Abstract
Post-transcriptional regulation and the formation of mRNA 3′ ends are crucial for gene expression in eukaryotes. Interspecies conservation of many sequences within 3′UTRs reveals selective constraint due to similar function. To study the pattern of conservation within 3′UTRs, we compiled and aligned 50 sets of complete orthologous 3′UTRs from four orders of mammals. We observed a mosaic pattern of conservation, with alternating regions of high (phylogenetic footprints) and low similarity. Conservation in 3′UTRs correlates with their base composition and also with the synonymous substitution rate in corresponding coding regions. The non-uniform distribution of conservation is more pronounced for 3′UTRs with a moderate or low level of overall conservation, where invariant nucleotides are more numerous, and their runs of lengths 4–7 occur more frequently than if conservation were random. Many runs of invariant nucleotides are AU-rich or pyrimidine-rich. Some of these runs coincide with known functional cis- elements of eukaryotic mRNAs, such as the U-rich upstream element, polyadenylation signal and DICE regulatory signal. More divergent regions of multiple alignments of 3′UTRs are often more G- and/or C-rich. Our results provide evidence on the importance of moderately conserved regions in 3′UTRs and suggest that regulatory functions of 3′UTRs might utilize gene-specific information in these regions.
INTRODUCTION
Selective constraint affects both coding and non-coding DNA sequences (1,2). On average, 5′-untranslated regions (UTRs) and 3′UTRs of protein-coding genes are less conservative across species than protein-coding DNA sequences (CDSs), but more conservative than untranscribed sequences (3,4). Still, conservation of UTRs is quite substantial and sometimes even exceeds the conservation of the corresponding coding regions (5).
Patterns in conservation of coding sequences offer important clues on protein structure and function. Highly conservative amino acids are crucial for protein folding or catalytic activity (6,7). A similar, although much less studied, correspondence between conservation and function also exists in UTRs (8). Thus, comparing homologous UTRs can shed light on their function, which is still rather poorly studied.
There is evidence that conserved elements in 3′UTRs control post-transcriptional gene regulation and the stability of mRNAs (9). A remarkable finding is the existence of highly conserved regions within 3′UTRs (10,11). However, even moderately conservative stem–loop structures in 3′UTRs can possess crucial, well defined functions (12).
We studied the extent of interspecies conservation of orthologous 3′UTRs from four orders of mammals. We found a mosaic pattern with regions of high similarity (phylogenetic footprints, further referred to as footprints) interspersed with non-alignable sequences, and detected some shared sequence motifs among footprints. Conservation in 3′UTRs correlates with their base composition and also with the synonymous substitution rate in the corresponding coding regions. Our results show that functional regions of 3′UTRs may include longer sequences than those covered by the known short conservative cis-elements. Although motif sequences are likely to act by similar mechanisms, our computer analysis can be used to identify footprints and to search for common new motifs within UTRs, but not to predict their functions.
MATERIALS AND METHODS
Fifty orthologous complete 3′UTR sequences from four mammalian orders (Primates, Rodentia, Carnivora and Artiodactyla) were selected from GeneBank, HOVERGEN and UTRDB databases using standard searching procedures (see ftp://ftp.ncbi.nih.gov/pub/kondrashov/utr) and aligned by CLUSTAL W using the default parameters (13).
Footprints were isolated from CLUSTAL W alignments using the following heuristic. We first isolated a kernel defined as a 15 nt frame with at least nine matches giving >60% similarity. Each kernel was extended in both directions with 7 nt frames. The ends were trimmed so that the final similarity of the extended regions was >50% and the boundaries were a match. These parameters are rather conservative; a simple argument is as follows. The lowest significantly non-random level of similarity for two-sequence alignments (A ∼30%, U ∼30%, G ∼20% and C ∼20%) is 42% (2). In the case of four-sequence alignments, 42% four-way matches would also be significant, 50 and 60% are even more so. Footprints, in which the Karlin–Altschul p-value was below 0.01 (match = 1, mismatch = –1, gap = –1) for at least one of the six pairs of sequences, were selected (14). The final set contains 261 footprints that cover ∼50% of the UTR sequences.
We distinguish two types of matches in the alignments. Multiple matches are sites occupied by the same nucleotide in all the four sequences (also known as invariant nucleotides). A pairwise match corresponds to a match between any two sequences. For example, a nucleotide column AUUU in a multiple alignment contains three pairwise U-U matches. We analyzed the ratios of pairwise and multiple matches in the multiple alignments and their pairwise sub-alignments. To estimate the expected numbers of multiple matches for each multiple alignment, we created randomly simulated sets of multiple alignments with the observed number of matches between all six pairs of sequences.
The consensus sequence in the conservative positions of alignments consists of real nucleotides A, U, G and C, if >50% of the same nucleotide is present in a position. Statistical analyses were conducted using Excel (Microsoft, USA) and our statistical tools. The levels of synonymous and non-synonymous divergence (KS and KA, respectively) were calculated with the PAML program (ftp://abacus.gene.ucl. ac.uk/pub/paml/) using the default parameters and the yn00 estimation method (15).
We identified common fragments of runs of invariant nucleotides (further referred to as runs) and footprints. We required common fragments to be at least 6 nt long, since most regulatory signals have conservative cores of six or more nucleotides. Fragments also had to occur in at least eight alignments. We performed single-linking clustering on these fragments using the HistogramAC program (16).
RESULTS
Multiple alignment statistics
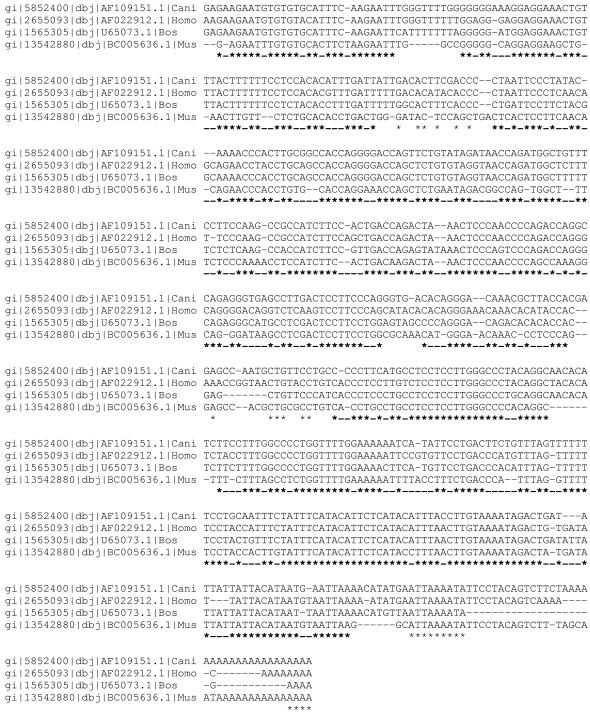
We aligned 3′UTR sequences from four species of mammals and isolated footprints within alignments, as described in Materials and Methods. A sample alignment is presented in Figure 1. Alignments of footprints mostly contain unambiguously aligned, highly similar segments of sequences. More than 97% of runs of multiple matches group together with pairwise matches to form footprints. Footprints alternate with relatively short inter-footprints, which are mostly composed of low similarity segments or are aligned against gaps (Table 1 and Fig. 1). The lengths of orthologous UTRs in the four species are highly similar (t = 6.2, r ranges from 0.82 to 0.52, P < 0.001). There is a positive correlation (r = 0.92, P < 0.0001) between the length of the UTR and the number of conserved nucleotides in it. On average, footprints are 70% similar in our set of UTRs; they are significantly more common within nucleotides 45–65 and 120–220 after the stop codon. Multiple alignment statistics are presented in Table 1; the general pattern of similarity in footprints is different from that of 3′UTRs. 3′UTRs can be classified into three classes: those with low overall conservation (<0.4), group A; with an average level of conservation (≥0.4 and <0.6), group B; and highly conserved (≥0.6), group C.
Figure 1.
Sample multiple alignment of 3′UTRs of cGMP phosphodiesterase delta subunit (PDE6D) mRNAs. Multiple matches conserved for all four sequences are indicated with asterisks. Footprints are shown in bold.
Table 1. Average nucleotide length of characteristic features in alignments of orthologous mammalian 3′UTRs, phylogenetic footprints and regions between footprints (inter-footprints).
| Primates | Rodentia | Carnivora | Ruminantia | |||||
|---|---|---|---|---|---|---|---|---|
| nt | % | nt | % | nt | % | nt | % | |
| 3′UTR length | 510.8 | 553.7 | 478.2 | 469.3 | ||||
| Alignment length | 542.4 | 100.0 | 614.7 | 100.0 | 512.4 | 100.0 | 509.7 | 100.0 |
| Multiple matches | 233.5 | 43.1 | 259.6 | 42.2 | 223.9 | 43.7 | 222.0 | 43.6 |
| Multiple mismatches | 277.3 | 51.1 | 294.1 | 47.8 | 254.3 | 49.6 | 247.3 | 48.5 |
| Insertion/deletion (indels) | 31.6 | 5.8 | 61.0 | 9.9 | 34.3 | 6.7 | 40.5 | 7.9 |
| Pairwise matches | 353.6 | 65.2 | 365.4 | 59.4 | 334.0 | 65.2 | 328.8 | 64.5 |
| Pairwise mismatches | 142.8 | 26.3 | 203.0 | 33.0 | 138.2 | 27.0 | 142.7 | 28.0 |
| Pairwise indels | 46.0 | 8.5 | 46.3 | 8.5 | 40.3 | 7.4 | 38.2 | 7.0 |
| Footprints | 261.5 | 48.2 | 287.5 | 46.8 | 251.3 | 49.0 | 250.8 | 49.2 |
| Multiple matches | 178.8 | 68.4 | 197.7 | 68.8 | 171.5 | 68.2 | 170.7 | 68.1 |
| Multiple mismatches | 76.9 | 29.4 | 81.9 | 28.5 | 75.3 | 29.9 | 74.5 | 29.7 |
| Indels | 5.8 | 2.2 | 7.9 | 2.8 | 4.6 | 1.8 | 5.5 | 2.2 |
| Pairwise matches | 215.0 | 82.2 | 228.7 | 79.6 | 206.2 | 82.1 | 205.0 | 81.7 |
| Pairwise mismatches | 40.3 | 15.4 | 52.5 | 18.3 | 38.6 | 15.4 | 40.1 | 16.0 |
| Pairwise indels | 6.2 | 2.4 | 6.3 | 2.2 | 6.5 | 2.6 | 5.7 | 2.3 |
| Inter-footprints | 280.9 | 51.8 | 327.2 | 53.2 | 261.2 | 51.0 | 259.0 | 50.8 |
| Multiple matches | 54.8 | 19.5 | 61.9 | 18.9 | 52.4 | 20.1 | 51.3 | 19.8 |
| Multiple mismatches | 200.4 | 71.3 | 212.2 | 64.9 | 179.1 | 68.6 | 172.8 | 66.7 |
| Indels | 25.8 | 9.2 | 53.1 | 16.2 | 29.7 | 11.4 | 34.9 | 13.5 |
| Pairwise matches | 138.6 | 49.3 | 136.7 | 41.8 | 127.8 | 48.9 | 123.8 | 47.8 |
| Pairwise mismatches | 102.5 | 36.5 | 150.5 | 46.0 | 99.6 | 38.1 | 102.6 | 39.6 |
| Pairwise indels | 39.9 | 14.2 | 40.0 | 12.2 | 33.8 | 12.9 | 32.6 | 12.6 |
Base composition and 3′UTR conservation
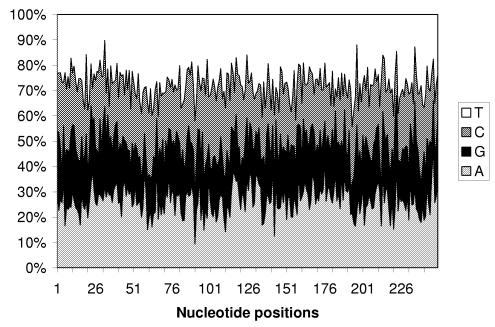
As seen from the overall base composition for the first 250 nt of 3′UTRs, the frequency of G is low (∼18%) throughout this region, and there are some sequence fragments with a low frequency of C (Fig. 2). Our results indicate that there is no position-specific conservation within the first 250 nt after the stop codon, despite the high interspecies conservation in the region. Runs in 3′UTRs are AU-rich; UTRs are depleted in G nucleotides, but GC content is higher in footprints than in runs (lengths 6 nt or longer) (Table 2). There are significant positive correlations between complementary G and C nucleotides (r = 0.55, P < 0.0001) in 3′UTRs. Positive correlation between complementary nucleotides A and U is somewhat lower (r = 0.41, P < 0.005).
Figure 2.
The overall base composition for the first 250 nt of 3′UTRs.
Table 2. Base composition of runs with lengths of 6 nt or longer, footprints and total 3′UTRs.
| Runs | Footprints | Overall | ||||
|---|---|---|---|---|---|---|
| A | 4996 | 30.8% | 13 352 | 28.0% | 28 062 | 27.8% |
| C | 2964 | 18.3% | 10 408 | 21.8% | 21 602 | 21.4% |
| G | 2760 | 17.0% | 9620 | 20.2% | 20 109 | 19.9% |
| T | 5476 | 33.8% | 14 264 | 29.9% | 29 978 | 29.7% |
| A+T | 10 472 | 64.7% | 27 616 | 58.0% | 58 040 | 57.5% |
| C+G | 13 436 | 35.3% | 38 024 | 42.0% | 79 642 | 41.3% |
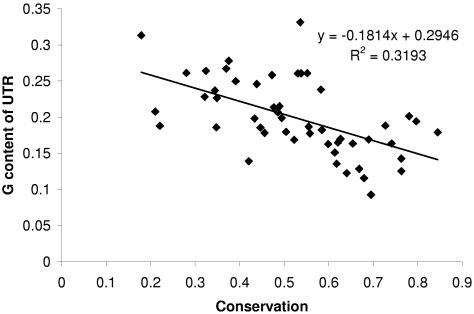
Correlation coefficients for GC(G) content and overall conservation in 3′UTRs, KS values (the level of synonymous divergence), and GC(G) content in the 4-fold degenerate sites (GC4 and G4) of corresponding coding regions are presented in Table 3. There is a significant negative correlation between the frequency of G and the overall conservation in 3′UTRs (Fig. 3). The correlation between GC content and overall conservation is somewhat lower. The GC content in the 4-fold degenerate sites of coding regions is correlated with GC content in 3′UTRs (r2 = 23%). KS values in coding regions and the 3′UTR conservation are negatively correlated (r2 = 22%). The average KS value for mRNAs in group A (low conservation in 3′UTR) is 0.68; it is 0.55 in group B and 0.43 in group C. We found a significant positive correlation between GC(G) content in UTRs and the level of synonymous divergence (KS values) for the corresponding protein-coding regions; there was no significant correlation between KA and GC(G) content. GC4 and KS are correlated better than GC4 and GC content in 3′UTRs, and better than KS and GC content in 3′UTRs.
Table 3. Correlation coefficients for GC(G) content and conservation in 3′UTRs, and GC4(G4) content and KS in the corresponding coding regions.
| GC4(G4) CDS | Conservation 3′UTRs | KS CDS | |
|---|---|---|---|
| GC(G) 3′UTRs | 0.48 (0.42), P < 0.005 | –0.49 (–0.56), P < 0.0005 | 0.38 (0.43), P < 0.01 |
| GC(G4) CDS | –0.48 (–0.46), P < 0.001 | 0.54 (0.47), P < 0.001 | |
| Conservation 3′UTRs | –0.47, P < 0.001 |
Figure 3.
Regression of the G content of the 3′UTR against the level of overall 3′UTR conservation.
Patterns of similarity and variability in 3′UTRs
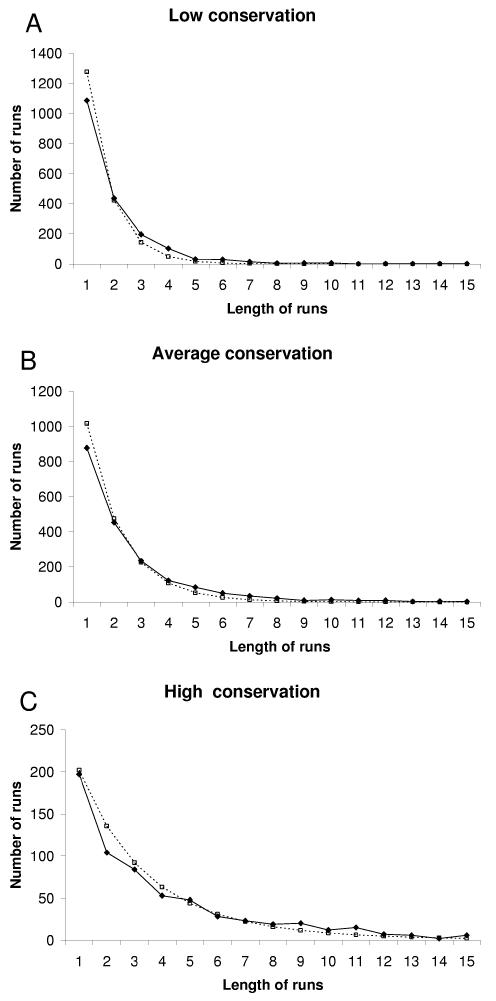
Assuming that matches are independent events occurring with frequency p, the probability of a run of n matches is (1 – p)p(n – 1). We compared this theoretical distribution with the distributions of runs of matches in our multiple alignments for the three different groups of 3′UTRs. These distributions of runs show that the length scale is no longer than ∼20 nt (Fig. 4). Frequencies of runs of 3–7 matches for group A, of 4–8 matches for group B, and of 8–12 matches for group C, were above expectation. On average, runs of 1–3 matches are under-represented, and runs of 6–10 matches are over-represented. These results indicate that some matches are clumped. Although the frequency of runs of matches is in good agreement with what is expected under random mutations with a uniform rate, the parameter of the distribution (p = 0.66) is higher than the baseline probability of a match (p = 0.44). The following conserved oligonucleotide motifs in the 3′UTRs were revealed by the HistogramAC clusterization program: AAUAAA, AUUUAu/a, UUAUUU, c/uCAGAA, GGCCc/uC, CUCCCa/c, UUCUU, c/uUUUc/u, and other CU-rich motifs, which resemble DICE motifs.
Figure 4.
(A–C) The distributions of theoretically predicted (dashed lines) and calculated (solid lines) numbers of runs in 3′UTRs with different overall conservation: low, <40% (A); average, ≥40% and <60% (B); high, ≥60% (C).
In looking for characteristic features and structure of similarity in 3′UTRs, we estimated the expected number of multiple matches for each 3′UTR alignment. Theoretical frequencies of multiple matches for each multiple alignment were calculated using a Monte Carlo simulation procedure with the same numbers of pairwise matches (see Materials and Methods). We estimated the over-representation of multiple matches for each set of UTRs based on the predicted average frequencies of multiple matches (Table 4). Bold numbers are significantly higher (<1%) than the average number of multiple matches in the simulated alignments. The differences between the real and predicted numbers of multiple matches have a significant negative correlation with the overall conservation of 3′UTRs (r = –0.55, P < 0.0001), a significant positive correlation with G and GC content in 3′UTRs (r = 0.49 and 0.38, P < 0.0001), and a significant positive correlation with KS values for the corresponding protein-coding regions (r = 0.42, P < 0.0001).
Table 4. Calculated pairwise and multiple matches, and theoretical average of multiple matches in simulated multiple alignments of 3′UTRs.
| Name | Pairwise matches | Multiple matches | Theoretical average | SD |
|---|---|---|---|---|
| 11-HSD2 | 2405 | 310 | 296.1 | 3.44 |
| 3b_HSD | 1353 | 130 | 106.9 | 3.82 |
| adrenomedullin | 2693 | 330 | 312.6 | 3.77 |
| AF298813 | 1040 | 117 | 109.1 | 2.76 |
| ATPaseNaK | 954 | 127 | 125.7 | 1.23 |
| AVPR2 | 702 | 50 | 29.5 | 3.36 |
| B3AR | 2110 | 205 | 174.8 | 5.14 |
| BIGLYCAN | 895 | 87 | 73.5 | 3.14 |
| CAT | 2694 | 317 | 304.8 | 4.04 |
| CCALAC | 1119 | 129 | 124.2 | 2.72 |
| CD34 | 4401 | 425 | 322.9 | 7.39 |
| CD9 | 1682 | 243 | 237.7 | 2.06 |
| collagentypeII | 2325 | 348 | 346.2 | 1.97 |
| DECORIN | 378 | 53 | 51.1 | 0.96 |
| G_CSF | 1847 | 213 | 182.8 | 4.07 |
| GAMMA | 1801 | 233 | 219.4 | 2.86 |
| glut_transp | 6039 | 650 | 583.8 | 7.25 |
| G-protein | 1471 | 211 | 205.6 | 2.11 |
| ICAM_1 | 4599 | 515 | 454.9 | 5.89 |
| IL-10 | 831 | 107 | 101.6 | 2.09 |
| IL2 | 1267 | 171 | 166.2 | 2.25 |
| IL6 | 1966 | 260 | 252.0 | 2.81 |
| K_channel | 1359 | 187 | 180.4 | 2.00 |
| mcp1 | 805 | 90 | 79.0 | 2.23 |
| MHC_DQB1 | 1002 | 71 | 48.2 | 3.81 |
| MMP13 | 4228 | 463 | 431.7 | 5.70 |
| MOTILIN | 652 | 81 | 78.2 | 1.95 |
| Na_Caexchanger | 1208 | 179 | 177.0 | 1.55 |
| NHE_1A | 2817 | 264 | 195.1 | 5.82 |
| NPC-1A | 1931 | 241 | 216.5 | 3.55 |
| OB1 | 8845 | 902 | 767.5 | 9.91 |
| P53A | 2866 | 262 | 218.7 | 6.01 |
| PAI_1 | 6193 | 655 | 538.4 | 7.84 |
| PDE6 | 2439 | 324 | 310.8 | 3.19 |
| PHOSDUCI | 1020 | 115 | 106.9 | 2.78 |
| PININ | 1572 | 228 | 226.2 | 1.58 |
| PP2A | 3695 | 565 | 562.3 | 2.12 |
| Prolactin_a | 609 | 74 | 70.6 | 1.84 |
| protocollagen | 1635 | 242 | 234.4 | 2.02 |
| Pselectin | 550 | 75 | 76.0 | 0.93 |
| PTH1 | 873 | 91 | 80.9 | 2.68 |
| S_albumin | 889 | 114 | 113.5 | 1.85 |
| SF-1 | 1887 | 232 | 213.3 | 3.39 |
| SP17 | 783 | 99 | 92.6 | 2.07 |
| STAR | 2204 | 209 | 185.1 | 4.87 |
| STS1 | 712 | 98 | 99.3 | 1.32 |
| Sum_tranth | 588 | 69 | 63.8 | 2.01 |
| THYRO | 1104 | 136 | 131.9 | 2.52 |
| TIMP | 466 | 60 | 61.4 | 1.37 |
| VCAM | 1247 | 144 | 140.1 | 2.71 |
DISCUSSION
There are two possible explanations for the presence of runs and footprints in 3′UTRs: they are either mutational cold spots or are under selective constraint due to conservative function. The second is more likely in view of the observed nucleotide frequency patterns and moderate non-random clumping of matches in UTRs with low and average overall conservation, as well as experimental studies that link runs and footprints with regulatory signals (8,9,17,18). Although interspecies conservation is substantially elevated within the first 250 nt after the stop codon, we did not find any universal regulatory signals or instances of position-specific consensus in this region. Most likely, these conservative sequences contain gene-specific information.
The order of nucleotide preference for 3′UTRs, A > U > C > G, is the same as for cleavage site nucleotides (19). We showed that runs in 3′UTRs are AU-rich and have a lower GC content than footprints. Since most of the runs are located within footprints, moderately conserved sequences in footprints have a higher GC content than UTRs on average (see Tables 2 and 3). These GC-rich areas within footprints could be structurally important.
Most mammalian genomes consist of discrete regions of distinct G and C content, so-called isochores (20). Local similarities of GC content in linked genes in rodents and humans supports the existence of such a pattern (21). The above-mentioned data agree with our results on positive correlation between GC contents in 3′UTRs and in the corresponding coding regions, and negative correlation between conservation of 3′UTRs and KS in the corresponding CDSs (Table 3). These data may indicate different selection in different parts of genomes. The existence or the absence of correlation between KS and GC4 is important for understanding the forces responsible for the evolution of isochores, which is one of the unresolved problems debated between neutralists and selectionists (22). The KS–GC4 correlation and correlation between GC base composition and conservation in 3′UTRs (Table 3) suggest the existence of a general constraint that is acting on both 3′UTRs and coding sequences. Partially, this constraint may be due to the necessity to maintain base composition of mRNA sequences. Because KS in the coding regions and conservation in 3′UTRs, GC4 and KS are correlated better than one could expect solely due to the compositional constraint in 3′UTRs, additional constraints may be acting on these sequences. Correlation between KS in coding regions and conservation in 3′UTRs and the similarity of KS and K values (1) may imply that not all synonymous sites in CDSs are neutral. Such an interpretation agrees with the results of the analysis of orthologous human–mouse–rat mRNAs done by Makalowski and Boguski (1) and with a recently proposed differential CpG content hypothesis (23). GC content in 3′UTRs cannot explain much of the variance in KS (r2 ∼ 16%), and there is neither a significant correlation between KA and G(GC) content nor between KA and overall UTR conservation. We cannot answer definitely the question whether it is possible to consider GC content and conservation of 3′UTRs as one of the real predictors of KS in mammalian genes. Additional methodological studies are needed to solve the problem of the proper determination of KS (22).
A positive correlation between frequencies of complementary nucleotides in 3′UTRs and a significant inverse correlation between the GC content of 3′UTRs and their lengths (3) probably indicates the importance of stable secondary structure formation of mRNA. Since the energy of interaction for G-C base pairs is approximately three times higher than for A-U pairs (24), G-C interaction may be more important for the secondary structure stability in 3′UTRs. There is evidence that moderately conserved GC-rich regions in 3′UTRs have many non-random invariant positions: the differences between real and predicted numbers of multiple matches negatively correlate with overall conservation of 3′UTRs and positively correlate with G and GC content. These results also provide evidence on the importance of moderately conserved regions in 3′UTRs.
The distribution of runs (Fig. 4) and conserved oligonucleotide motifs provide evidence on the existence of selective constraint specific for 3′UTRs. Although runs of matches are distributed exponentially, the parameter of this distribution is higher than the baseline probability. Lengths of conserved runs (4–7 nt) correspond to those of protein-binding sites, conserved elements of RNA secondary structure and functional elements that influence the UTR stability and regulate the formation of mRNA 3′ ends. Higher than expected frequencies of runs suggest a biological function (25,26), or can be explained by local sequence complexity and mutable contexts (fuzzy hot spots) (27,28).
Some runs of invariant AU-rich or pyrimidine-rich nucleotides in 3′UTRs have homology to known functional cis-elements of eukaryotic mRNAs, such as the U-rich upstream element, the poly(A) site (AAUAAA) and DICE regulatory signals. Most of the AU-rich runs and motifs are functionally important for RNA–protein interactions, and are possibly involved in the control of mRNA stability (5,8,29). Two AU-rich elements (AUUUA or UUAUUU) are the most frequent motifs in our set of UTRs. These oligonucleotides are present in one or several copies within A/U-rich elements (AREs), which are responsible for ARE-mediated mRNA decay. They also have been found in numerous short-lived mRNAs encoding regulatory proteins, such as growth factors and their receptors, inflammatory mediators and cytokines (30,31). In mammalian cells, one of the three important elements that specify transcript cleavage and polyadenylation, is the highly conserved hexanucleotide AAUAAA (18).
We also found some protein-binding CU-rich motifs, which may be involved in UTR regulation. The key element of the translation silencing is the presence of repetitive CU-rich motifs, called DICE (differentiation control elements) (32). mRNA–protein complex formed between DICE in the 3′UTR and hnRNPs K and E1 prevents 15-lipoxygenase translation. One possible explanation of the abundance of conserved pyrimidine-rich sites, and of the low G content of 3′UTRs is their complementary interaction with different RNAs, for example, with G-rich clinger fragments in 18S rRNAs (33,34), with G-rich 5′UTRs, with the coding regions of the same mRNA forming secondary structures, or with microRNAs. Nucleotide G,C-rich motifs that are less common in the 3′UTRs may interact with specific RNA-binding proteins, as well as perform a structural function. For example, the G,C-rich cis-regulatory element of the glucose transporter (GLUT1) mRNA has been shown to contribute to both rapid GLUT1 mRNA decay as well as to its stabilization in response to TNF treatment (35).
In contrast with DNA regulatory signals, the biological function of RNA regulatory signals depends on a combination of the primary and secondary structures, i.e. on both the nucleotide sequence and the surrounding stem–loop elements. Some secondary structure elements in the UTRs of mRNAs (selenocestein and iron-responsive stem–loops) are particularly conservative and play an important functional role (12,36).
Distinct 3′UTR cis-elements, such as the U-rich upstream element, the poly(A) site, may be too short to precisely define the 3′ end cleavage and formation. We suggest that moderately conserved elements with enhanced GC content probably work as structural or additional functional elements in the regulation of these processes. Our results provide evidence on the importance of moderately conserved regions in 3′UTRs and suggest that regulatory functions of 3′UTRs might utilize gene-specific information in these regions.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Igor Rogozin and Alexey Spiridonov for helpful discussions and comments. We also thank Nikolay Spiridonov for reading the manuscript and two anonymous referees for helpful comments on an earlier version.
REFERENCES
- 1.Makalowski W. and Boguski,M.S. (1998) Evolutionary parameters of the transcribed mammalian genome: an analysis of 2,820 orthologous rodent and human sequences. Proc. Natl Acad. Sci. USA, 95, 9407–9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shabalina S.A. and Kondrashov,A.S. (1999) Pattern of selective constraint in C. elegans and C. briggsae genomes. Genet. Res., 74, 23–30. [DOI] [PubMed] [Google Scholar]
- 3.Persole G., Mignone,F., Gissi,C., Grillo,G., Licciulli,F. and Liuni,S. (2001) Structural and functional features of eukaryotic mRNA untranslated regions. Gene, 276, 73–81. [DOI] [PubMed] [Google Scholar]
- 4.Larizza A., Makalowski,W., Pesole,G. and Saccone,C. (2002) Evolutionary dynamics of mammalian mRNA untranslated regions by comparative analysis of orthologous human, artiodactyls and rodent gene pairs. Comput. Chem., 26, 479–490. [DOI] [PubMed] [Google Scholar]
- 5.Spicher A., Guicherit,O.M., Duret,L., Aslanian,A., Sanjines,E.M., Denko,N.C., Giaccia,A.J. and Blau,H.M. (1998) Highly conserved RNA sequences that are sensors of environmental stress. Mol. Cell. Biol., 18, 7371–7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li W.-H. (1997) Molecular Evolution. Sinauer Associates, Inc., Sunderland, MA. [Google Scholar]
- 7.Koonin E. and Galperin,M. (2001) Sequence–Evolution–Function. Computational Approaches in Comparative Genomics. Kluwer Academic Publishers, Boston, MA. [PubMed] [Google Scholar]
- 8.Grzybowska E.A., Wilczynska,A. and Siedlecki,J.A. (2001) Regulatory functions of 3′UTRs. Biochem. Biophys. Res. Commun., 288, 291–295. [DOI] [PubMed] [Google Scholar]
- 9.Mignone F., Gissi,C., Liuni,S. and Pesole,G. (2002) Untranslated regions of mRNAs. Genome Biol., 3, REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duret L. and Bucher,P. (1997) Searching for regulatory elements in human noncoding sequences. Curr. Opin. Struct. Biol., 7, 399–406. [DOI] [PubMed] [Google Scholar]
- 11.Duret L., Dorkeld,F. and Gautier,C. (1993) Strong conservation of non-coding sequences during vertebrates evolution: potential involvement in post-transcriptional regulation of gene expression. Nucleic Acids Res., 21, 2315–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kryukov G.V. and Gladyshev,V.N. (2002) Mammalian selenoprotein gene signature: identification and functional analysis of selenoprotein genes using bioinformatics methods. Methods Enzymol., 347, 84–100. [DOI] [PubMed] [Google Scholar]
- 13.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlin S. and Altschul,S.F. (1993) Applications and statistics for multiple high-scoring segments in molecular sequences. Proc. Natl Acad. Sci. USA, 90, 5873–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z. (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci., 13, 555–556. [DOI] [PubMed] [Google Scholar]
- 16.Kondrashov A.S. and Shabalina,S.A. (2002) Classification of common conserved sequences in mammalian intergenic regions. Hum. Mol. Genet., 11, 669–674. [DOI] [PubMed] [Google Scholar]
- 17.Pesole G., Liuni,S., Grillo,G., Licciulli,F., Mignone,F., Gissi,C. and Saccone,C. (2002) UTRdb and UTRsite: specialized databases of sequences and functional elements of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Update 2002. Nucleic Acids Res., 30, 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao J., Hyman,L. and Moore,C. (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev., 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen F., MacDonald,C.C. and Wilusz,J. (1995) Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res., 23, 2614–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernardi G., Olofsson,B., Filipski,J., Zerial,M., Salinas,J., Cuny,G., Meunier-Rotival,M. and Rodier,F. (1985) The mosaic genome of warm-blooded vertebrates. Science, 228, 953–958. [DOI] [PubMed] [Google Scholar]
- 21.Williams E.J and Hurst,L.D. (2000) The proteins of linked genes evolve at similar rates. Nature, 407, 900–903. [DOI] [PubMed] [Google Scholar]
- 22.Hurst L.D. and Williams,E.J. (2000) Covariation of GC content and the silent site substitution rate in rodents: implications for methodology and for the evolution of isochores. Gene, 261, 107–114. [DOI] [PubMed] [Google Scholar]
- 23.Subramanian S. and Kumar,S. (2003) Neutral substitutions occur at a faster rate in exons than in noncoding DNA in primate genomes. Genome Res., 13, 838–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freier S.M. and Altmann,K.H. (1997) The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically modified DNA:RNA duplexes. Nucleic Acids Res., 25, 4429–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silva J.C. and Kondrashov,A.S. (2002) Patterns in spontaneous mutation revealed by human–baboon sequence comparison. Trends Genet., 18, 544–547. [DOI] [PubMed] [Google Scholar]
- 26.Webb C.T., Shabalina,S.A., Ogurtsov,A.Y. and Kondrashov,A.S. (2002) Analysis of similarity within 142 pairs of orthologous intergenic regions of Caenorhabditis elegans and Caenorhabditis briggsae. Nucleic Acids Res., 30, 1233–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogozin I.B. and Pavlov,Y.I. (2003) Theoretical analysis of mutation hotspots and their DNA sequence context specificity. Mutat. Res., 544, 65–85. [DOI] [PubMed] [Google Scholar]
- 28.Krawczak M., Ball,E.V. and Cooper,D.N. (1998) Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. Am. J. Hum. Genet., 63, 474–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zubiaga A.M., Belasco,J.G. and Greenberg,M.E. (1995) The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol. Cell. Biol., 15, 2219–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sirenko O., Bocker,U., Morris,J.S., Haskill,J.S. and Watson,J.M. (2002) IL-1 beta transcript stability in monocytes is linked to cytoskeletal reorganization and the availability of mRNA degradation factors. Immunol. Cell Biol., 80, 328–339. [DOI] [PubMed] [Google Scholar]
- 31.Laroia G., Cuesta,R., Brewer,G. and Schneider,R.J. (1999) Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science, 284, 499–502. [DOI] [PubMed] [Google Scholar]
- 32.Ostareck D.H., Ostareck-Lederer,A., Wilm,M., Thiele,B.J., Mann,M. and Hentze,M.W. (1997) mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell, 89, 597–606. [DOI] [PubMed] [Google Scholar]
- 33.Mauro V.P. and Edelman,G.M. (2002) The ribosome filter hypothesis. Proc. Natl Acad. Sci. USA, 99, 12031–12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matveeva O.V. and Shabalina,S.A. (1993) Intermolecular mRNA–rRNA hybridization and the distribution of potential interaction regions in murine 18S rRNA. Nucleic Acids Res., 21, 1007–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGowan K.M., Police,S., Winslow,J.B. and Pekala,P.H. (1997) Tumor necrosis factor-alpha regulation of glucose transporter (GLUT1) mRNA turnover. Contribution of the 3′-untranslated region of the GLUT1 message. J. Biol. Chem., 272, 1331–1337. [DOI] [PubMed] [Google Scholar]
- 36.Rouault T. and Klausner,R. (1997) Regulation of iron metabolism in eukaryotes. Curr. Top. Cell. Regul., 35, 1–19. [DOI] [PubMed] [Google Scholar]