Abstract
A self-assembly monolayer (SAM) of hairpin DNA can be formed on a gold substrate in order to make special biosensors. Labeling the hairpin loop probes with electroactive compositions rather than a fluorophore illustrates interesting profiles of redox current versus temperature. For a biosensor interacting with perfectly complementary targets, the profile shows a characteristic plateau, which disappears when the targets have a single base variation. The plateau is split into multiple steps by tuning the hybridization temperature. We propose that the phenomena are due to hairpin loop compartmentalization. The novel characteristics lead to a thermal gradient detection method that permits perfect discrimination of a target sequence from single nucleotide mismatches.
INTRODUCTION
Sequence-selective DNA detection is becoming increasingly important as scientists unravel the genetic basis of disease. Optical (1), electrochemical (2–9), microgravimetric (10) and quartz crystal microbalance (10,11) methods have been reported for detection of DNA hybridization events. One recent development is molecular beacons (12), which were reported for highly selective oligonucleotide recognition. Molecular beacons possess a hairpin loop structure. A fluorophore is attached to one end of its stem and a quencher is attached to the other end, which quenches the fluorescence of the fluorophore when the two arms hybridize with each other. The hybridization can be visualized through the detection of fluorescence on stem opening. Studies on the unique thermodynamics and specificity of molecular beacons (13–15) have demonstrated excellent sensitivity and selectivity in the detection of a single mismatch in a nucleic acid sequence; they also have the advantage of easy direct detection of unlabeled oligonucleotides. In another biotechnology line of research, the development of bioelectronic DNA analysis has attracted substantial effort in the area of DNA biosensors (5–8), which have applications in gene analysis, detection of genetic disorders, tissue matching, forensic applications and DNA computers (16–18). Although the study of molecular beacons has yielded new technologies based on optical detection methods in areas such as genomics (19), DNA chips (20), quantitative PCR (21–23) and RNA detection (24,25), harnessing the characteristics of molecular beacons in the development of an electronic transduction detection method is still at an early stage.
Here we report our studies of temperature-dependent electronic transduction based on a new type of biosensor. The core of the biosensor is a hairpin loop-modified gold electrode. The hairpin oligonucleotide has the same structure as a molecular beacon, as shown in Figure 1A. The loop consists of a probe sequence that is complementary to a target sequence; the stem sequence is unrelated to the target sequence. An electroactive moiety is attached to the 3′-end of the oligonucleotide and the 5′-end is covalently bound to the surface of a gold electrode through an alkanethiol group. The electroactive group offers free electrons that are able to form a microcurrent pulse along the electrode under a suitable applied electric potential. Because of the relative rigidity of double helices, the formation of a probe–target hybrid inhibits simultaneous existence of the hairpin structure. Thus a difference in electron transfer pattern exists between the zipped and unzipped states of the hairpin loop. In this paper we study the different electron transfer patterns by plotting thermal characteristics of the biosensor system.
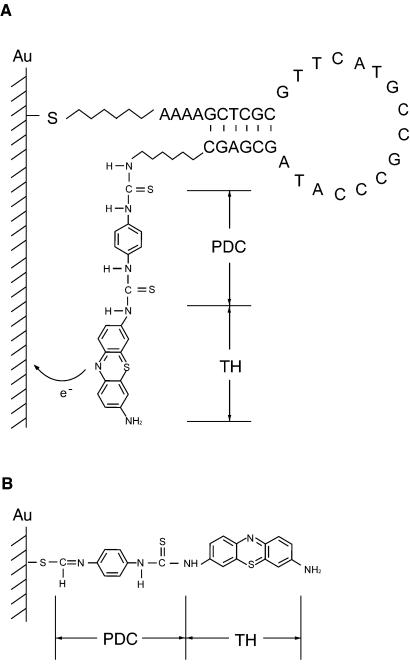
Figure 1.
(A) The scheme shows the structure of the hairpin oligonucleotide immobilized on the gold surface. The redox-active moiety TH was modified on the 3′-end by PDC. The 5′-end was prolonged using four adenosines. (B) Self-assembly of PDC-TH on the gold surface.
MATERIALS AND METHODS
Design and synthesis of the hairpin DNA
Three synthetic oligonucleotide hairpin loops (Sangon Bioengineering Inc., China), modified with a primary amino group at its 3′-end and an alkanethiol group at its 5′-end, were used for all experiments reported here. We selected segments of 16 nt as target sequences extracted from the human gene for the p53 tumor suppressor (26). A typical oligonucleotide hairpin loop is shown in Figure 1A. The loop portion of the hairpin oligonucleotides is perfectly complementary to the target sequences. The design of the arm sequences is optimized in order to maximize the separation between the two arms when probes are hybridized to targets. Thionine (TH) was modified at the 3′-end through a 1,4-phenylene diisothiocyanate (PDC) linker. In addition, PDC is also able to self-assemble on the gold surface via the thiol moiety (Fig. 1B). Therefore, the molecules covalently immobilized on the electrode surface consist of both hairpin DNA and PDC-TH products. The sequences of the hairpin oligonucleotide used in all tests were: 5′-AAAAGCTCGCGTTCATG CCGCCCATAGCGAGC-3′; 5′-AAAACGACGCAGGTCT TGGCCAGTCGTCG-3′; 5′-AAAAGCACGCTGAGGAGG GGCCAGACGTGC-3′. The underlined bases are the arm sequences. A thiol group (-SH) is covalently linked to the 5′ phosphate via a (CH2)6 spacer; an amino group (-NH2) is linked to the 3′ hydroxyl via a (CH2)6 spacer.
Self-assembly of hairpin oligonucleotide-modified gold electrodes
Each prepared gold electrode was immersed in 50 µl of a 50 nM solution of hairpin oligonucleotides in a 0.5 ml tube for 24 h at 4°C. Then it was washed with a pure solution of dimethyl formamide (DMF) and put into 0.5 ml tubes containing 50 µl of 0.2% PDC solution in 10% pyridine/DMF, which was deoxygenated with pure N2. At this step, the PDC molecules were attached to the gold surface. After ∼4–5 h, the electrode was taken out, rinsed with ethanol and double distilled water and placed in 0.5 ml tubes containing 50 µl Tris buffer (20 mM Tris–HCl, pH 8, containing 1 mM MgCl2 and 10 µM thionine) for at least 4–5 h at room temperature. The probe electrode prepared in this way could be stored in Tris buffer at 4°C for a long time.
To self-assemble PDC-modified electrodes, which acted as controls of our experiments, each prepared gold electrode was exposed to 50 µl of 0.2% PDC solution in 10% pyridine/DMF for 12 h and deoxygenated with pure N2.
Hybridization
The prepared biosensors were incubated in a 50 µl solution containing 50 nM target oligonucleotides, 1 mM MgCl2, 10 µM thionine and 20 mM Tris–HCl (pH 8). The sequences of perfect complements for three kinds of probes were: (a) 5′-ATGGGCGGCATGAAC-3′; (b) 5′-ACTGGCCAAGAC CTG-3′; (c) 5′-TCTGGCCACTCCTCAG-3′.
The experiments were repeated with imperfect targets, which included a single nucleotide mismatch, and an unrelated oligonucleotide. The mismatches for different probes were, respectively: (a) 5′-ATGGGCGTCATGAAC-3′, 5′-ATGGG CGACATGAAC-3′ and 5′-ATGGGCGCCATGAAC-3′; (b) 5′-ACTGGCCGAGACCTG-3′; (c) 5′-TCTGGCCCCTCC TCAG-3′. The unrelated sequence was 5′-CGCAATAA TCGACCT-3′. The identity of the mismatches is indicated by underlining. The position of the mismatches is a natural frequent mutation site in the human p53 gene.
Temperature-dependent voltammetry detection
Electrochemical detection was conducted in a three-electrode system using a model 660A electrochemical workstation (CH Instruments Inc., USA), with an Ag/AgCl reference electrode, a platinum counter electrode and a modified biosensor as the working electrode. The electrolyte was 20 mM Tris–HCl (pH 8) containing 1 mM MgCl2. In cyclic voltammetry, the potential versus Ag/AgCl was swept from –0.4 to 0.0 V, with a scan rate of 0.1 V/s. The gold electrode was picked out from a target solution and detected by linear sweep voltammetry or single-step cyclic voltammetry. The voltammetric detection was repeated as the temperature decreased from 60 to 5°C at a rate of 5°C/25 min. After each voltammetry cycle, the gold electrodes were returned to the target solution for recovery of electroactivity and stability of performance.
RESULTS AND DISCUSSION
Electrocatalysis on hairpin loop-modified electrodes
The TH molecule covalently tethered to the 3′-end of a hairpin loop has been widely used as an electron transfer mediator (27–30). At neutral pH, the redox reaction of the molecule is known to be (30):
TH+ + 2e– + H+ ↔ leucothionine (LTH) 1
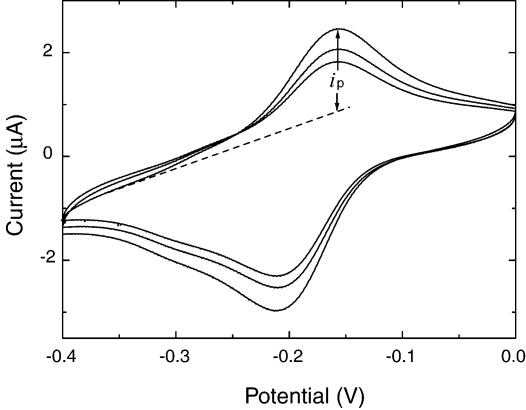
A typical successive cyclic voltammogram on a well-modified electrode is shown in Figure 2. Our interest focused on the peak current ip in the voltammogram, defined as the difference between the maximum current and baseline current (dashed line in Fig. 2). The reduction peak current is located at about –1.6 V, identical to the reduction potential of TH bound to a bare gold electrode. Previous works have shown that there is no shift in redox potential for an electroactive intercalator on a DNA-modified electrode (31,32); our results are in good agreement with these reports. The first three cycles without a change in redox potential show that the TH groups modified on the electrode surface are steady and stable within the present potential limits. In the case of unhybridized biosensor, hybridized biosensor and PDC-TH-modified electrode, the voltammograms show no obvious differences in the redox potential.
Figure 2.
Cyclic voltammogram for hairpin DNA biosensor versus Ag/AgCl. The initial voltage was –0.4 V and the end voltage was 0, the scan rate was 0.1 V/s, with a sample interval of 0.001 V and sensitivity of 10–6 A/V. The value of ip was defined as the difference between the reduction peak current and the baseline current (dashed line).
Distance-dependent electron transfer through a hairpin loop sensor
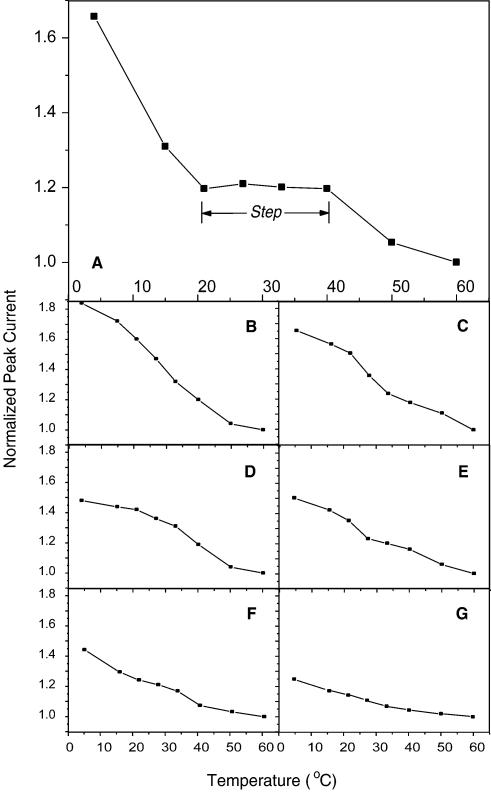
To observe the effect of distance-dependent electron transfer during the process of hairpin loop unzipping, we first measured ip as a function of temperature when oligonucleotides were not hybridized to any target and the electrodes were just modified with PDC-TH. The ip–T curves of these two cases are presented in Figure 4F and G, respectively. The measurements on PDC-TH-modified electrodes give the baseline of zero distance electron transfer in the system. Figure 4G indicates that the PDC self-assembly monolayer (SAM) releases TH with increasing temperature and restores TH with decreasing temperature.
Figure 4.
ip–T profiles of the hairpin loop biosensors detecting different targets. The experimental data were all normalized by means of setting the value of ip at 60°C to 1. (A) Profile of the biosensor after hybridization with perfectly complementary targets. A step is apparent in the profile. (B–D) Profiles of the biosensors for the strands with a single mutation. The mutations are: (B) G→T; (C), G→A; (D) G→C. (E) Profile of probe electrode interacting with unrelated oligonucleotides. (F) ip–T curve of the biosensor without detection of any targets. (G) ip–T curve of PDC-TH-modified electrode.
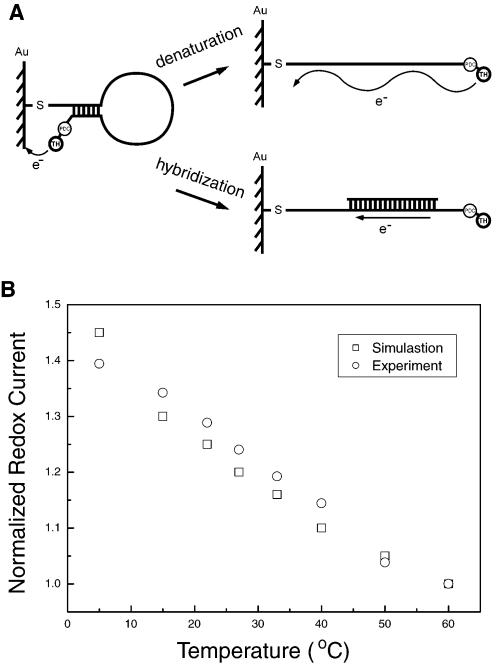
The biosensor surface covered by SAMs is composed of two elements that possess different structures and properties. One is a SAM formed by a DNA hairpin loop and the other is a SAM formed by PDC-TH directly immobilized on a gold surface. The thickness of the latter is much less than that of the former. The reduction current results from electron tunneling through the monolayer of oligonucleotides (31,32) (Fig. 3A). The redox current at any potential should decrease exponentially with the barrier thickness according to the expression (27–30):
Figure 3.
(A) Electron transfer derived from the redox activity of TH. Electron transfer produces a tunneling current when the probes are not hybridized to targets. When the hairpin is opened by heating, the tunneling current will be greatly weakened. However, when probes are coupled with targets, the electrons transfer along the double-stranded DNA as along a wire. (B) Comparison of experiments (circle) and simulation (square) for the redox current on the electrode in the process of hairpin loop unzipping.
i(T) = i0(T)e–βd(T) 2
where T is temperature, i0(T) is the current measured for a bare electrode, β is the potential-independent electron tunneling coefficient and d(T) is the thickness between the redox center and the bare surface of the electrode. Assuming that f(T) describes the percentage of unzipped hairpin loop, the reduction current provided by the redox reaction of TH at the 3′-end of hairpin loops should be:
i(T) = i0(T)[1 – f(T)] + i0(T)f(T) e–βdm 3
where dm represents the distance between TH on the 3′-end of the hairpin loop and the bare electrode surface when the hairpin loop is unzipped. Equation 3 describes the difference between Figure 4F and G, i.e. the redox current in the process of hairpin loop unzipping. The simulation results shown in Figure 3B were basically consistent with the experimental data. This analysis indicates that in the case of the unhybridized biosensor, the distance dependence of electron transfer increases the slope of the ip–T curve relative to that of the PDC-TH-modified electrode (compare Fig. 4F and G).
Influence of hybridization on ip–T curves
After the oligonucleotide hairpin loop hybridized to its perfect complement at 37°C, we profiled the ip–T relationship. The result is shown in Figure 4. One notices that a step exists from ∼20 to ∼40°C in the ip–T curves. The step is located below the melting temperature of the probe–target hybrids. The step from ∼20 to ∼40°C is reproducible and robust in repeated experiments and in the case of changing the sequence setting (with length fixed). The variation between experiments causes small changes in the current value of the step, however, the characteristic of the plateau persists in different situations. The step disappears when the target sequence has a single base mismatch, as shown in Figure 4B–D. These profiles look much like that measured from biosensors interacting with unrelated strands (Fig. 4E). This characteristic contrast between wild-type and mutants was clear cut and invariant with the variations of probe sequence, which thereby raised the question of the mechanism beneath the phenomenon.
It has been reported that the coverage of an oligonucleotide attached to a surface is related to its attachment concentration and reaction duration (9,33). We could therefore change the surface coverage by the hairpin loop by tuning its attachment concentration or duration to test the performance of ip–T curves. We found that the step in the ip–T profiles existed only when the attachment concentration was within the range 50–200 nM. With the same surface density of probe, we decreased the concentration of perfect target to test the performance of the biosensor. The step totally vanished when the ratio of target to probe concentration was decreased to 1/10. These results indicate that a certain range of coverage of double-stranded DNA (dsDNA) on the electrode surface is a necessary condition for appearance of the step. To clearly determine the coverage of dsDNA, we recently studied the topology of a dsDNA SAM on a gold surface by atomic force microscopy (AFM). Our study showed that the saturation coverage of the dsDNA SAM under our experimental conditions was about 5.4 ± 0.8 × 1012 molecules/cm2. This observation implies that when the attachment concentration was >50 nM, self-assembly achieves saturation coverage. The AFM study also showed that when the attachment concentration is >0.5 µM, multilayer structures form on the SAM, which can cause problems in hybridization. A detailed analysis of the AFM study will appear elsewhere.
Mechanism of the observed ip–T curves
To understand the observed phenomena, we propose a scheme that explains the step effect observed in our experiments. Our previous work demonstrated that moderately packed DNA molecules on a surface could form a reversibly switchable nanocompartment. In this study, the DNA density on the gold surface meets the needs for nanocompartment formation, which was proved by our AFM observations. Our AFM studies and many other works (8,31,34) all showed that oligonucleotides immobilized on a gold surface basically orient vertical to the surface when they are packed close to the saturation coverage. This may be due to the strong electrostatic repulsion forces between the negatively charged sugar–phosphate backbones of DNA. When the hairpin loops are completely opened through hybridization with perfectly complementary targets, the double helices would pack densely into a membrane. Therefore, a compartment with nanoscale height is formed between the DNA membrane and bare gold surface. The dense DNA membrane blocks TH molecules from diffusing out of the compartment. The amount of TH in the compartment is thus retained at the level after the nanocompartment was closed. Detailed calculations show that the ip–T relationship in the case of perfect hybridization is given by the following expressions:
where σ is the number of charges per single-stranded probe oligonucleotide, fdsDNA(T) is the percentage of dsDNA on the electrode, σinner is the area density of PDC-TH, Cth is the concentration of TH in solution, d0 is the effective height of the DNA compartment and A is the area of electrode surface. The first term, σe–βd(T), of equation 4 represents the contribution of TH attached at the 3′-end of hairpin loops; the second term, σinner(T), represents the contribution of PDC-TH directly immobilized on the gold surface; the third term, Cthd0, represents the contribution of the free TH encapsulated in the DNA compartment. For a detailed deduction of the results, please see Supplementary Material. Because of a lack of information on the necessary parameters, we can only qualitatively compare equation 4 with our experimental data. We notice that the equation describes the main characteristics of experimental ip–T profiles: ip is nearly a constant when T1 < T < T2; it increases exponentially with decreasing temperature under T < T1. The analysis gives an explanation for the appearance of the step in the ip–T profiles for perfect targets.
Tuning steps in ip–T profiles
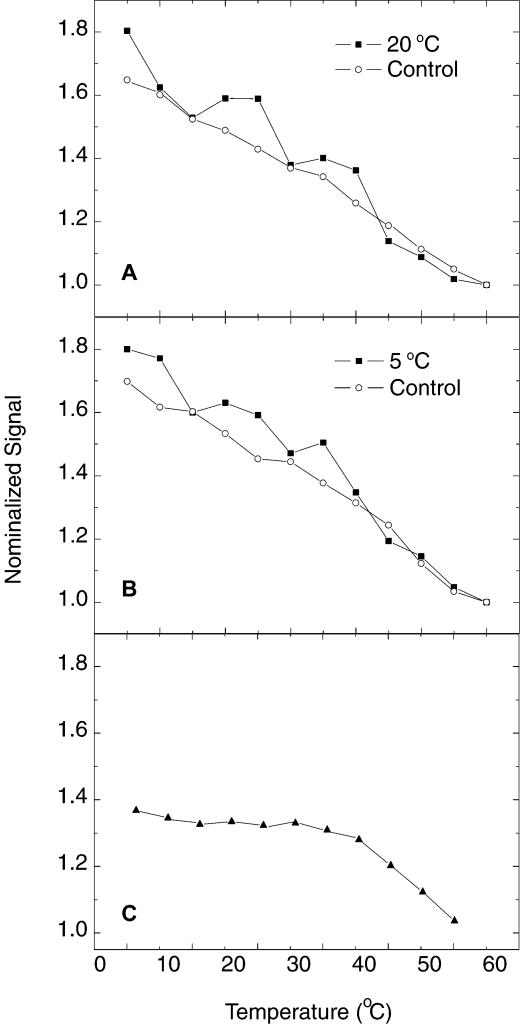
The aforementioned hybridization experiments were performed at 37°C. When the hybridization temperature was tuned to 20 or 5°C, the original single step in ip–T profiles was split into multiple steps, as shown in Figure 5. The ip–T curve for the perfect target at 20°C (Fig. 5A) gives two obvious steps (solid squares), compared to its control experiments (open circles). When the hybridization temperature was decreased to 5°C, the number of steps (see Fig. 5B) seemed to increase to three. Numerous repeated experiments showed that the multistep effect was robust and reproducible. However, the number of steps showed a small variation under the same experimental configuration. The control experiments (open circle in Fig. 5A and B) were performed on electrodes hybridized with single base mismatches. They are comparable to their counterparts at 37°C hybridization temperature (see Fig. 4B–D), demonstrating continuous diffusion of TH from the surface with increasing temperature. The observed multistep effect can also be explained by the mechanism of hairpin loop compartmentalization. A possible scenario explaining the multiple steps is proposed as follows. The increase in temperature causes the PDC-TH SAM to release TH so that its concentration in the nanocompartment increases. When the osmotic pressure difference for TH surpasses a certain threshold, the dsDNA membrane can no longer hold TH and allows TH to flow between the nanocompartment and the external solution, resulting in the transition from one step to the next in the ip–T curves. Similar phenomena have been investigated in an analogous system (data to be published).
Figure 5.
ip–T curves for (A) 20°C and (B) 5°C hybridization temperature. The data represented by solid squares correspond to the hybridization with perfectly complementary targets. The control experiments in (A) and (B) (open circles) correspond to hybridization with single mismatches. (C) ip–T curves for deletion of the modification of PDC at the 3′-end of the hairpin loop. The data were obtained from 37°C hybridizations with perfectly complementary targets.
To test the model proposed above, we deleted the modification of PDC at the 3′-end of the hairpin loop. The data obtained in the case of a perfect target, shown in Figure 5C, demonstrate that the current signal in the temperature range from 0 to 20°C is greatly diminished compared to the signal obtained from the original biosensors (Fig. 4A). The comparison indicates that the TH covalently bound to the 3′-end of the hairpin loop corresponds to the item σe–βd(T) in equation 4, which does not exist in the case of deletion of PDC at the 3′-end of the hairpin loop. The results partially confirm equation 4 and the mechanism proposed above.
Thermal gradient detection assay
Our experimental studies imply that voltammetry at two temperatures in the range of one step is enough to discriminate targets from single base mismatches. We tested this idea by performing voltammetry detection at room temperature (20°C) and 40°C. The hybridization events can be signaled by:
Δip/ip = [ip(20°C) – ip(40°C)]/ip(40°C). 5
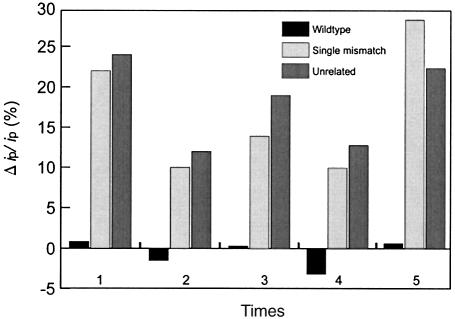
Typical results are shown in Figure 6. The relative difference Δip/ip for strands with mismatches is more than 10% for all cases. In the case of perfect hybridization it fluctuates around 0. The thermal gradient detection method is thereby an easy assay to obtain information on DNA sequence. An approximately 0 Δip/ip means successful sequence recognition and an Δip/ip value of >10% means failed recognition. The selection criterion is so clear cut that we believe the selectivity is nearly perfect. The biosensors could be reused in this approach for at least five successive experiments over several days, if they were washed with a high concentration of urea (30% in H2O) after each hybridization experiment and stored in a solution of 20 mM Tris–HCl (pH 8) at 4°C.
Figure 6.
Δip/ip comparison with thermal gradient detection. Δip/ip from hybridization between probe and wild-type target fluctuated slightly around 0 in the range of measuring error. However, the values in the case of one mismatch oligonucleotides or unrelated oligonucleotides are all >10%.
Conclusion
Our experimental studies show that the biosensors can exist in two functional states at given temperatures, depending on whether the oligonucleotide probes on the gold surface are hybridized to their targets. Hybridization causes a spontaneous conformational change of the hairpin loop SAM, which in turn prevents the change in redox capability of the system. The most significant elements that affect the performance of the biosensors lie in two aspects. One is the binding energy between the two arms; the other is the area density of the dsDNA on the electrode surface. The former decides whether the arms of the oligonucleotides could be opened by perfect targets; the latter guarantees the necessary structure of SAMs on the electrode surface. In comparison with our general studies of DNA nanocompartments on a surface, we conclude that the characteristics of the biosensor studied in this work are due to the DNA nanocompartment structure, which shows potential applications in molecular sensing, recognition and mutation detection.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Wei Deng and Jun Feng for insightful discussions. This work was supported by grants from the Research Fund of Peking University.
REFERENCES
- 1.Jordan C.E., Smith,L.M. and Corn,R.M. (1997) Surface plasmon resonance imaging measurements of DNA hybridization adsorption and streptavidin/DNA multilayer formation at chemically modified gold surfaces. Anal. Chem., 69, 4939–4947. [DOI] [PubMed] [Google Scholar]
- 2.Hashimoto K., Ito,K. and Ishimori,Y. (1994) Sequence-specific gene detection with a gold electrode modified with DNA probes and an electrochemically active dye. Anal. Chem., 66, 3830–3833. [DOI] [PubMed] [Google Scholar]
- 3.Kelley S.O., Boon,E.M., Barton,J.K., Jackson,N.M. and Hill,M.G. (1999) Single-base mismatch detection based on charge transduction through DNA. Nucleic Acids Res., 27, 4830–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardea A., Dagan,A. and Willner,I. (1999) Amplified electronic transduction of oligonucleotide interactions: novel routes for Tay-Sachs biosensors. Anal. Chim. Acta, 385, 33–43. [Google Scholar]
- 5.Wang J., Palecek,E., Nielsen,P.E., Rivas,G., Cai,X., Shiraishi,H., Dontha,N., Luo,D. and Farias,P.A.M. (1996) Peptide nucleic acid probes for sequence-specific DNA biosensors. J. Am. Chem. Soc., 118, 7667–7670. [Google Scholar]
- 6.Hashimoto K., Ito,K. and Ishimori,Y. (1994) Novel DNA sensor for electrochemical gene detection. Anal. Chim. Acta, 286, 219–224. [Google Scholar]
- 7.Patolsky F., Lichtenstein,A. and Willner,I. (2001) Detection of single-base DNA mutations by enzyme-amplified electronic transduction. Nat. Biotechnol., 19, 253–257. [DOI] [PubMed] [Google Scholar]
- 8.Boon E.M., Ceres,D.M., Drummond,T.G., Hill,M.G. and Barton,J.K. (2000) Mutation detection by electrocatalysis at DNA-modified electrodes. Nat. Biotechnol., 18, 1096–1100. [DOI] [PubMed] [Google Scholar]
- 9.Steel A.B., Herne,T.M. and Tarlov,M.J. (1998) Electrochemical quantitation of DNA immobilized on gold. Anal. Chem., 70, 4670–4677. [DOI] [PubMed] [Google Scholar]
- 10.Bardea A., Dagan,A., Ben-Dov,I., Amit,B. and Willner,I. (1998) Amplified microgravimetric quartz crystal microbalance analysis of oligonucleotide complexes: a route to Tay-Sachs biosensor device. Chem. Commun., 839–840. [Google Scholar]
- 11.Okahata Y., Kawase,M., Niikura,K., Ohtake,F., Furusawa,H. and Ebara,Y. (1998) Kinetic measurements of DNA hybridization on an oligonucleotide-immobilized 27-MHz quartz crystal microbalance. Anal. Chem., 70, 1288–1296. [DOI] [PubMed] [Google Scholar]
- 12.Tyagi S. and Kramer,F.R. (1996) Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol., 14, 303–308. [DOI] [PubMed] [Google Scholar]
- 13.Bonnet G. and Libchaber,A. (1999) Optimal sensitivity in molecular recognition. Physica, A263, 68–77. [Google Scholar]
- 14.Bonnet G., Krichevsky,O. and Libchaber,A. (1998) Kinetics of conformational fluctuations in DNA hairpin-loops. Proc. Natl Acad. Sci. USA, 95, 8602–8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonnet G., Tyagi,S., Libchaber,A. and Kramer,F.R. (1999) Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl Acad. Sci. USA, 96, 6171–6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adleman L.M. (1994) Molecular computation of solutions to combinatorial problems. Science, 266, 1021–1024. [DOI] [PubMed] [Google Scholar]
- 17.Liu Q.H., Frutos,A.G., Thiel,A.J., Corn,R.M. and Smith,L.M. (1998) DNA computing on surfaces: encoding information at the single base level. J. Comput. Biol., 5, 269–278. [DOI] [PubMed] [Google Scholar]
- 18.Frutos A.G., Smith,L.M. and Corn,R.M. (1998) Enzymatic ligation reactions of DNA ‘words’ on surfaces for DNA computing. J. Am. Chem. Soc., 120, 10277–10282. [Google Scholar]
- 19.Marras S.A.E., Kramer,F.R. and Tyagi,S. (1999) Multiplex detection of single-nucleotide variation using molecular beacons. Genet. Anal. Biomol. E, 14, 151–156. [DOI] [PubMed] [Google Scholar]
- 20.Steemers F.J., Ferguson,J.A. and Walt,D.R. (2000) Screening unlabled DNA targets with randomly ordered fiber-optic gene array. Nat. Biotechnol., 18, 91–94. [DOI] [PubMed] [Google Scholar]
- 21.Tyagi S., Bratu,D.P. and Kramer,F.R. (1997) Multicolor molecular beacons for allele discrimination. Nat. Biotechnol., 16, 49–53. [DOI] [PubMed] [Google Scholar]
- 22.Vet J.A., Majithia,A.R., Marras,S.A., Tyagi,S., Dube,S., Poiesz,B.J. and Kramer,F.R. (1999) Multiples detection of four pathogenic retroviruses using molecular beacons. Proc. Natl Acad. Sci. USA, 96, 6394–6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitcombe D., Theaker,J., Guy,S.P., Brown,T. and Little,S. (1999) Detection of PCR products using self-probing amplicons and fluorescence. Nat. Biotechnol., 17, 804–807. [DOI] [PubMed] [Google Scholar]
- 24.Sokol D.L., Zhang,X., Lu,P. and Gewirtz,A.M. (1998) Real-time detection of DNA·RNA hybridization in living cells. Proc. Natl Acad. Sci. USA, 95, 11538–11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leone G., Schijndei,H., Gemen,B., Kramer,F.R. and Schoen,C.D. (1998) Molecular beacon probes combined with amplification by NASBA enable homogeneous, real-time detection of RNA. Nucleic Acids Res., 26, 2150–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yunje C., Sveltlana,G., Philip,D.J. and Nikola,P.P. (1994) Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorgenic mutations. Science, 265, 346–355. [DOI] [PubMed] [Google Scholar]
- 27.Ruan C., Yang,F., Lei,C. and Deng,J. (1998) Thionine covalently tethered to multilayer horseradish peroxidase in a self-assembled monolayer as an electron-transfer mediator. Anal. Chem., 70, 1721–1725. [DOI] [PubMed] [Google Scholar]
- 28.Jean C., Vesna S. and Vera,Z. (1995) Thionine self-assembly on polyoriented gold and sulphur-modified gold electrodes. J. Electroanal. Chem., 386, 157–163. [Google Scholar]
- 29.Masayuki O., Susumu,K. and Hiroshi,Y. (1997) Electrochemical oxidation of reduced nicotinamide coenzymes at Au electrodes modified with phenothiazine derivative monolayers. J. Electroanal. Chem., 422, 45–54. [Google Scholar]
- 30.Chen H.Y., Zhou,D.M., Xu,J.J. and Fang,H.Q. (1997) Electrocatalytic oxidation of NADH at a gold electrode modified by thionine covalently bound to self-assembled cysteamine monolayers. J. Electroanal. Chem., 422, 21–25. [Google Scholar]
- 31.Kelley S.O. and Barton,J.K. (1997) Electrochemistry of methylene blue bound to a DNA-modified electrode. Bioconjugate Chem., 8, 31–37. [DOI] [PubMed] [Google Scholar]
- 32.Hartwich G.H., Caruana,D.J., de Lumley-Woodyear,T., Wu,Y., Campbell,C.N. and Heller,A. (1999) Electrochemical study of electron transport through thin DNA films. J. Am. Chem. Soc., 121, 10803–10812. [Google Scholar]
- 33.Herne T.M. and Tarlov,M.J. (1997) Characterization of DNA probes immobilized on gold surfaces. J. Am. Chem. Soc., 119, 8916–8920. [Google Scholar]
- 34.Kelley S.O., Barton,J.K., Jackson,N.M., McPherson,L.D., Potter,A.B., Spain,E.M., Allen,M.J. and Hill,M.G. (1998) Orienting DNA helices on gold using applied electric fields. Langmuir, 14, 6781–6784. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.