Abstract
Glucose-6-phosphatase (Glc6Pase), the last enzyme of gluconeogenesis, is only expressed in the liver, kidney and small intestine. The expression of the Glc6Pase gene exhibits marked specificities in the three tissues in various situations, but the molecular basis of the tissue specificity is not known. The presence of a consensus binding site of CDX proteins in the minimal Glc6Pase gene promoter has led us to consider the hypothesis that these intestine-specific CDX factors could be involved in the Glc6Pase-specific expression in the small intestine. We first show that the Glc6Pase promoter is active in both hepatic HepG2 and intestinal CaCo2 cells. Using gel shift mobility assay, mutagenesis and competition experiments, we show that both CDX1 and CDX2 can bind the minimal promoter, but only CDX1 can transactivate it. Consistently, intestinal IEC6 cells stably overexpressing CDX1 exhibit induced expression of the Glc6Pase protein. We demonstrate that a TATAAAA sequence, located in position –31/–25 relating to the transcription start site, exhibits separable functions in the preinitiation of transcription and the transactivation by CDX1. Disruption of this site dramatically suppresses both basal transcription and the CDX1 effect. The latter may be restored by inserting a couple of CDX- binding sites in opposite orientation similar to that found in the sucrase-isomaltase promoter. We also report that the specific stimulatory effect of CDX1 on the Glc6Pase TATA-box, compared to CDX2, is related to the fact that CDX1, but not CDX2, can interact with the TATA-binding protein. Together, these data strongly suggest that CDX proteins could play a crucial role in the specific expression of the Glc6Pase gene in the small intestine. They also suggest that CDX transactivation might be essential for intestine gene expression, irrespective of the presence of a functional TATA box.
INTRODUCTION
Glucose-6-phosphatase (Glc6Pase, EC. 3.1.3.9) is a key enzyme involved in blood glucose homeostasis. Until recently, it has been an accepted view that Glc6Pase gene expression is restricted to the liver and the kidney and confers on these tissues only the capacity to release glucose in blood (1). However, we have now shown that the rat small intestine constitutes a third gluconeogenic organ, which is able to produce glucose in insulinopenic states, such as diabetes or fasting (2,3). In the three tissues, the expression of the Glc6Pase gene is increased in insulinopenia but it is noteworthy that tissue-specific regulations exist. For example, during development, the changes in expression are more marked in the liver and the small intestine than in the kidney, and the time courses are different in the three tissues (4). In the intestine, the Glc6Pase expression is strongly induced after birth, and a dramatic decrease, but not a total suppression, in Glc6Pase expression occurs around the suckling–weaning transition (4). In adults, the Glc6Pase gene is expressed in the duodenum and jejunum in normal fed rats and in the duodenum, jejunum and ileum in humans (5). Glc6Pase gene expression is increased in the duodenum and the jejunum in diabetic or fasted rats and is normalized upon insulin treatment or refeeding, respectively (5). In addition, Glc6Pase mRNA and activity are expressed in the ileum in fasted rats and during development but not in fed diabetic rats (5). This points out that specificity in expression may also exist within the small intestine along the anterioposterior axis.
The tissue-specific expression of genes is directed by the combinatorial effects of ubiquitous and tissue-restricted transcription factors. In the liver, kidney and small intestine, tissue-specific factors particularly include hepatocyte nuclear factor (HNF) families (HNF1, HNF3, HNF4 and HNF6) (6). An additional specificity in the intestine may be conferred by the expression of specific-intestine factors named CDX1 and CDX2, which are not expressed in the liver and the kidney (7). CDX1 and CDX2 proteins are members of the caudal-related homeobox gene family and are involved in the early differentiation, proliferation and maintenance of intestinal epithelial cells, and in intestine-specific gene transcription (7–9). The comparison of specific intestinal promoters, such as that of sucrase-isomaltase (SI), intestinal phospholipase A/lysophospholipase, lactase-phlorizin hydrolase, claudin-2, has suggested a common structure for enterocyte-specific promoters involving both HNF1 and CDX binding sites (7,10–13). Noteworthy, the characterization of the Glc6Pase promoter has already shown that several HNF factors, and especially HNF1α and HNF1β, are essential for the expression of this gene (14–19). That the Glc6Pase promoter may also bind CDX1/CDX2 has constituted an attractive hypothesis. CDX1 and CDX2 proteins bind to a binding site (CDX-BS) rich in A/T-rich whose consensus sequence is C/TATAAAT/G in direct or reverse orientation (20). In some instances, the CDX-BS presents high homology with the canonical TATA-box sequence, and indeed the CDX1 and/or CDX2 homeoproteins revealed able to bind to TATA-boxes of some intestinal genes, such as that of the calbindin-D9 gene (21,22) and the clusterin gene (23).
In this study, we have investigated whether the Glc6Pase promoter could be regulated by CDX proteins, with a particular interest in the ATATAAAATG sequence located –31/–25 bp upstream of the transcription start site, previously hypothesized to constitute the TATA box of the gene (24). We demonstrate that this sequence indeed has separable functions in basal transcription and CDX1-stimulated transcription of the Glc6Pase promoter.
MATERIALS AND METHODS
Reporter plasmids and expression vectors
The rat Glc6Pase promoter constructs containing regions up to nucleotide –1640, relative to the transcription start site, and cloned into the ‘pGL2 basic’ vector (denoted by B in construct names, Promega) upstream of a luciferase reporter gene, were previously described (14). The recombinant expression vectors pCDX1-S and pCDX2-S encode the murine CDX1 and CDX2 homeoproteins, respectively, under the control of the cytomegalovirus promoter (25). The plasmids pCdx1-S and pCdx2-S were modified by mutagenesis (GeneEditor™ in vitro Site-directed Mutagenesis System, Promega) to insert a NheI site immediately downstream of the ATG initiation codon using, respectively, the oligonucleotides 5′-GCGGGGGACCTGCGGTCACCATGCTAGCTTACGTGGGCTATGTGCCTGGA-3′ and 5′-GATAAGCTTGATCCACCATGCTAGC TTACGTGAGCTACCTTCTGGA-3′. The double strand DNA encoding the HA-tag formed by hybridization of the sense and antisense oligonucleotides (respectively 5′-CTAGGATATCCCTATGACGTCCCAGACTATGCC-3′ and 5′-CTAGGGCATAGTCTGGGACGTCATAGGGATACT-3′) was inserted into the NheI site giving rise to the recombinant expression vectors pCB6-HA-CDX1 and pCB6-HA-CDX2, respectively. The recombinant expression vector pXJ41-hTBP encodes the human TBP and is a gift from L. Tora (IGBMC, Strasbourg) (26).
Mutant forms of the Glc6Pase promoters, as well as swapped versions of the expression plasmids encoding the murine CDX1 and CDX2 homeoproteins were generated by site-directed mutagenesis using the GeneEditor™ in vitro Site-directed Mutagenesis System (Promega). Details about the mutagenic oligonucleotides are available upon request. All mutant constructs were confirmed by sequencing. Plasmids used for transfection were purified using the Plasmid maxi kit (Jet Star, Genomed).
Cell culture and transfection
HepG2 human hepatoma cells, HeLa human epithelial cervical carcinoma cells, and Caco-2 human colon carcinoma cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 6 (for HepG2 cells), 10 (for HeLa cells) or 15% (for Caco-2 cells) fetal bovine serum, 5 mM glutamine, streptomicin (1 µg/ml) and penicillin (1 U/ml) at 37°C in a humidified 5% CO2/95% air atmosphere. For transient transfection, 1 day (or 2 days for Caco-2 cells) before the transfection, 200 000 cells were plated out in 35-mm wells in six-well cell culture plates. The complete medium was refreshed 1 h prior to transfection. HepG2 and HeLa cells were transfected by the calcium-phosphate transfection method as previously described (14) and Caco-2 cells were transfected using Exgen reagent (Euromedex) with 1 µg of the Glc6Pase-LUC plasmid, 2 ng of pCMV-RL (Promega) to correct for transfection efficiency and variable amount (1 to 200 ng) of expression vectors as indicated in the figures. The total amount of DNA (2.2 µg) was kept constant by addition of pBluescript SK+ plasmid. The cells were then washed three times with phosphate-buffered saline and lysed with passive lysis buffer (Promega). After 15 min incubation, cells were scraped and centrifuged at 10 000 g for 5 min at 4°C to eliminate cell debris. Renilla luciferase (RL) and firefly luciferase (LUC) activities were determined with a BCLBook luminometer (Promega) using the Dual-Luciferase kit Assay Reagent (Promega). The levels of LUC activities were normalized by means of the RL activities. Statistical analyses were performed using the Student’s t-test for unpaired data.
Gel shift mobility assays
Gel mobility shift assays were performed with whole cell protein extracts (27) from HeLa cells, CDX1-expressing HeLa cells, CDX2-expressing HeLa cells and differentiated Caco-2 cells (obtained after 1 week of post-confluence culture). Oligonucleotide sequences were as follows: Glc6Pase TATAAAA top strand: 5′-ATCCAGGGCATATAAAATGGGCAAGGCACA-3′, Glc6Pase TATAAAA bottom strand: 5′-TGTGCCTTGCCCATTTTATATGCCCTGGAT-3′, consTATA box top strand: 5′-GGATGAAGAATAAAAGGAAGCACCCT-3′, and consTATA box bottom strand: 5′-AGG GTGCTTCCTTTTATTCTTCATCC-3′. Binding reactions (20 µl) contained 5 or 15 µg of whole cell extract, end-labelled double-stranded oligonucleotide probes (0.1 ng; 30 000–50 000 c.p.m.), 1 µg of polydI.dC in binding buffer (12.5 mM HEPES, 100 mM KCL, 1 mM EDTA, 1 mM dithiothreitol, 100 µg/ml bovine serum albumin, pH 7.6, 10% glycerol). Reaction mixtures were incubated for 20 min at room temperature. For supershift assay, antibodies against TBP (Santa Cruz Biotechnology) were added and incubated 10 min at room temperature before the addition of the probe. Free DNA and DNA–protein complexes were separated on a 6% non-denaturing polyacrylamide gel (acrylamide- bisacrylamide, 19/1) in 0.5× TBE buffer (45 mM Tris–borate, 1 mM EDTA, pH 8.3). In competition experiments, the competitor DNA was incubated in the mixture prior to the addition of the probe. After electrophoresis, gels were dried and analysed on a PhosphorImager SI (Molecular Dynamics).
Immunoprecipitation assays
HTC 116 colon carcinoma cells (2 × 106) were co-transfected using Jet-PEI (Polyplus Transfection) with 2 µg of vehicle pCB6, pCB6-HA-mCDX1 or pCB6-HA-mCDX2 plasmids and 2 µg of pXJ41-hTBP (26). Cells were lysed 48 h later for whole protein extracts in 1 ml of 50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% Igepal (Sigma) and protease inhibitor cocktail (Sigma). Immunoprecipitation assays were performed using the µMACS Epitope Tagged Protein Isolation kit (Miltenyi Biotec). Protein extract was incubated overnight at 4°C with the magnetic anti-HA coupled beads, beads were then washed five times with washing buffer and proteins were eluted in SDS-loading buffer. The immunoprecipitate was separated by electrophoresis (SDS–PAGE 12%) and blots revealed using the monoclonal anti-HA (Babco, 1/1000) or the anti-TBP (3G3 obtained from L. Tora, IGBMC, Strasbourg, France) (28) antibodies.
Western blotting
Whole protein extracts were prepared from 4-day-old culture of IEC6-CAT or IEC6-CDX1 cells [described by Soubeyran et al. (29)] lysed in SDS-loading buffer, sonicated and boiled for 5 min. Thirty micrograms of the cell extract were separated by electrophoresis (SDS–PAGE), and the CDX1, the Glc6Pase and the actin proteins were detected by western blot using rabbit polyclonal anti-CDX1 (30), rabbit polyclonal anti-Glc6Pase (1/2000) or mouse monoclonal anti-actin (Chemicon, 1/15 000) antibodies.
RESULTS
The Glc6Pase promoter is active in intestinal Caco-2 cells
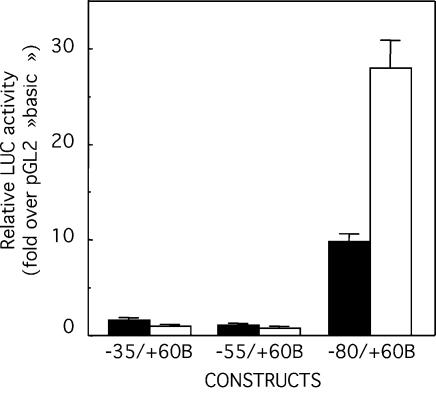
To demonstrate the functionality of the Glc6Pase promoter in an intestinal context, we used the Caco-2 cell line, derived from a human colonic adenocarcinoma, that spontaneously differentiates in enterocyte-like cells after post-confluence (31) with concomitant induction of endogenous Glc6Pase expression (5). Three Glc6Pase promoter constructs overlapping the putative Glc6Pase TATA box were transiently transfected in intestinal Caco-2 cells and in hepatic HepG2 cells as a positive control. The shortest constructs –35/+60B and –55/+60B had no activity in either cell type (Fig. 1). The –80/+60B conferred the minimal promoter activity in both HepG2 and Caco-2 cells, consistent with the fact that Glc6Pase is expressed in liver and gut (Fig. 1). This expression reflected tissue specificity since none of these constructs had activity in cervical HeLa cells (data not shown).
Figure 1.
Glc6Pase promoter activity in Caco-2 and HepG2 cells. Caco-2 cells (black bars) and HepG2 (white bars) were transiently transfected with each luciferase reporter plasmid containing different fragments of the Glc6Pase promoter (1 µg) together with pCMV-RL plasmid (2 ng) as a control for correction for transfection efficiency. LUC activity was determined 48 h after transfection and was normalized relative to the level of RL activity. The transcriptional activity of each construct is expressed relative to the LUC activity of pGL2‘basic’ and is the mean ± S.E.M. of at least three independent experiments performed in duplicate.
CDX1, but not CDX2, has transcriptional effect on the Glc6Pase gene
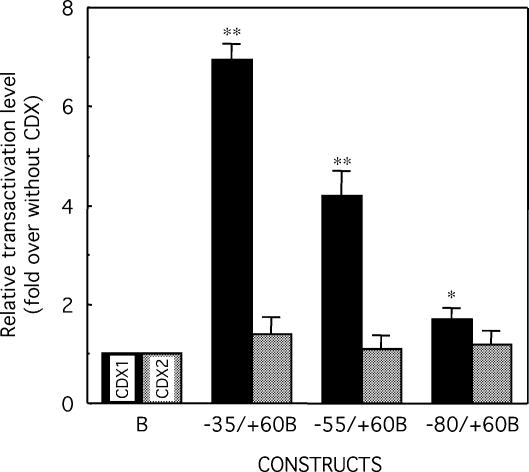
In order to analyse the putative effect of CDX proteins in the transactivation of the Glc6Pase promoter, we co-transfected HepG2 cells, which do not express CDX genes, with the expression vectors pCDX1-S and pCDX2-S that respectively encode the murine CDX1 and CDX2 proteins, together with the Glc6Pase-LUC constructs. The expression of CDX2 in HepG2 cells did not affect the Glc6Pase promoter activity of any construct (Fig. 2). In contrast, the CDX1 expression in HepG2 cells resulted in the activation of the reporter gene expression by about seven times, four times and twice for the –35/+60B, –55/+60B and –80/+60B constructs, respectively (Fig. 2). Noteworthy, the highest induction by CDX1 was observed with the shortest fragment (–35/+60), overlapping the putative TATA-box (TATAAAA) located near its 5′-end (–31/–25bp).
Figure 2.
Transactivation of the Glc6Pase promoter by CDX1 and CDX2 in HepG2 cells. HepG2 cells were transiently transfected with the 5′-end deleted Glc6Pase promoter fragments fused to a LUC reporter gene in pGL2‘basic’ (1 µg), pCMV-RL (2 ng, used as internal control), without or with pCDX1-S (100 ng) or pCDX2-S (100 ng). LUC activities were determined 48 h after transfection and were normalized relatively to the level of RL activities. The transactivation level of each construct by CDX1 (black bars) and CDX2 (grey bars) is expressed as fold of induction over the basal condition (without CDX) relatively of pGL2‘basic’. Results are the mean ± S.E.M. of at least three independent experiments performed in duplicate. * and **, significantly different from without CDX, P < 0.05 and P < 0.01, respectively.
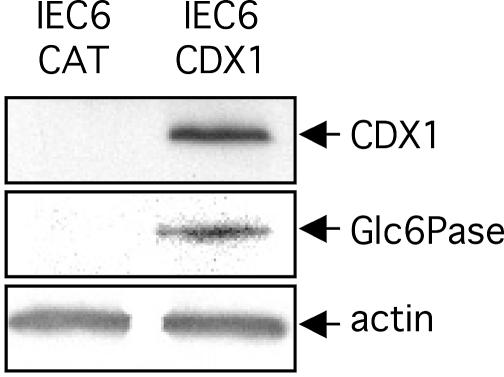
Overexpression of CDX1 in IEC6 cells induces the expression of the Glc6Pase gene
The potential function of CDX1 in the Glc6Pase gene expression was analysed in stable-transfected intestinal IEC6 cell lines (29). Control IEC6 cells transfected to express bacterial CAT did neither synthetize CDX1 nor the Glc6Pase protein (Fig. 3). In contrast, IEC6 cells overexpressing CDX1 expressed Glc6Pase protein (Fig. 3).
Figure 3.
The overexpression of CDX1 in IEC6 cells induces the Glc6Pase expression. The protein expression was analysed by western blotting on whole protein extracts from IEC6-CAT cells or IEC6-CDX1 cells. Blots were revealed with anti-CDX1 (upper panel), anti-Glc6Pase (middle panel) or anti-actin (lower panel).
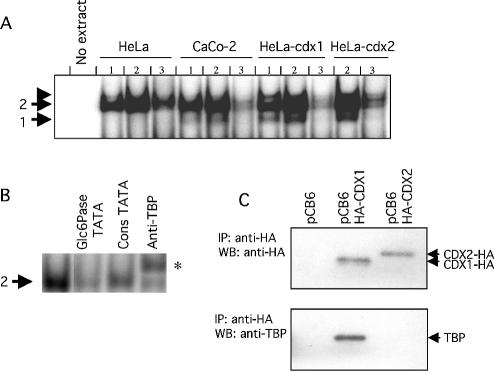
Both CDX1 and CDX2 are able to bind the Glc6Pase TATAAAA (–31/–25) but only CDX1 specifically interacts with the TATA-binding protein
Noteworthy, this TATAAAA (–31/–25) sequence exhibits obvious sequence similarity with the consensus CDX-BS (C/TATAAAG/T), while no additional putative CDX-BS could be found elsewhere in this promoter fragment. To examine whether CDX proteins could bind to this TATAAAA region, we designed a double-stranded oligonucleotide spanning the region –42/–12 of the Glc6Pase promoter. Gel mobility shift assays were performed using cellular extracts from differentiated Caco-2 cells, which express a low amount of CDX2, from CDX1- or CDX2-transfected HeLa cells and from control HeLa cells not expressing CDX proteins. Gel shift experiments showed that the binding to the Glc6Pase TATAAAA region led to the formation of two complexes. One complex (referred to as complex 1) was obtained only with cell extracts containing the CDX1 and/or CDX2 proteins, namely the differentiated Caco-2 cells and CDX-transfected HeLa cells, but not with the control HeLa cell extracts (Fig. 4A, lanes 1 and 2). The formation of the latter complex was competed by an excess of unlabelled oligonucleotide (Fig. 4A, lanes 3). These results strongly suggested that both CDX1 and CDX2 were able to bind the TATAAAA region of the Glc6Pase gene. A second complex (complex 2) was obtained in the presence of all cell extracts (Fig. 4A). The formation of this complex was competed by an excess of unlabelled oligonucleotide (Fig. 4A, lanes 3, and 4B). It was also competed by an excess of unlabelled consensus TATA probe, and shifted upon preincubation of the protein extracts with an antibody against TBP (Fig. 4B). This strongly suggested that complex 2 could be identified as TBP. Since both CDX1 and CDX2 as well as TBP bind the Glc6Pase TATA-box containing oligonucleotide, we have investigated whether any of the CDX proteins can interact with TBP. For this purpose, co-immunoprecipitation assays using anti-HA antibody were performed with protein extracts of transfected cells co-expressing TBP proteins and either CDX1-HA or CDX2-HA. Western blots performed with the anti-HA antibody confirmed the presence of CDX1-HA or CDX2-HA in precipitates of cells transfected with the respective plasmids (Fig. 4C, upper panel). Western blots performed with the anti-TBP antibody showed that TBP protein was present only in the precipitate from CDX1-HA expressing cells (Fig. 4C, lower panel). On the contrary, we failed to detect TBP in the CDX2-containing co-immunoprecipitate. These results strongly suggested that CDX1, but not CDX2, was able to interact with TBP directly.
Figure 4.
Specific binding of CDX1 to the TATA box of the Glc6Pase promoter and interaction with the TATA-binding protein. (A) Gel shift assays were carried out with 5 (lanes 1) or 15 µg (lanes 2 and 3) of whole cell extracts from HeLa cells, differentiated Caco-2 cells, CDX1-expressing HeLa cells, or CDX2-expressing HeLa cells and double-strand radiolabelled oligonucleotide matching the Glc6Pase TATAAAA sequence. Control was performed without protein (no extract). Competition experiments were performed in the presence of 100-fold excess of unlabelled Glc6Pase TATAAAA oligonucleotide (lanes 3). ‘1’ and ‘2’ on the left refer to DNA–protein complexes. The arrowhead above 2 indicates the presence of a non-reproducible complex. (B) Gel shift assays were carried out with 15 µg of whole cell extract from HeLa cells and double-strand radiolabelled oligonucleotide matching the Glc6Pase TATAAAA sequence. Competition experiments were performed with 100-fold excess of unlabelled oligonucleotides (Glc6Pase TATAAAA and consTATA box). Supershift experiment was performed with anti-TBP antibody, and indicated by an asterisk. (C) CDX1 and TBP interaction. HTC116 cells were co-transfected with vehicle alone (pCB6 vector), HA-CDX1 expressing vector (pCB6-HA-CDX1) or HA-CDX2 expressing vector (pCB6-HA-CDX2) and TBP expressing vector (pXJ41-hTBP plasmid). Immunoprecipitation (IP) was performed with anti-HA antibody and revealed by western blotting (WB) with anti-HA (upper panel) or anti-TBP (lower panel).
The CDX1 transactivation domain rather than the homeodomain provides specific effects of CDX1 protein
Because both CDX-proteins appeared able to bind the TATAAAA (–31/–25) sequence, we constructed two chimerical proteins with swapped homeodomains to ascertain the specificity of the CDX1 effect on the Glc6Pase promoter compared to CDX2. Sequence alignment of the CDX1 and CDX2 homeodomains revealed three differences between the murine proteins, but only two differences between the human proteins (Fig. 5A). Therefore, CDX1HD2 was constructed by changing N190 and T194 of CDX1 respectively into T and S, while CDX2HD1 was constructed by changing T221 and S225 of CDX2 respectively into N and T. Thus, the protein CDX1HD2 contained the DNA binding homeodomain (HD) of CDX2 in the context of CDX1, while reciprocally the protein CDX2HD1 contained the CDX1 homeodomain in the context of CDX2. As shown in Figure 5B, the CDX2HD1 chimerical protein, like CDX2, failed to activate the –35/+60B Glc6Pase promoter in HepG2. In contrast, the CDX1HD2 chimerical protein induced the activity of the –35/+60B promoter in the same range as did CDX1, by ∼6-fold. These results indicated that the protein domains outside the DNA-binding homeodomains were responsible for the specificity of action of CDX1 and CDX2 on the Glc6Pase promoter.
Figure 5.
Specific transactivation and binding competition of CDX proteins on the Glc6Pase promoter. (A) Amino acid sequence of the homeodomains of the murine and human CDX1 and CDX2 proteins. Uppercase letters indicate the differences between these proteins. (B) HepG2 cells were transiently transfected with the –35/+60B construct (1 µg) together with pCMV-RL (2 ng, used as internal control), without or with 100 ng of pCDX1-S or pCDX2-S, or pCDX1HD2, or pCDX2HD1. (C) HepG2 cells were transiently transfected with the –35/+60B construct (1 µg) together with pCMV-RL (2 ng), in the presence of pCDX1-S (100 ng) and pCDX2-S (1–200 ng, black circles) or pCDX2-HD1 (1–200 ng, black triangles), or pCDX1-HD2 (1–200 ng, black squares) expression vectors. LUC activities were determined 48 h after transfection and were normalized with regards to the level of RL activities. The transactivation rate is expressed as fold of induction over the basal condition. Results are the mean ± S.E.M. of at least three independent experiments performed in duplicate.*, significantly different from without CDX, P < 0.01.
Since both CDX1 and CDX2 were capable of binding the TATA-box of the Glc6Pase promoter, while only CDX1 transactivated this promoter, we carried out competition assays by co-expressing CDX1 in the presence of CDX2, or CDX1HD2 or CDX2HD1 in HepG2 cells. The stimulatory effect of CDX1 on the –35/+60B promoter activity (obtained in the presence of 100 ng of pCDX1-S expression vector) was progressively reduced by increasing the amount of CDX2 (from 10 to 200 ng of expression vector) and even better by increasing the amount of CDX2HD1 (Fig. 5C). On the contrary, the stimulatory effect of CDX1 was enhanced by increasing the amount of CDX1HD2 (Fig. 5C). Together, these results strongly suggested that, although the CDX1, CDX2, CDX1HD2 and CDX2HD1 proteins were all able to bind the –35/+60 promoter region of the Glc6Pase gene, only proteins containing the CDX1 transactivation domain (CDX1 or CDX1HD2) might transactivate this promoter, whereas proteins containing the CDX2 transactivating domain (CDX2 and CDX2HD1) prevented transactivation by CDX1.
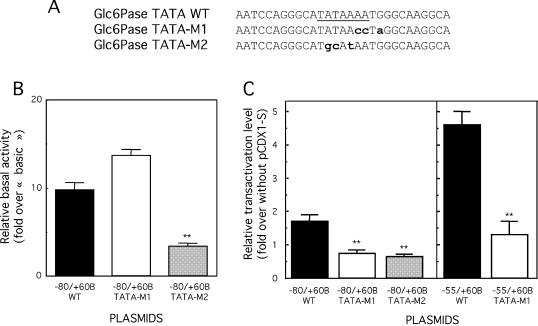
The Glc6Pase TATAAAA (–31/–25) is crucial for both the preinitiation of transcription and the transactivation by CDX1
We then investigated whether the TATAAAA sequence exhibited a single or separate functions involved in the preinitiation of transcription and in transactivation by CDX1. For this purpose, two different mutants were constructed in the TATAAAA region (Fig. 6A). In the first mutant, TATA-M1, the TATAAAA sequence was changed into TATAACC with the aim of preserving a canonical TATA-box, but altering the CDX-BS. In the second mutant, TATA-M2, the sequence was changed into TGCATAA, in order to disrupt both the TATA-box and the CDX-BS. Both mutations were inserted in two constructs of the Glc6Pase promoter, i.e. –80/+60B and –55/+60B. Figure 6B shows that the TATA-M2 mutation resulted in a dramatic decrease in the basal promoter activity of the –80/+60B construct (by ∼70%). In contrast, the TATA-M1 mutation did not modify the basal promoter activity (Fig. 6B). This strongly suggested that the TATAA sequence at the 5′-side of the Glc6Pase TATAAAA region was necessary and sufficient to support basal promoter activity and recognition by the TATA binding protein (TBP)/preinitiation complex.
Figure 6.
Mutation of the TATA box alters the CDX1 transactivation of the Glc6Pase promoter. (A) Mutations (M1 and M2) introduced in the TATAAAA region of the Glc6Pase promoter. Wild type (WT) TATA-box site is underlined. Mutated bases are in lowercase and bold. (B and C) HepG2 cells were transiently transfected with the –80/+60B and –55/+60B wild type (WT) or mutated constructs (1 µg) together with pCMV-RL (2 ng, used as internal control), without or with 100 ng of pCDX1-S. LUC activities were determined 48 h after transfection and were normalized with regards to the level of RL activities. (B) Basal transcription activity of the wild type and mutated forms of the –80/+60B constructs. The transcriptional activity of each construct is expressed relative to the LUC activity of pGL2‘basic’ and is the mean ± S.E.M. of at least three independent experiments performed in duplicate. **, significantly different from pGL2basic activity, P < 0.01. (C) Transactivation level by CDX1, expressed as fold of induction over the basal condition (without CDX1). Results are the mean ± S.E.M. of at least three independent experiments performed in duplicate. **, significantly different from without pCDX1-S, P < 0.01.
Next, we analysed the effect of the TATA-M1 and -M2 mutations on the transactivation by CDX1 (Fig. 6C). The residual promoter activity resulting from the TATA-M2 mutation in either –80/+60B and –55/+60B could not be stimulated by CDX1. Strikingly, the TATA-M1 mutation, although not altering the basal promoter activity, prevented any stimulation by CDX1, which indicated that an intact sequence at the 3′-side of the Glc6Pase TATAAAA region was required for transactivation by the homeoprotein. Taken together, these results showed that the TATAAAA region exhibited two separable functions involved in both basal transcriptional activity and stimulation by the CDX1 homeoprotein.
Transactivation by CDX1 can be restored by two CDX-BS head-to-tail in the absence of a canonical TATA-box
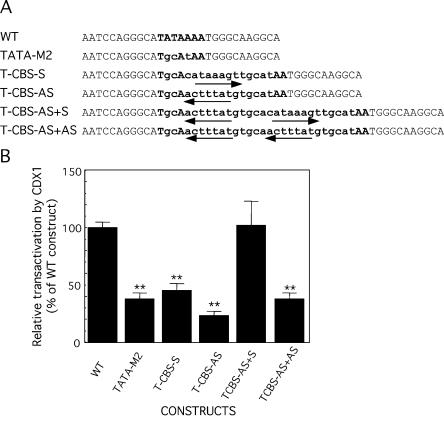
The above results suggested that the stimulatory effect of CDX1 required an intact CDX-BS in the context of a canonical TATA-box. We then raised the question to know if a CDX-BS could support transcriptional activation by CDX1 in the absence of a TATA-box. With this aim, we constructed a series of mutants in which the TATAAAA region of the Glc6Pase promoter was replaced by one or two copies of a consensus CDX-BS whose sequence was chosen so that it did not correspond to a typical TATA-box (Fig. 7A). The consensus CDX-BS sequence 5′-CATAAAGT-3′ was inserted in sense or antisense orientation (5′-ACTTTATG-3′). Transfection experiments conducted in HepG2 revealed that all the mutants had a very low basal promoter activity (not shown), indicating that a CDX-BS different in sequence from the wild type TATAAAA (–31/–25) hardly recruits the basal transcriptional machinery in the absence of CDX factors. In cotransfection experiments with the CDX1-expressing vector, CDX1 failed to transactivate the mutants containing one copy of CDX-BS, in either sense or antisense orientation (Fig. 7B). In contrast, the mutant in which two copies of CDX-BS were inserted head-to-tail, was stimulated by CDX1 as efficiently as the promoter construct containing the wild type TATAAAA sequence (Fig. 7B). Noteworthy, the head-to-tail organization was required since two CDX-BS in-tandem-repeats did not allow similar transcriptional activation (Fig. 7B). This result indicated that, although the structure formed by two CDX-BS head-to-tail was not able to overcome the deficiency of a canonical TATA-box in the absence of CDX1, it could overcome such a deficiency and support significant transcriptional activity in the presence of this homeodomain protein.
Figure 7.
Insertion of CDX-BS to overcome the lack of functional TATA box. (A) The mutations introduced to replace the TATA box of the Glc6Pase promoter in the –80/+60B TATAM2 construct. Wild-type (WT) TATA box site is in bold uppercase. Mutated bases are in lowercase and bold. The mutants contain one CDX-BS (T-CBS-S, T-CBS-AS) or two CDX-BS (T-CBS-AS+S and T-CBS-AS+AS) instead of the TATA-box. Arrows indicate the CDX-BS in sense and antisense orientation. Sense orientation corresponds to the CDX-BS formed by the TATA-box. (B) Relative transactivation level by CDX1 of the mutants compared to the wild type construct. HepG2 cells were transiently transfected with the –80/+60B wild type (WT) or mutated constructs (1 µg) together with pCMV-RL (2 ng, used as internal control), without or with 100 ng of pCDX1-S. LUC activities were determined 48 h after transfection and were normalized with regards to the level of RL activities. Transactivation level by CDX1 was expressed as fold of induction over the basal condition (without CDX1) and compared with that obtained with the WT construct (100%). Results are the mean ± S.E.M. of at least three independent experiments performed in duplicate. **, significantly different from WT construct transactivation, P < 0.01.
DISCUSSION
Several transcription factors expressed in the three gluconeogenic tissues, i.e. the liver, kidney and small intestine, have been shown to be involved in the regulation of the Glc6Pase gene, e.g. HNF1, HNF4 and HNF6 (14,15,17,18). However, the molecular mechanisms underlying tissue-specific expression of this gene remain to be specified. Here we show that the two related CDX1 and CDX2 transcription factors, which are intestine-specific [with a low expression of CDX2 also in the endocrine pancreas (32)], may participate in the control of the specific expression of Glc6Pase in the gut. An intriguing result has been that CDX1 efficiently and specifically transactivates the minimal Glc6Pase promoter, whereas CDX2 has no effect alone and even can antagonize the effect of CDX1. Distinct effects of CDX1 and CDX2 have already been reported in the case of the PCNA promoter (33), but the present study is the first example of a direct antagonistic role of both homeoproteins. CDX1 and CDX2 exhibit strong sequence homology between their homeodomains, which serve in DNA binding. On the contrary, they only share limited stretches of common amino acids outside the homeodomain. Swapping experiments at the level of the CDX1 and CDX2 homeodomains indicate that the respective effects of these proteins on the Glc6Pase promoter are not dependent on the homeodomain itself, but instead on the region(s) outside the homeodomain. Consistent with the latter, the stimulatory effect of CDX1, with regard to the absence of effect of CDX2, should likely be associated to its specific ability to interact with the TBP, even in the absence of DNA. Moreover, it can be inferred from band shifts and transactivation competition experiments that the suppressor role of CDX2 on the CDX1 effect could be related to its capacity to compete for the binding to the same DNA site. The alternative hypothesis of a repressor role of CDX2 by itself is unlikely because CDX2 has no effect on the basal transcription in the absence of CDX1 (results of Fig. 2).
The crucial role in vivo of the Glc6Pase promoter induction by CDX1 is strongly suggested by the observation that the overexpression of CDX1 in IEC6 cells reveals sufficient to trigger the expression of the endogenous Glc6Pase gene. Noteworthy, the CDX1–CDX2 duality of action might have a crucial role to play in the versatility of Glc6Pase expression in CDX-expressing tissues, i.e. the small and large intestines and pancreatic islets. In pancreas and isolated pancreatic islets, the Glc6Pase mRNA has been reported to be absent (34,35), which is in line with the fact that only CDX2, but not CDX1, is expressed in the endocrine pancreas (32). It is also worth noting that the CDX1/CDX2 ratio changes along the anterior-to-posterior axis of the gut (8), which may account for the specific pattern of Glc6Pase expression in the consecutive regions along the gut in the standard situation, fasting and refeeding (5). An additional level of complexity is provided by the fact that the CDX2 protein can modulate its own promoter as well as the Cdx1 gene promoter (36,37), and by the fact that CDX2, at least, can be the target of post-translational modifications that either increase or reduce its activity (38,39). So far, data on the transcriptional and post-translational regulation of CDX1 and CDX2 during fasting/refeeding and in diabetes are lacking. Although not investigated so far, the link between CDX transcription factors and Glc6Pase may also be important for pathological cases characterized by ectopic expression of the Cdx1 and/or Cdx2 genes, like in some gastric cancers and cholangiocarcinomas (40,41).
A clear-cut finding in this work has been the unravelling of the role of the TATAAAA (–31/–25) sequence of the Glc6Pase promoter, first as the actual TATA-box of the gene and second as the site of action of CDX1. Moreover, we have shown that these two functions are functionally separable by site-directed mutagenesis. Indeed, the two A nucleotides at the 3′-end of this sequence are not required for basal transcription, but they are required to confer on this sequence the capacity to be regulated by CDX1/CDX2. On the other hand, disrupting the sequence at the 5′-end alters both the basal transcription and the regulation by CDX proteins. Noteworthy, the existence of functional interrelationships between CDX-proteins and the basal transcription machinery could be a general feature in the regulation of intestine genes. Indeed, most of the known target genes of CDX proteins contain one CDX-BS in the very proximal promoter region, which sometimes overlaps the TATA-box like in the calbindin-9 and the clusterin genes (21–23), and the Glc6Pase gene (present study). Using mutated forms of the Glc6Pase promoter in which the endogenous TATA box was deleted, we demonstrate that the incorporation of a single consensus CDX-BS in place of the functional TATA-box fails to overcome the lack of the TATA-box. In contrast, two CDX-BS, specifically in opposite orientation, can rescue transcription activity if CDX1 is present, suggesting that pairs of CDX factors can actively recruit the transcriptional machinery in the absence of a functional TATA box. This conclusion is supported by the structure of the proximal promoter of two other intestine genes, which also contain pairs of CDX-BS. One gene is the claudin-2 gene, and its promoter is TATA-less (12). The second gene, encoding SI, has two CDX-BS in opposite orientation in the proximal promoter, but it also contains a TATATA sequence at position –29/–24 presumed to represent the TATA box. Disruption of one of the CDX-BS significantly reduces and disruption of the two sites almost totally inhibits, both the CDX-stimulated and the basal transcriptional activity of the SI promoter, providing evidence of the importance of these sites for the function of the promoter (42). In separate experiments, we have addressed the role of the TATATA sequence of the SI promoter by changing it into TGCATA (similar to TATA-M2, refer to Fig. 6). We found that this mutation did neither affect the basal transcriptional activity of the SI promoter, nor its stimulation by CDX factors, demonstrating that the TATATA sequence has no TATA box function, and hence that the SI proximal promoter with two CDX-BS is also TATA-less (unpublished results). Thus, it comes out from these results that two CDX-BS in a head-to-tail organization can recruit the basal transcription machinery in the presence of CDX proteins, even in absence of typical TATA-box. Considering the number of intestinal genes exhibiting either one CDX-BS in close vicinity from the TATA-box (7,10,11,13), or a CDX-BS overlapping the TATA-box (21–23; Glc6Pase), or even two CDX-BS replacing the TATA-box (12,42), it might be suggested that two related mechanisms of recruitment of TBP could have a specific role in the expression of intestinal genes. Hence, the association of TBP to target genes would require two points of contact, which could be either two CDX proteins, or a TATA-DNA sequence and a CDX protein bound to it or in close vicinity. This may provide a molecular basis to the homeotic function of CDX proteins during the development of the gastrointestinal tract, given that these genes deliver the positional information needed for the gut specification (43–46).
In summary, the results presented here constitute the first report that a crucial gluconeogenic gene, i.e. Glc6Pase, whose expression is restricted to the liver, kidney and small intestine (5), is a direct target for intestine-specific transcription factors, namely the factors of the CDX family. More specifically, we have provided evidence that the Glc6Pase TATAAAA sequence (–31/–25) has separate functions involved in basal transcription and in regulation, and that CDX1 and CDX2 exert antagonist effects on the Glc6Pase TATA-box. Based on the sequence conservation of the CDX proteins and the Glc6Pase promoter between mouse, rat and human, these data may be highly relevant for human physiology.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank V. Moucadel (INSERM U. 315, Marseille, France), for providing us the human CDX1 and CDX2 expression vectors and helpful discussion at the beginning of the work, J. Iovanna (INSERM U. 315, Marseille, France) for providing the anti-CDX1 antibodies and the IEC6-CAT and IEC6-CDX1 cells and L. Tora (IGBMC, Strasbourg, France) for providing the pXJ41-hTBP plasmid and the anti-TBP antibodies. We are also grateful for A. Stefanutti for precious help in some experiments. C.D.-D. was a recipient of a grant from the Ipsen Fundation and A.C. was a recipient of the grant of the Région-Alsace.
REFERENCES
- 1.Mithieux G. (1997) New knowledge regarding glucose-6 phosphatase gene and protein and their roles in the regulation of glucose metabolism. Eur. J. Endocrinol., 136, 137–145. [DOI] [PubMed] [Google Scholar]
- 2.Croset M., Rajas,F., Zitoun,C., Hurot,J.M., Montano,S. and Mithieux,G. (2001) Rat small intestine is an insulin-sensitive gluconeogenic organ. Diabetes, 50, 740–746. [DOI] [PubMed] [Google Scholar]
- 3.Mithieux G. (2001) New data and concepts on glutamine and glucose metabolism in the gut. Curr. Opin. Clin. Nutr. Metab. Care, 4, 267–271. [DOI] [PubMed] [Google Scholar]
- 4.Chatelain F., Pegorier,J.P., Minassian,C., Bruni,N., Tarpin,S., Girard,J. and Mithieux,G. (1998) Development and regulation of glucose-6-phosphatase gene expression in rat liver, intestine and kidney: in vivo and in vitro studies in cultured fetal hepatocytes. Diabetes, 47, 882–889. [DOI] [PubMed] [Google Scholar]
- 5.Rajas F., Bruni,N., Montano,S., Zitoun,C. and Mithieux,G. (1999) The glucose-6 phosphatase gene is expressed in human and rat small intestine: regulation of expression in fasted and diabetic rats. Gastroenterology, 117, 132–139. [DOI] [PubMed] [Google Scholar]
- 6.Cereghini S. (1996) Liver-enriched transcription factors and hepatocyte differentiation. Faseb J., 10, 267–282. [PubMed] [Google Scholar]
- 7.Traber P.G. and Silberg,D.G. (1996) Intestine-specific gene transcription. Annu. Rev. Physiol., 58, 275–297. [DOI] [PubMed] [Google Scholar]
- 8.Silberg D.G., Swain,G.P., Suh,E.R. and Traber,P.G. (2000) Cdx1 and cdx2 expression during intestinal development. Gastroenterology, 119, 961–971. [DOI] [PubMed] [Google Scholar]
- 9.Freund J.N., Domon-Dell,C., Kedinger,M. and Duluc,I. (1998) The Cdx-1 and Cdx-2 homeobox genes in the intestine. Biochem. Cell Biol., 76, 957–969. [DOI] [PubMed] [Google Scholar]
- 10.Taylor J.K., Boll,W., Levy,T., Suh,E., Siang,S., Mantei,N. and Traber,P.G. (1997) Comparison of intestinal phospholipase A/lysophospholipase and sucrase-isomaltase genes suggest a common structure for enterocyte-specific promoters. DNA Cell Biol., 16, 1419–1428. [DOI] [PubMed] [Google Scholar]
- 11.Mitchelmore C., Troelsen,J.T., Spodsberg,N., Sjostrom,H. and Noren,O. (2000) Interaction between the homeodomain proteins Cdx2 and HNF1alpha mediates expression of the lactase-phlorizin hydrolase gene. Biochem. J., 346, 529–535. [PMC free article] [PubMed] [Google Scholar]
- 12.Sakaguchi T., Gu,X., Golden,H.M., Suh,E., Rhoads,D.B. and Reinecker,H.C. (2002) Cloning of the human claudin-2 5′-flanking region revealed a TATA-less promoter with conserved binding sites in mouse and human for caudal-related homeodomain proteins and hepatocyte nuclear factor-1alpha. J. Biol. Chem., 277, 21361–21370. [DOI] [PubMed] [Google Scholar]
- 13.Krasinski S.D., Van Wering,H.M., Tannemaat,M.R. and Grand,R.J. (2001) Differential activation of intestinal gene promoters: functional interactions between GATA-5 and HNF-1 alpha. Am. J. Physiol. Gastrointest. Liver Physiol., 281, G69–G84. [DOI] [PubMed] [Google Scholar]
- 14.Rajas F., Gautier,A., Bady,I., Montano,S. and Mithieux,G. (2002) Polyunsaturated fatty acyl coenzyme A suppress the glucose-6-phosphatase promoter activity by modulating the DNA binding of hepatocyte nuclear factor 4 alpha. J. Biol. Chem., 277, 15736–15744. [DOI] [PubMed] [Google Scholar]
- 15.Streeper R.S., Hornbuckle,L.A., Svitek,C.A., Goldman,J.K., Oeser,J.K. and O’Brien,R.M. (2001) Protein kinase A phosphorylates hepatocyte nuclear factor-6 and stimulates glucose-6-phosphatase catalytic subunit gene transcription. J. Biol. Chem., 276, 19111–19118. [DOI] [PubMed] [Google Scholar]
- 16.Lin B., Morris,D.W. and Chou,J.Y. (1997) The role of HNF1alpha, HNF3gamma and cyclic AMP in glucose-6-phosphatase gene activation. Biochemistry, 36, 14096–14106. [DOI] [PubMed] [Google Scholar]
- 17.Streeper R.S., Svitek,C.A., Goldman,J.K. and O’Brien,R.M. (2000) Differential role of hepatocyte nuclear factor-1 in the regulation of glucose-6-phosphatase catalytic subunit gene transcription by cAMP in liver- and kidney-derived cell lines. J. Biol. Chem., 275, 12108–12118. [DOI] [PubMed] [Google Scholar]
- 18.Lin B., Morris,D.W. and Chou,J.Y. (1998) Hepatocyte nuclear factor 1alpha is an accessory factor required for activation of glucose-6-phosphatase gene transcription by glucocorticoids. DNA Cell Biol., 17, 967–974. [DOI] [PubMed] [Google Scholar]
- 19.Streeper R.S., Eaton,E.M., Ebert,D.H., Chapman,S.C., Svitek,C.A. and O’Brien,R.M. (1998) Hepatocyte nuclear factor-1 acts as an accessory factor to enhance the inhibitory action of insulin on mouse glucose-6-phosphatase gene transcription. Proc. Natl Acad. Sci. USA, 95, 9208–9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Margalit Y., Yarus,S., Shapira,E., Gruenbaum,Y. and Fainsod,A. (1993) Isolation and characterization of target sequences of the chicken CdxA homeobox gene. Nucleic Acids Res., 21, 4915–4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambert M., Colnot,S., Suh,E., L’Horset,F., Blin,C., Calliot,M.E., Raymondjean,M., Thomasset,M., Traber,P.G. and Perret,C. (1996) cis-Acting elements and transcription factors involved in the intestinal specific expression of the rat calbindin-D9K gene: binding of the intestine-specific transcription factor Cdx-2 to the TATA box. Eur. J. Biochem., 236, 778–788. [DOI] [PubMed] [Google Scholar]
- 22.Barley N.F., Prathalingam,S.R., Zhi,P., Legon,S., Howard,A. and Walters,J.R. (1999) Factors involved in the duodenal expression of the human calbindin-D9k gene. Biochem. J., 341, 491–500. [PMC free article] [PubMed] [Google Scholar]
- 23.Suh E., Wang,Z., Swain,G.P., Tenniswood,M. and Traber,P.G. (2001) Clusterin gene transcription is activated by caudal-related homeobox genes in intestinal epithelium. Am. J. Physiol. Gastrointest. Liver Physiol., 280, G149–G156. [DOI] [PubMed] [Google Scholar]
- 24.Argaud D., Zhang,Q., Pan,W., Maitra,S., Pilkis,S.J. and Lange,A.J. (1996) Regulation of rat liver glucose-6-phosphatase gene expression in different nutritional and hormonal states: gene structure and 5′-flanking sequence. Diabetes, 45, 1563–1571. [DOI] [PubMed] [Google Scholar]
- 25.Lorentz O., Duluc,I., Arcangelis,A.D., Simon-Assmann,P., Kedinger,M. and Freund,J.N. (1997) Key role of the Cdx2 homeobox gene in extracellular matrix-mediated intestinal cell differentiation. J. Cell Biol., 139, 1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.May M., Mengus,G., Lavigne,A.C., Chambon,P. and Davidson,I. (1996) Human TAF(II28) promotes transcriptional stimulation by activation function 2 of the retinoid X receptors. EMBO J., 15, 3093–3104. [PMC free article] [PubMed] [Google Scholar]
- 27.Manley J.L., Fire,A., Samuels,M. and Sharp,P.A. (1983) In vitro transcription: whole-cell extract. Methods Enzymol., 101, 568–582. [DOI] [PubMed] [Google Scholar]
- 28.Brou C., Chaudhary,S., Davidson,I., Lutz,Y., Wu,J., Egly,J.M., Tora,L. and Chambon,P. (1993) Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J., 12, 489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soubeyran P., Andre,F., Lissitzky,J.C., Mallo,G.V., Moucadel,V., Roccabianca,M., Rechreche,H., Marvaldi,J., Dikic,I., Dagorn,J.C. et al. (1999) Cdx1 promotes differentiation in a rat intestinal epithelial cell line. Gastroenterology, 117, 1326–1338. [DOI] [PubMed] [Google Scholar]
- 30.Domon-Dell C., Schneider,A., Moucadel,V., Guérin,E., Guenat,D., Aguillon,S., Duluc,I., Martin,E., Iovanna,J.L., Launay,J.-F. et al. (2003) The Cdx1 homeobox gene during human colon cancer progression. Oncogene, 22, ONC 12531. [DOI] [PubMed] [Google Scholar]
- 31.Zweibaum A., Triadou,N., Kedinger,M., Augeron,C., Robine-Leon,S., Pinto,M., Rousset,M. and Haffen,K. (1983) Sucrase-isomaltase: a marker of foetal and malignant epithelial cells of the human colon. Int. J. Cancer, 32, 407–412. [DOI] [PubMed] [Google Scholar]
- 32.Laser B., Meda,P., Constant,I. and Philippe,J. (1996) The caudal-related homeodomain protein Cdx-2/3 regulates glucagon gene expression in islet cells. J. Biol. Chem., 271, 28984–28994. [DOI] [PubMed] [Google Scholar]
- 33.Oh E.J., Park,J.H., Cho,M., Lee,W.J., Choi,Y.H. and Yoo,M.A. (2002) The caudal-related homeodomain protein CDX1 activates proliferating cell nuclear antigen expression in hepatocellular and colorectal carcinoma cells. Int. J. Oncol., 20, 23–29. [PubMed] [Google Scholar]
- 34.Mithieux G., Vidal,H., Zitoun,C., Bruni,N., Daniele,N. and Minassian,C. (1996) Glucose-6-phosphatase mRNA and activity are increased to the same extent in kidney and liver of diabetic rats. Diabetes, 45, 891–896. [DOI] [PubMed] [Google Scholar]
- 35.Portha B., Giroix,M.H., Serradas,P., Gangnerau,M.N., Movassat,J., Rajas,F., Bailbe,D., Plachot,C., Mithieux,G. and Marie,J.C. (2001) Beta-cell function and viability in the spontaneously diabetic GK rat: information from the GK/Par colony. Diabetes, 50, S89–S93. [DOI] [PubMed] [Google Scholar]
- 36.Xu F., Li,H. and Jin,T. (1999) Cell type-specific autoregulation of the Caudal-related homeobox gene Cdx-2/3. J. Biol. Chem., 274, 34310–34316. [DOI] [PubMed] [Google Scholar]
- 37.Domon-Dell C. and Freund,J.N. (2002) Stimulation of Cdx1 by oncogenic beta-catenin/Tcf4 in colon cancer cells; opposite effect of the CDX2 homeoprotein. FEBS Lett., 518, 83–87. [DOI] [PubMed] [Google Scholar]
- 38.Houde M., Laprise,P., Jean,D., Blais,M., Asselin,C. and Rivard,N. (2001) Intestinal epithelial cell differentiation involves activation of p38 mitogen-activated protein kinase that regulates the homeobox transcription factor CDX2. J. Biol. Chem., 276, 21885–21894. [DOI] [PubMed] [Google Scholar]
- 39.Rings E.H., Boudreau,F., Taylor,J.K., Moffett,J., Suh,E.R. and Traber,P.G. (2001) Phosphorylation of the serine 60 residue within the Cdx2 activation domain mediates its transactivation capacity. Gastroenterology, 121, 1437–1450. [DOI] [PubMed] [Google Scholar]
- 40.Ren P., Silberg,D.G. and Sirica,A.E. (2000) Expression of an intestine-specific transcription factor (CDX1) in intestinal metaplasia and in subsequently developed intestinal type of cholangiocarcinoma in rat liver. Am. J. Pathol., 156, 621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silberg D.G., Furth,E.E., Taylor,J.K., Schuck,T., Chiou,T. and Traber,P.G. (1997) CDX1 protein expression in normal, metaplastic and neoplastic human alimentary tract epithelium. Gastroenterology, 113, 478–486. [DOI] [PubMed] [Google Scholar]
- 42.Suh E., Chen,L., Taylor,J. and Traber,P.G. (1994) A homeodomain protein related to caudal regulates intestine-specific gene transcription. Mol. Cell. Biol., 14, 7340–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamai Y., Nakajima,R., Ishikawa,T., Takaku,K., Seldin,M.F. and Taketo,M.M. (1999) Colonic hamartoma development by anomalous duplication in Cdx2 knockout mice. Cancer Res., 59, 2965–2970. [PubMed] [Google Scholar]
- 44.Silberg D.G., Sullivan,J., Kang,E., Swain,G.P., Moffett,J., Sund,N.J., Sackett,S.D. and Kaestner,K.H. (2002) Cdx2 ectopic expression induces gastric intestinal metaplasia in transgenic mice. Gastroenterology, 122, 689–696. [DOI] [PubMed] [Google Scholar]
- 45.Mutoh H., Hakamata,Y., Sato,K., Eda,A., Yanaka,I., Honda,S., Osawa,H., Kaneko,Y. and Sugano,K. (2002) Conversion of gastric mucosa to intestinal metaplasia in Cdx2-expressing transgenic mice. Biochem. Biophys. Res. Commun., 294, 470–479. [DOI] [PubMed] [Google Scholar]
- 46.Beck F. (2002) Homeobox genes in gut development. Gut, 51, 450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]