum. Genet., 107, 362–365. [DOI] [PubMed] [Google Scholar]
- 5.Hess S.T., Blake,J.D. and Blake,R.D. (1994) Wide variations in neighbor-dependent substitution rates. J. Mol. Biol., 236, 1022–1033. [DOI] [PubMed] [Google Scholar]
- 6.Gu X. and Li,W.H. (1995) The size distribution of insertions and deletions in human and rodent pseudogenes suggests the logarithmic gap penalty for sequence alignment. J. Mol. Evol., 40, 464–473. [DOI] [PubMed] [Google Scholar]
- 7.Ophir R. and Graur,D. (1997) Patterns and rates of indel evolution in processed pseudogenes from humans and murids. Gene, 205, 191–202. [DOI] [PubMed] [Google Scholar]
- 8.Petrov D.A. and Hartl,D.L. (1999) Patterns of nucleotide substitution in Drosophila and mammalian genomes. Proc. Natl Acad. Sci. USA, 96, 1475–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saitou N. and Ueda,S. (1994) Evolutionary rates of insertion and deletion in noncoding nucleotide sequences of primates. Mol. Biol. Evol., 11, 504–512. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z., Harrison,P. and Gerstein,M. (2002) Identification and analysis of over 2000 ribosomal protein pseudogenes in the human genome. Genome Res., 12, 1466–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wool I.G., Chan,Y.L. and Gluck,A. (1995) Structure and evolution of mammalian ribosomal proteins. Biochem. Cell Biol., 73, 933–947. [DOI] [PubMed] [Google Scholar]
- 12.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearson W.R. (1997) Comparison of DNA sequences with protein sequences. Genomics, 46, 24–36. [DOI] [PubMed] [Google Scholar]
- 14.Gojobori T., Li,W.H. and Graur,D. (1982) Patterns of nucleotide substitution in pseudogenes and functional genes. J. Mol. Evol., 18, 360–369. [DOI] [PubMed] [Google Scholar]
- 15.Li W.H., Wu,C.I. and Luo,C.C. (1984) Nonrandomness of point mutation as reflected in nucleotide substitutions in pseudogenes and its evolutionary implications. J. Mol. Evol., 21, 58–71. [DOI] [PubMed] [Google Scholar]
- 16.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 17.Russell G.J., Walker,P.M., Elton,R.A. and Subak-Sharpe,J.H. (1976) Doublet frequency analysis of fractionated vertebrate nuclear DNA. J. Mol. Biol., 108, 1–23. [DOI] [PubMed] [Google Scholar]
- 18.Gentles A.J. and Karlin,S. (2001) Genome-scale compositional comparisons in eukaryotes. Genome Res., 11, 540–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulmer M. (1986) Neighboring base effects on substitution rates in pseudogenes. Mol. Biol. Evol., 3, 322–329. [DOI] [PubMed] [Google Scholar]
- 20.Karlin S. and Mrazek,J. (1997) Compositional differences within and between eukaryotic genomes. Proc. Natl Acad. Sci. USA, 94, 10227–10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duret L. and Galtier,N. (2000) The covariation between tpa deficiency, cpg deficiency and g+c content of human isochores is due to a mathematical artifact. Mol. Biol. Evol., 17, 1620–1625. [DOI] [PubMed] [Google Scholar]
- 22.Luscombe N.M., Qian,J., Zhang,Z., Johnson,T. and Gerstein,M. (2002) The dominance of the population by a selected few: power–law behaviour applies to a wide variety of genomic properties. Genome Biol., 3, RESEARCH0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waterston R.H., Lindblad-Toh,K., Birney,E., Rogers,J., Abril,J.F., Agarwal,P., Agarwala,R., Ainscough,R., Alexandersson,M., An,P. et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature, 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 24.Toth G., Gaspari,Z. and Jurka,J. (2000) Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res., 10, 967–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Capy P. (2000) Perspectives: evolution. Is bigger better in cricket? Science, 287, 985–986. [DOI] [PubMed] [Google Scholar]
- 26.Levinson G. and Gutman,G.A. (1987) Slipped-strand mispairing: A major mechanism for DNA sequence evolution. Mol. Biol. Evol., 4, 203–221. [DOI] [PubMed] [Google Scholar]
- 27.Pearson C.E. and Sinden,R.R. (1998) Trinucleotide repeat DNA structures: Dynamic mutations from dynamic DNA. Curr. Opin. Struct. Biol., 8, 321–330. [DOI] [PubMed] [Google Scholar]