Abstract
Tubby, an autosomal recessive mutation, mapping to mouse chromosome 7, was recently found to be the result of a splicing defect in a novel gene with unknown function. Database searches revealed that sequences corresponding to the C terminus of the tub protein were highly conserved across a number of species including humans, mice, Caenorhabditis elegans, Arabidopsis, rice, and maize, and that tub was a member of a gene family. We describe here, TUB, the human homolog of mouse tub, and two newly characterized family members, TULP1 for tubby like protein 1 and TULP2. These three family members, which differ in the N-terminal half of the protein, share 60–90% amino acid identity across their conserved C-terminal region and have distinct tissue expression patterns. Alternatively spliced transcripts with 5′ variable sequences, three of which have been identified for the tubby gene, may mediate tissue specific expression. We also report that TUB, TULP1, and TULP2 map to human chromosomes 11p15.4, 6p21.3, and 19q13.1, respectively. TULP1 and TULP2 map within the minimal intervals identified for retinitis pigmentosa 14 on chromosome 6p21.3 and cone-rod dystrophy on chromosome 19q13.1. TULP1 and TULP2, which are expressed in the retina, make excellent candidates for these ocular diseases as a mutation within the tub gene is known to lead to early progressive retinal degeneration.
Mice homozygous for a splicing defect in the C terminus of the tub gene are characterized by progressive retinal and cochlear degeneration (1, 2), maturity-onset obesity with insulin resistance, and impaired glucose tolerance (3). We recently identified tub as a novel gene with high expression in the retina, brain, and testis (4). Analysis of the tub gene revealed sequence similarity to the p46 mouse cDNA (5) and an integrated molecular analysis of genomes and their expression (I.M.A.G.E.) expressed sequence tag (EST) clone from a human retinal cDNA library, suggesting that tub is a member of a family of genes. To further characterize this family and explore the possibility that its members may be candidate genes for diseases that exhibit similar phenotypes as tubby, we obtained and characterized human full-length cDNAs for TUB, for the gene corresponding to the human EST, now referred to as TULP1 for tubby like protein 1, as well as for a new member, TULP2.
MATERIALS AND METHODS
Cloning of TUB cDNA.
32P-labeled hybridization probes were prepared from two tub exon trap products, ET-3636.p01.a04 (nucleotides 1422–1593, 171 bp, GenBank accession no. U52433U52433) and ET-3636.p01.d01 (nucleotides 1323–1421, 99 bp) by random hexamer priming, as described (6). These probes were used to screen ≈1.2 × 106 plaque forming units of a human adult brain cDNA library in λgt11, plated according to the manufacturer’s instructions (CLONTECH). Duplicate membrane filters were hybridized with labeled probe in 10% dextran sulfate, 1% SDS, 1 M NaCl, 100 μg/ml of salmon testes DNA at 65°C for 18 hr. After hybridization, filters were washed at 65°C in 2× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7)/0.1% SDS for 45 min and twice in 0.2× SSC/0.1% SDS for 45 min each and positive plaques identified by autoradiography. Following plaque purification, cDNA inserts were PCR amplified using λgt11 primers (BRL) and directly cloned into pCR2.1 for sequencing, according to the manufacturer’s instructions (Invitrogen). Automated fluorescence sequencing was used to characterize the cDNA clones (Prism, Applied Biosystems).
Cloning of TULP1 cDNA.
Approximately 1 × 106 plaque-forming units of human retinal cDNA λgt11 library (CLONTECH) were hybridized with a 32P-labeled EcoRI/SacII fragment (1–962 bp) of I.M.A.G.E. EST clone 221670 (Research Genetics, Huntsville, AL) at 65°C overnight. The membranes were washed sequentially for 1 hr each with 2× SSC/0.1% SDS at 50°C, 1× SSC/0.1% SDS at 50°C, and 0.5× SSC/0.1% SDS at 60°C. Thirty positive plaques were purified and phage positives of insert size >1 kb were digested with EcoRI and subcloned into pUC9. One of these clones was a full-length cDNA containing the entire ORF. Independently, to isolate the flanking 5′ sequences, nested oligonucleotide primer pairs AP1 and hMan2-R3 5′-AGCTCCTCGGAGCCCTAGC-3′ (nucleotides 2017–2035 of the TULP1 sequence), as well as AP2 and hMan2-R1 5′-ACATGCCTCGATCCATGCC-3′ (nucleotides 979–997), were used in subsequent rounds of amplification of Marathon–Ready brain cDNA (CLONTECH), according to manufacturer’s protocol. Amplification products were gel purified (Qiagen, Chatsworth, CA) and sequenced automatically (Prism, Applied Biosystems) or manually by dideoxy cycle sequencing (Sequitherm, Epicentre Technologies, Madison, WI). Alternately, gel purified products were subcloned into the TA-cloning vector according to the manufacturer’s instruction (Invitrogen) and plasmids were isolated by a standard protocol (6) prior to sequencing.
Cloning of TULP2 cDNA.
A PCR product amplified from a mouse testis cDNA library using primers MP46.1 5′-TCTACAGAGACAAACTATGCCC-3′ and MP46.2 5′-GGAAATGTGCTACACCATC CTC-3′ was used as a probe to screen 1 × 106 plaque-forming units of human testis cDNA library in λDR2 (CLONTECH). Hybridization, washes, and radiography were carried out as described for cloning of the TUB cDNAs. Thirty-four positive clones were detected after overnight exposure and after tertiary screening, 28 positives were isolated and converted to plasmid DNA in pDR2. The C-terminal intron of TULP2 was PCR amplified and sequenced from DNA of 10 unrelated individuals using primers TULP2-F1 5′-ATCGTGGATCCCAAACACC-3′ and TULP2-R1 5′-GCTGGCAAGGGTATGGTATT-3′.
Southern Blot Analysis.
Southern blots of EcoRI-digested genomic DNA from a number of animal species were obtained from CLONTECH. They were hybridized with a 32P-labeled HindIII fragment (281–1833 bp) of TUB cDNA and a 32P-labeled EcoRI/BstXI TULP1 fragment (the first 365 bp of 5′ end of clone 221670). Blots were washed in 2× SSC/0.05% SDS at room temperature two times for 10 min and at 60°C for 20 min, then twice in 0.2× SSC/0.1% SDS at 60°C for 20 min each.
Northern Blot Analysis.
Human multiple tissue Northern blots (MTN I, II, and III; CLONTECH) were hybridized with the probes described above for Southern blot analyses, in 10× Denhardt’s/2% SDS, 100 μg/ml of sheared salmon sperm DNA, and 50% formamide at 42°C for 18 hr, then washed in 2× SSC/0.05% SDS at room temperature three times for 10 min and in 0.1× SSC/0.1% SDS at 50°C two times for 20 min.
The same blots were hybridized with the 32P-labeled product of TULP2 (amplified using primers HP46.F1 5′-CCACTAAATGAACAGGAGTCGC-3′ and HP46.R1 5′-GAAACTGGACAAGCAGATGCTG-3′). The hybridization was done in ExpressHyb solution (CLONTECH) at 60°C for 2 hr, according to the manufacturer’s instructions. Initially, the blots were washed three times in 2× SSC/0.05% SDS at room temperature, followed by washing in 0.1× SSC/0.1% SDS at 55°C for 2 × 40 min and in 0.1× SSC/0.1% SDS at 65°C for 40 min.
Sequence Analyses.
Sequence assembly and analysis were performed using the programs assemblylign and macvector (Oxford). Multiple alignments and phylogenetic trees were generated with the program macaw (7). Database searches were conducted using the blast programs (8).
Radiation Hybrid Mapping.
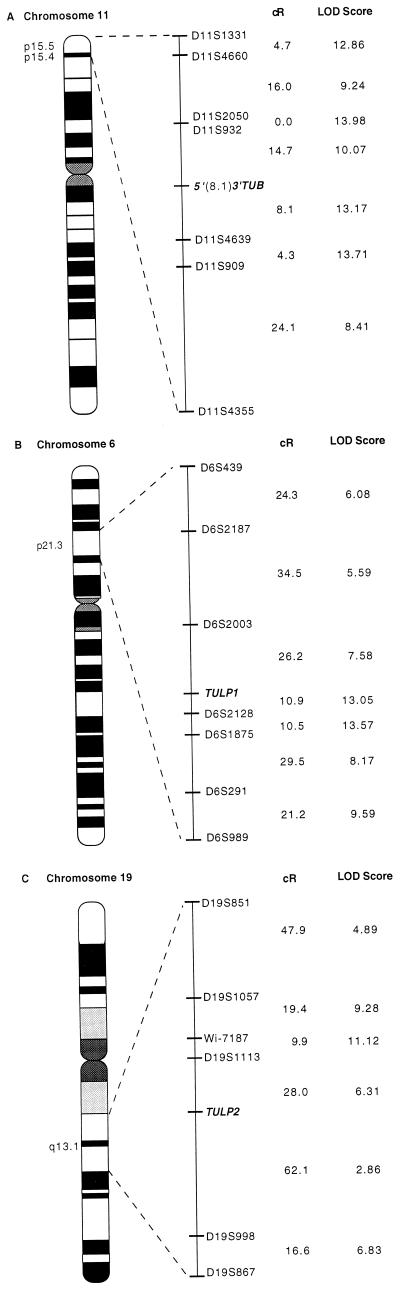
Oligonucleotide primers were constructed from the novel 5′ end of TUB (hMan1-F2 5′-CAGGACACCTTTGCCTTCT-3′, hMan1-R2 5′-GCGATCTCCCTTCCTTCCT-3′), the 3′ noncoding region for TUB (hMan6.4-F1 5′-TGCCTGGGAATCCTGCTGC-3′ and hMan6.4-R1 5′-TCCTAAGGGTCCTGCCACT-3′), and for TULP1 (hMan2-F7 5′-CGAAAACGGAGCAAGACAG-3′, hMan2-R7 5′-TATGAGGCTCTCCAGCGTC-3′), using the macvector computer program (Oxford). Primers for TULP2 were constructed from an exon in the 3′ portion of the gene (HP46.F1 5′-CCACTAAATGAACAGGAGTCGC-3′ and HP46.R2 5′-TTGGAAGTTCTTCACCGAAGCC-3′). After confirming by sequencing that the appropriate product was amplified, the retention patterns for each oligonucleotide pair were obtained by PCR assay in the Stanford G3 Radiation Hybrid panel (9) as previously described (10). hMan1-F2/R2 amplifies a band of ≈480 bp, constant hamster specific bands were observed at ≈150, 200, and 213 bp. hMan6.4-F1/R1 amplifies a 221-bp product with constant hamster DNA-specific bands at ≈290 and 320 bp. The product of hMan2-F7/R2 is 92 bp in length with a constant hamster DNA amplified band at ≈230 bp. HP46.F1/R2 amplifies a 162-bp fragment. Data entered into an online database (rhserver@shgc.stanford.edu) was analyzed by rhmap software developed by Boehnke et al. (11). Markers mapping in proximity to the TUB or TULP genes, available in the database, are included in the ideograms (Fig. 4).
Figure 4.
Radiation hybrid mapping (A) 5′ novel splice variant and the 3′ untranslated region of TUB to chromosome 11p15.4. (B) TULP1 to chromosome 6p21.3. (C) TULP2 to chromosome 19q13.1. Centiray distances (cR8,000) and lod scores are shown to the right of the ideogram. RH data of additional markers obtained from the Stanford Human Genome Center were included in the data analyses (rhmap computer software 2.01). The branch and bound approach was used to determine the most likely locus order (rhmaxlik). Two point analyses were done using rh2pt to obtain an estimated distance and the lod scores between markers.
RESULTS
Isolation and Characterization of the TUB, TULP1, and TULP2 cDNAs.
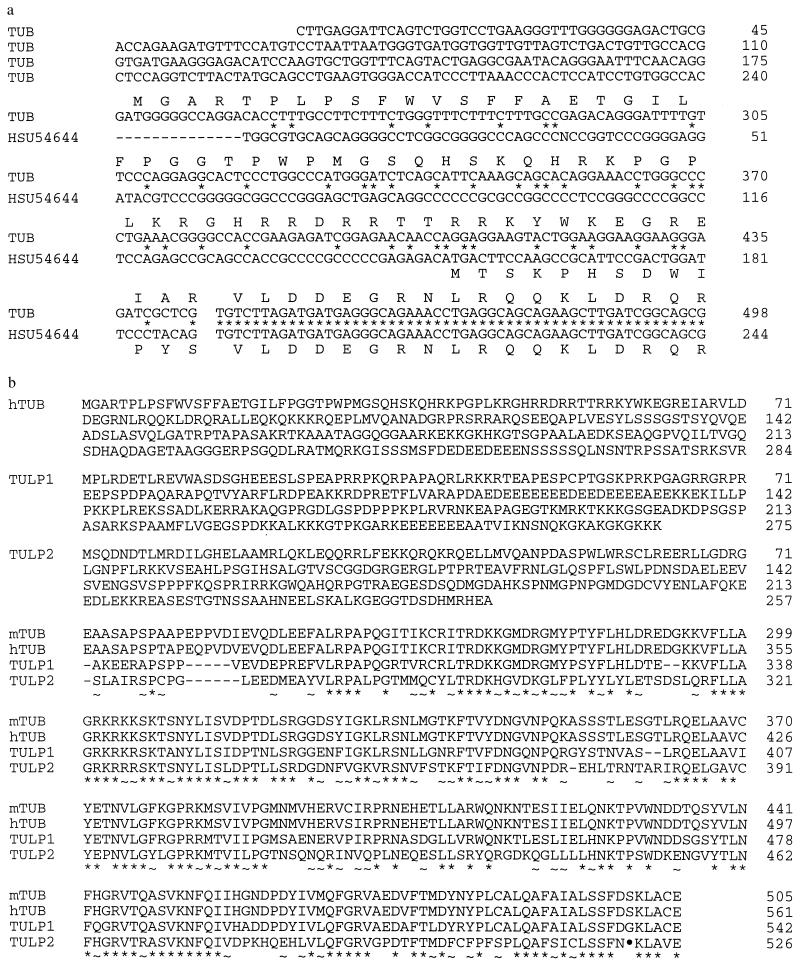
The human homolog of the mouse tubby gene was identified by screening an adult brain cDNA library (CLONTECH) with tub derived probes. The full-length cDNA is ≈7,500 bp long and can code for a 561-amino acid protein starting at the first methionine in the ORF. Comparison of the tubby mouse (U52433; refs. 4 and 12) and human brain cDNA sequences (HSU54644; ref. 12) with the TUB sequences obtained here, indicates that they differ in the first 444 bases (Fig. 1a). The position at which the human brain TUB sequences diverge coincides with an intron/exon splice junction, suggesting that these cDNAs derive from alternatively spliced mRNAs. To support this hypothesis, we determined the chromosomal location of the novel 5′ end of the TUB human brain sequence and found that it maps to the same location as TUB, 8.1 cR from the 3′ untranslated region (Lod score, 13.17). The novel 5′ sequence extends the ORF by 55 amino acids beyond the sequence reported for TUB (12). The first 32 amino acids of this ORF are predominantly hydrophobic, a distinct contrast to the overall highly hydrophilic nature of the predicted protein. A third 5′ splice variant of the tubby gene had been previously observed in mouse testis (U52824; ref. 4). The remaining TUB coding sequence, nucleotides 445-1924, is highly conserved between mouse and human with 89% nucleotide identity and 94% amino acid identity (Fig. 1b).
Figure 1.
(a) Comparison of nucleotide sequence and deduced amino acid sequence of the 5′ splice variants of TUB and HSU54644. Break in sequence denotes the intron/exon junction. (b) Comparison of mouse tub and human TUB, TULP1, and TULP2. ∗, Conserved identical amino acids; ~, conservative replacements. The alignments were produced with the computer program clustal w1.6 (13).
The EST clone 221670 was used to screen a human retinal cDNA library for TULP1. Multiple partial cDNA clones encompassing the 3′ portion of TULP1 and one full-length clone were obtained. 5′ rapid amplification of cDNA ends of human brain cDNA was performed to confirm the 5′ end of the cDNA sequence. The full-length cDNA is comprised of 2116 bp that potentially encodes a 542-amino acid protein (Fig. 1b).
A probe derived from amplification of mouse testis cDNA using oligonucleotide primers designed according to the published p46 mouse cDNA sequence (5) was used to screen a human testis cDNA library. A clone was isolated that contained a 1733-bp long cDNA insert ending in a Poly(A) tail. The cDNA can code for a 520-amino acid protein beginning at the first methionine in the ORF (Fig. 1b). The corresponding gene was named TULP2.
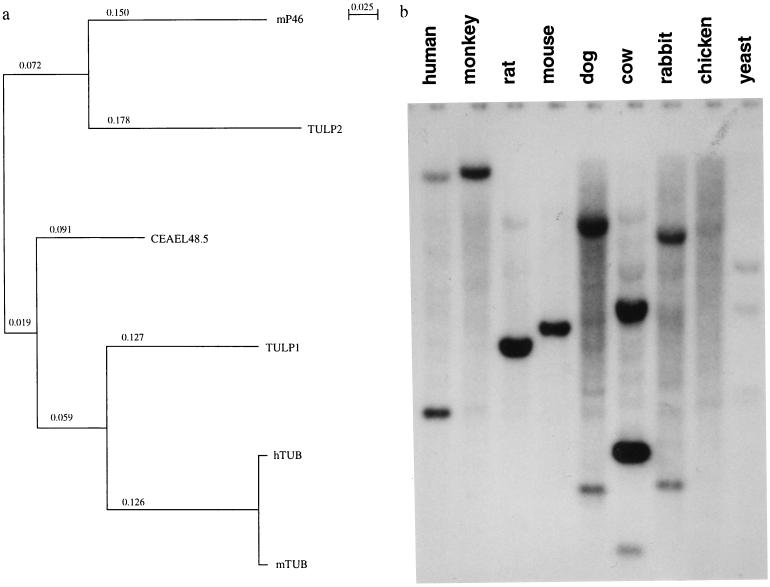
Phylogenetic analysis over the conserved 217-amino acid C terminus, indicates that TUB is more closely related to TULP1 than to the TULP2 protein (Fig. 2a). Ninety percent amino acid identity is observed between TUB and TULP1 while 66% amino acid identity is observed between TUB and TULP2 over the conserved region. TULP1 and TULP2 have 63% amino acid identity over the same region. The conservation of this gene family apparently extends across multiple animal species, as strongly hybridizing bands were detected in a high stringency genomic Southern blot (Fig. 2b). In addition, database searches reveal tubby-like proteins in Drosophila, Caenorhabditis elegans, Arabidopsis, rice, and maize. Such conservation across species suggests that tubby and tubby-like proteins may be fundamental to normal function of these organisms.
Figure 2.
(a) Phylogenetic tree of family members. (b) Hybridization of Southern blot of EcoRI-digested genomic DNAs from nine eukaryotic species with TULP1. Both TUB and TULP1 genes are conserved in all species including yeast (hybridization pattern of TULP1 shown).
Expression and Tissue Distribution of TUB, TULP1, and TULP2.
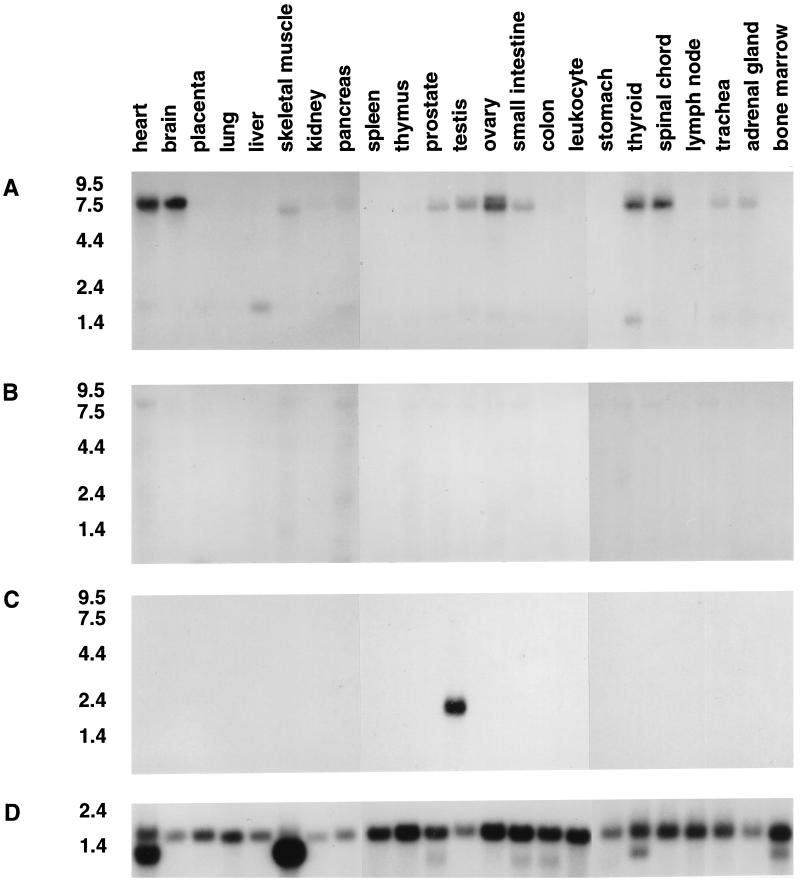
TUB gives rise to a major 7–7.5 kb transcript by Northern analysis (Fig. 3A). Like the mouse homolog, TUB is transcribed in brain and testis but in contrast to tub it is also highly expressed in liver, heart, small intestine, and ovary. Expression was also detected in skeletal muscle, thyroid, spinal cord, trachea, and adrenal gland—tissues not tested in the mouse. In addition to the major transcript, Northern analysis also revealed the presence of a faster migrating mRNA species, ≈2.4 kb, with highest expression in thyroid and liver.
Figure 3.
(A) Expression of TUB. Northern blot analysis of adult human tissues shows a ≈7–7.5-kb transcript with strong expression in heart, brain, testis, ovary, thyroid, and spinal cord after 48-hr exposure. TUB is also detected in skeletal muscle, prostate, small intestine, trachea, and adrenal gland. High expression of a 2.4-kb transcript is observed in liver and thyroid. (B) Expression of the TULP1 gene. No bands were observed on the same Northern blots when hybridized with a TULP1 probe after seven days of exposure. (C) Expression of the TULP2 gene. A ≈1.8-kb transcript is observed in testis. (D) Actin control.
In contrast to the high expression of TUB, multiple tissue Northern blots probed with TULP1 showed either faintly hybridizing or no bands (Fig. 3B). Probing the same blots with TULP2 showed a 1.8-kb transcript in testis (Fig. 3C).
Retinal mRNA was not included in the Northern tissue blots above. However, because mouse tub is also highly expressed in the eye, a human retinal cDNA library was screened with human derived TUB, TULP1, and TULP2 cDNA clones. TUB transcripts, confirmed by sequencing, comprised ≈0.005% of retinal cDNA clones. TULP1 had a 0.01% incidence in the retinal cDNA library, suggesting high expression of this gene in the retina. Murine multiple tissue Northern blots probed with human TULP1 confirmed the observation as only retinal mRNA showed a strongly hybridizing band of the correct transcript size (data not shown). TULP2 appears to be a very low abundance transcript in the retina as it comprised only 0.0001% of the total clones screened in the retinal cDNA library.
Mapping of TUB, TULP1, and TULP2 Genes.
Heckenlively et al. (1) and Ohlemiller et al. (2) have reported that the histopathological changes in the eyes and ears of tubby mice are similar to those seen in individuals with Usher syndrome. Because Usher syndrome, Type1C maps to human chromosome 11p15.1, the homologous region of tub on mouse chromosome 7, they suggested that tubby may be the Usher 1C homolog. The chromosomal location of TUB was determined in the Stanford G3 Radiation Hybrid panel with oligonucleotide primers specific to the 3′ noncoding region. TUB was localized to human chromosome 11p15.4 (Fig. 4A) in close proximity to D11S932 (14.7 cR). Since marker D11S932 resides outside the minimal region established for Usher syndrome, Type 1C (14), our mapping results exclude TUB as a candidate gene for this disease locus.
Gene specific PCR primers were also constructed for TULP1 and TULP2 to determine their chromosomal location in the Stanford G3 Radiation Hybrid panel. TULP1 localizes to chromosome 6p21.3 (Fig. 4B). Interestingly, two markers, D6S439 and D2S291, that flank TULP1 have been reported not to recombine with the retinitis pigmentosa 14 locus in a human kindred (15). TULP1 is tightly linked to the retinitis pigmentosa 14 locus and within the region of homozygosity of this autosomal recessive disease. TULP2 maps to human chromosome 19q13.1 (Fig. 4C) within the minimal region identified for a cone-rod retinal dystrophy locus (16).
DISCUSSION
In this communication, we have further characterized the human homolog of the mouse tub gene, and described two novel tubby gene family members. Our data indicate that TUB encodes a protein of 561 amino acids that is highly expressed in a number of tissues examined, including heart, brain, ovary, thyroid, spinal chord, and retina and maps to chromosome 11p15.4. TULP1, closely related to TUB, encodes a protein of 542 amino acids that is highly expressed in retina and maps to chromosome 6p21.3. TULP2, more distantly related to TULP1 and TUB, encodes a protein of 520 amino acids that is highly expressed in testis and maps to chromosome 19q13.1.
A striking feature among all known tubby related proteins is the high conservation observed in the ≈250 amino acids at the C terminus. Even the most distantly related members, TULP2 and the C. elegans 48.2K protein, show a 55% amino acid sequence identity in this region. This suggests that this conserved region may impart a common function to the gene family members. The functional importance of this conservation may not extend, however, throughout the C terminus. It appears that a mutation has been fixed in the TULP2 gene that introduced a premature stop codon in position 1654 of the cDNA sequence, truncating the protein by six amino acids. This truncated protein seems to predominate in humans since 10 of 10 sequenced DNAs from unrelated individuals have this form.
In contrast, the N-terminal portions of the tubby like proteins show little sequence similarity either among each other or with other unrelated proteins. The sequences diverge near an exon boundary present in TUB, TULP1, and TULP2..
The actual number of members in this novel tubby gene family is not known. However, low stringency Southern blot analysis indicates that there may be between six to ten members (data not shown). Although we screened for TULP2 in a human testis cDNA library with a probe from the mouse testis specific cDNA p46, it most probably is not the human homolog of this gene. The conserved C-terminal region of TULP2 and mouse p46 show only 68% amino acid identity, whereas the mouse and human TUB proteins are 99% identical over the same 273-amino acid region. Therefore, at least four unique family members have been identified, TUB, TULP1, TULP2, and mouse p46.
Unlike many housekeeping genes that are ubiquitously expressed, members of the tubby gene family show distinct tissue specific expression. Alternate 5′ splicing may additionally modulate expression in a tissue-specific manner. In at least one member, TUB, alternate splice variants predicting differing N termini were isolated. In mouse, one splice variant was observed only in testis, while two other variants are found in brain (4, 12). Whether they localize to different areas in the brain remains to be determined.
The function of this family of genes is also not known. However, a mutation in the highly conserved C-terminal portion of the tub gene leads to obesity, and cochlear and retinal degeneration. Therefore, tub and possibly related family members, must play a vital role in tissues in which they are expressed. Additionally, that their function is related to basic cellular processes is suggested by the fact that these family of genes are widely distributed among species, ranging from humans to maize.
Ultimately, the reason for studying mouse models is to understand human disease. We began with the cloning of mouse tub, and have subsequently identified two human tubby family members. We hypothesized that genes within the tub family must share similar functions and that mutations within some of the genes may lead to phenotypes in humans that are similar to those of tubby. Interestingly, TULP1, which is highly expressed in the retina, maps to human chromosome 6p21.3 in the same minimal interval as retinitis pigmentosa 14, and TULP2 maps within the minimal interval for rod-cone dystrophy on chromosome 19q13. Proof that the TULP genes are causative of these diseases will come from testing families segregating at those loci. The sequences presented here provide the means to test these hypotheses.
Acknowledgments
We thank Gayle Bouchard-Collin and Doug McMinimy for excellent technical assistance; Alan Buckler, Spencer Emtage, Nic Dracopoli, and Tim Harris for advice; and Barbara Knowles, Susan Ackerman, and Wayne Frankel for careful review of the manuscript. This work was supported by a grant from Sequana Therapeutics, Inc. (P.M.N.), National Retinitis Pigmentosa Foundation, Inc. (P.M.N.), and National Institutes of Health Grant R01DK46977 (J.K.N.). Institutional shared services are supported by National Cancer Institute Cancer Center Support Grant, CA-34196. K.N.T. is a recipient of a Deutsche Forschungs Gesellschaft fellowship.
ABBREVIATION
- EST
expressed sequence tag
Footnotes
References
- 1.Heckenlively J R, Chang B, Erway L C, Peng C, Hawes N L, Hageman G S, Roderick T H. Proc Natl Acad Sci USA. 1995;92:11100–11104. doi: 10.1073/pnas.92.24.11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohlemiller K K, Hughes R M, Mosinger-Olilvie J, Speck J D, Grosof D H, Silverman M S. NeuroReport. 1995;6:845–849. doi: 10.1097/00001756-199504190-00005. [DOI] [PubMed] [Google Scholar]
- 3.Coleman D L, Eicher E M. J Hered. 1990;81:424–427. doi: 10.1093/oxfordjournals.jhered.a111019. [DOI] [PubMed] [Google Scholar]
- 4.Noben-Trauth K, Naggert J K, North M A, Nishina P M. Nature (London) 1996;380:534–538. doi: 10.1038/380534a0. [DOI] [PubMed] [Google Scholar]
- 5.Vambutas V, Wolgemuth D J. Biochim Biophys Acta. 1994;1217:203–206. doi: 10.1016/0167-4781(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J B, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Greene/Wiley–Interscience; 1995. [Google Scholar]
- 7.Schuler G D, Altschul S F, Lipman D J. Proteins Struct Funct Genet. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 8.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 9.Cox D R, Burmeister M, Price E R, Kenn S, Myer R M. Science. 1990;250:245–250. doi: 10.1126/science.2218528. [DOI] [PubMed] [Google Scholar]
- 10.Collin G B, Münch A, Mu J-L, Naggert J K, Olsen A S, Nishina P M. Genomics. 1996;37:125–130. doi: 10.1006/geno.1996.0529. [DOI] [PubMed] [Google Scholar]
- 11.Boehnke M, Lange K, Cox D R. Am J Hum Genet. 1991;49:1174–1188. [PMC free article] [PubMed] [Google Scholar]
- 12.Kleyn P W, Fan W, Kovats S G, Lee J J, Pulido J C, et al. Cell. 1996;65:281–290. doi: 10.1016/s0092-8674(00)81104-6. [DOI] [PubMed] [Google Scholar]
- 13.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keats B J B, Nouri N, Pelias M Z, Deininger P L, Litt M. Am J Hum Genet. 1994;54:681–686. [PMC free article] [PubMed] [Google Scholar]
- 15.Shugart Y Y, Banerjee P, Knowles J A, Lewis C A, Jacobson S G, Matise T C, Penchaszadeh G, Gilliam T C, Ott J. Am J Hum Genet. 1995;57:499–502. [PMC free article] [PubMed] [Google Scholar]
- 16.Evans K, Fryer A, Inglehearn C, Duvall-Young J, Whittaker J L, Gregory C Y, Butler R, Ebenezer N, Hunt D M, Bhattacharya S. Nat Genet. 1994;6:210–213. doi: 10.1038/ng0294-210. [DOI] [PubMed] [Google Scholar]