Abstract
A novel experimental system to study mutation in starving bacteria was designed, relying on the activation of a promoterless phenol degradation operon of Pseudomonas putida. The Phe+ (phenol-utilizing) mutants accumulated in the starving culture of P. putida in the presence of phenol but not in the absence of it. We ruled out the possibility that the absence of phenol eliminates Phe+ mutants from the starving population. Sequence analysis of the Phe+ mutants revealed that base substitutions, deletions, and insertion of Tn4652 can result in creation of a sequence similar to the σ70-specific promoter consensus. One particular C → A transversion was predominant in the Phe+ mutants that arose in the starving population under selection for phenol use. In contrast, various deletions were the most frequent Phe+ mutants occurring in a culture growing without selection. The accumulation rate of the Phe+ mutants on selective plates was found to be higher for bacteria plated from stationary-phase culture than that from exponentially growing cells. This suggests that some specific processes, occurring predominantly in stationary-phase cells, facilitate generation and/or fixation of such mutations.
Mutational processes have been studied mostly in actively growing bacteria. Only in recent years have studies been focused on mutational mechanisms in bacteria that have been starved. In 1988, Cairns et al. (1) presented evidence that Lac+ revertants appeared in cultures of Lac− Escherichia coli starving on lactose-minimal plates, but not on plates lacking a carbon source. After this paper by Cairns et al., several similar cases were reported. Under starvation, bacterial cell division was prevented, but mutants escaping starvation continued to appear under selective conditions (2–8). This phenomenon has been called “directed” or “adaptive” mutation and has stimulated a lively scientific debate (9–17).
So far, mutation in starving bacteria has been studied mostly in E. coli, and the most thoroughly investigated model system for the directed mutations has been the reversion of a frameshift mutation in an episomal lac allele (5, 6, 18–24). Herein we describe a novel system for studying mutations in starving bacteria. This system reveals mutations that create promoters, permitting starving Pseudomonas putida cells to use phenol as a sole source of carbon. Besides characterizing the mutations arising under starvation, we have also studied effects of the physiological status of cells (growth phase) on the rate of occurrence of directed mutations.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, and Media.
E. coli strain HB101 (25) and P. putida strain PaW85 (26) were used as host strains. Complete medium was Luria–Bertani (LB; ref. 27) and minimal medium was M9 (28). Phenol-minimal plates with 1.5% Difco agar contained 2.5 mM phenol as the sole carbon and energy source. Antibiotics were added at the indicated final concentrations: ampicillin, 100 μg/ml for E. coli, and carbenicillin, 1500 μg/ml for P. putida. P. putida was grown at 30°C and E. coli was incubated at 37°C. Plasmid pEST1332 contains the phenol degradation genes pheBA without a promoter cloned into the RSF1010-derived vector plasmid pAYC32 (29, 30). Plasmid pEST1414 is a pEST1332 derivative that contains the 0.2-kb SacI–ClaI DNA fragment from the left hand of Tn4652 cloned upstream of the promoterless pheBA genes. P. putida strain PaW85 was transformed with plasmid pEST1414 by selecting transformants resistant to carbenicillin. Transformation of bacterial cells with plasmid DNA was done as described by Hanahan (31).
DNA Sequence Analysis and mRNA Mapping.
The nucleotide sequences were determined by the dideoxy chain-termination method (32) with T7 DNA polymerase (Promega). Oligonucleotides 5′-GTATGCTTGGCAGTCGT-3′ and 5′-TTTTAACAGTCATAATTACTCTCTC-3′, complementary to nucleotide sequences −120 to −136 and +13 to −12, relative to the start codon of pheB, respectively, were used to analyze the 250-bp region upstream of the pheB gene. The reverse transcriptase mapping of 5′ ends of mRNA was performed as described previously (33) by using 10 units of avian reverse transcriptase (Promega). Total RNA (20 μg), purified from P. putida PaW85 and E. coli HB101 cells according to Blomberg et al. (34), was used as the template.
Isolation of Independent Phe+ Mutants from the Growing Culture.
Independent cultures were generated by growing cells to saturation in LB medium, diluting this culture by 106 into fresh medium, dispensing 1-ml aliquots into 40 test tubes, and allowing the cells again to reach saturation. The cells were harvested by centrifugation, washed with M9 solution, and plated onto phenol-minimal plates. Independent Phe+ colonies were picked up from separate plates and were used for DNA sequence analysis of the plasmids constitutively expressing the pheBA genes.
Analysis of Insertions of Tn4652 into pEST1414.
Two oligonucleotides, 5′-CTCCTTGGCCGGTAGCTGACTGGGC-3′, complementary to the right end of Tn4652 at 186 to 211 nt from the right terminus of the transposon, and 5′-TGCGCTCAGCCCATC-3′, complementary to the pheA coding sequence at nucleotides +58 to +72, were used to amplify the Tn4652 insertion regions in Phe+ cells containing hybrid plasmids. During accumulation of Phe+ mutants on selective plates, ≈40 arising Phe+ colonies were tested for insertion of this transposon each day. We have previously shown (33) that fusion promoters are preferentially created by the right-end sequence of the transposon (18 of 19 insertions investigated). Thus, the detection scheme used here was designed to reveal promoters generated by fusion of the right end of the element with the upstream sequences of the pheA gene.
RESULTS
Isolation of Phe+ Mutants.
The phenol degradation genes pheA and pheB from Pseudomonas sp. strain EST1001 have been isolated and sequenced by us previously and are shown to be organized in a single operon (29, 30). These genes encode phenol monooxygenase and catechol 1,2-dioxygenase, respectively. Introduction of the pheBA- or pheA-expressing RSF1010-derived plasmid into the phenol nondegrading P. putida strain PaW85 conferred the ability to use phenol as a sole carbon source (29, 30).
Strain PaW85 carrying the promoterless pheBA genes on plasmid pEST1414 was grown in a liquid culture on M9 minimal medium (28), containing glucose as carbon source. Samples were taken from the culture, pelleted, and washed with M9 solution. Approximately 108 washed cells were spread onto phenol-minimal plates. Although a few Phe+ colonies appeared on phenol-minimal plates on day 2, the majority of the colonies began to appear on day 3 and continued to accumulate during the next 7 days. The accumulation rate of Phe+ colonies on selective plates will be discussed further.
Sequence Analysis of the Plasmid DNA That Constitutively Expressed the pheBA Genes.
We have previously described constitutively expressed fusion promoters created due to the insertion of transposon Tn4652 upstream of the pheA coding sequence (33). However, not all plasmids that constitutively expressed the pheBA genes contained insertions of the Tn4652. We collected several independent Phe+ mutants on days 3–7 after plating of cells on phenol-minimal plates and analyzed the plasmid DNA from these mutants. To be sure that the genetic rearrangements that permitted expression of the phenol degradation pathway in P. putida affected plasmids carrying the pheBA genes, we isolated plasmid DNA from different Phe+ colonies, transformed E. coli HB101 selecting for resistance to ampicillin, and assayed transformants for expression of the pheBA genes. Most AmpR transformants expressed the pheBA genes constitutively. However, in most cases, a small number of transformants did not express the pheBA genes. This suggests that descendent copies of the original wild-type (Phe−) plasmid were present in the Phe+ colonies.
The DNA sequence upstream of the pheBA operon was characterized in 54 pheBA-expressing E. coli HB101 transformants that did not contain insertions of Tn4652 into plasmid pEST1414 (each transformant was a derivative of plasmid DNA isolated from individual Phe+ clones). In 41 cases, we observed a base substitution (C → A) 96 nt upstream of the ATG initiation codon of the pheB gene. In five cases, a G → T substitution occurred 200 nt upstream of this gene (Fig. 1A and Table 1). In the remaining eight cases (seven of them were unique events), deletions from 2 to 23 bp were found in the sequenced DNA (Fig. 1A). It should be noted that we did not find any mutations in the sequenced DNA except those that caused the Phe+ phenotype.
Figure 1.
(A). Sequence analysis of the upstream region of the pheBA genes in mutant plasmids constitutively expressing the pheBA genes. Data on independent Phe+ mutants accumulated in starving cell populations under selective conditions on phenol-minimal plates (postplating mutants) and on the Phe+ mutants obtained from independent cultures grown without the selection (preexisting mutants) are summarized. Sequence of the wild-type DNA in pEST1414 (an ≈250-bp DNA region upstream from the pheB gene) is shown. Sequence at the left of the ClaI site is derived from transposon Tn4652 (29). The potential −35 and −10 hexamers of promoters are framed, −35 hexamers by dotted lines, and −10 hexamers by continuous lines. The base substitutions are marked by arrows without any numbering. Deletions are indicated by sloping lines and are numbered; the insertion sites are shown by arrows with underlined italic numbers. (B) Different newly created promoters of the pheBA genes are listed. Two hexamers homologous to the E. coli σ70-dependent promoters −35 and −10 consensus sequences are boxed. The promoter sequences for postplating mutants were verified by mapping of the transcription initiation site of the pheBA genes in mutant plasmids as described. Dots mark transcription starting points of the promoters. The different deletion mutants, marked above by numbers at sloping lines, are designated as PDEL2–PDEL45, where the numbers indicate the number of base pairs deleted. The mutants containing base substitutions are designated as PPM. Only one 2-bp insertion mutant, PINS2-AC (marked by 19), is listed. The 70-bp insertion (marked by 20 in A) is of unknown origin.
Table 1.
Spectrum of Phe+ mutants (insertions of Tn4652 excluded)
| Mutation | Starving cultures on selective plates*
|
Growing cultures without selection†
|
|
|---|---|---|---|
| Days 3–7 | Day 4 | Day 2 | |
| Deletions | 8 | 1 | 19 |
| Insertions | 0 | 0 | 2 |
| One-base pair substitutions | |||
| C → A | 41 | 23 | 8 |
| G → T | 5 | 2 | 0 |
| C → T | 0 | 1 | 0 |
| Undefined | 0 | 5 | 3 |
| Total | 54 | 32 | 32 |
Independent Phe+ mutants on days 3–7 (exp. 1) and on day 4 (exp. 2) after plating of cells of P. putida strain PaW85[pEST1414] onto phenol-minimal plates were collected, and the plasmid DNA from these mutants was analyzed by sequencing (described in Materials and Methods).
Phe+ mutants collected on day 2 were derived from independent 1-ml cultures plated onto different phenol-minimal plates. Colonies were picked from plates originating from separate cultures.
Sequence analysis of mutant plasmids and mapping of transcription start points of the pheBA genes in these plasmids revealed that point mutations and deletions had created promoter sequences similar to E. coli RNA polymerase σ70 promoter consensus sequence TTGACAN16–18TATAAT (Fig. 1B). Promoters were generated in two ways: (i) by base substitutions in the potential −10 regions, which made these sequences more similar to the −10 consensus TATAAT for E. coli σ70-specific promoters; and (ii) by deletions ranging from 2 to 23 bp that trimmed the space between −35 and −10 hexamers present in wild-type DNA to 17 or 18 bp—i.e., to the optimal spacing for this type of promoter (35). All de novo-created promoters except PPM-CA gave three to four matches to E. coli −35 consensus and three to six matches to E. coli consensus in the −10 region. Transcription of reporter genes was found to start at 6–8 nt downstream from the −10 hexamers of the de novo-generated promoters and was initiated at the same point in both E. coli and P. putida cells (Fig. 1B). These findings are in agreement with data showing that the RNA polymerases from Pseudomonas and E. coli are biochemically equivalent (36) and that RNA polymerases of E. coli and P. putida display identical contact with promoters (37).
Do the Phe+ Mutations Occur in a Growing Culture?
The Luria and Delbrück fluctuation test (38) was carried out to examine occurrence of the Phe+ mutants in cultures of P. putida growing without phenol selection. Phe+ colonies were counted on day 2 from platings of several 1-ml LB cultures onto phenol-minimal plates. Fluctuation in colony numbers was observed. The numbers of mutants that appeared on different phenol plates on days 3–7 were considerably less variable. Calculations of the ratio of the variance to the mean show that the ratio is high (37.3) at day 2 and is lower (6.7) for colonies arising on subsequent days. These data show that Phe+ colonies that emerge on selective plates on day 3 and later contain mutations occurring after the cells were plated. Therefore we call them postplating mutants. Mutants counted on day 2 are considered to exist before the exposure to the selecting agent, phenol.
The Effect of Growth Phase of Bacteria on Accumulation of Phe+ Mutants.
Because Phe+ mutants were seen to arise in starving cultures on selective plates, we investigated the effect of the growth phase of the plated cells on the accumulation of Phe+ mutations.
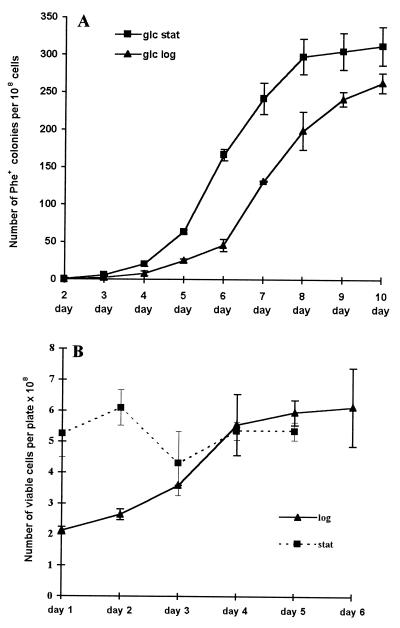
Cells from an exponentially growing culture (glc log) and a stationary-phase culture (glc stat) were plated onto selective medium. We found that during the first days after plating, the rate of appearance of Phe+ mutants was lower for the exponentially grown cells (Fig. 2A). Only later, after day 6, were the rates of accumulation of Phe+ colonies of glc log cells and glc stat cells equal.
Figure 2.
(A) Accumulation of Phe+ mutants on phenol-minimal plates as a function of time for two physiological states of the plated culture. For glc log plating, cells of the P. putida strain PaW85[pEST1414] from exponentially growing culture were used, and, for glc stat, the culture was grown to full density before plating. Each point represents the mean and standard deviation of at least four independent determinations. To calculate the number of Phe+ colonies appearing per 108 plated cells, the number of Phe+ colonies was divided by the number of viable cells present per plate 2 days earlier. (B) Viability of the PaW85[pEST1414] cells on phenol-minimal plates.
To prove that the different rates of accumulation of Phe+ mutants were not caused by different viability of cells from exponential and stationary-phase cultures on selective plates, cells either from exponential or stationary-phase cultures were plated on phenol-minimal plates (Fig. 2A), and small plugs were cut from the plates avoiding Phe+ colonies. Bacteria from these plugs were suspended in M9 medium, and dilutions were plated onto glucose-minimal plates containing carbenicillin to determine the number of colony-forming units. Means were calculated from three independent cultures plated onto four parallel plates. We found that the number of viable cells plated from glc stat culture onto phenol-minimal plates remained constant for the first 5 days (Fig. 2B). However, there was a slight increase in the number of viable cells on selective plates when glc log culture was plated (Fig. 2B). These data clearly show that the lower rate of accumulation of Phe+ mutants on selective plates seeded with exponentially grown cells does not reflect reduced viability of these cells on phenol-minimal plates. Therefore, the data in Fig. 2A imply that the frequency of occurrence of mutations leading to the activation of the phenol degradation genes is initially higher for bacteria sampled from the stationary growth phase.
Are the Preexisting and Postplating Phe+ Mutants Different?
Prival and Cebula (39) found striking differences in the distribution of particular transversions at the hisG428 locus in revertants arising after prolonged histidine starvation, as compared with those arising during growth in the presence of histidine. Also, the spectrum of Lac+ revertants that arise on lactose-minimal media after plating (postplating mutants) is different from that occurring during growth under nonselective conditions (preexisting mutants; refs. 19 and 20). Our results (Fig. 2) also indicate that stationary-phase cells may have some specific mechanisms for generation of mutations following their plating on a selective medium, and this mechanism might be expected to generate specific mutations.
To examine the differences in mutation types, we plated 40 independent cultures of P. putida PaW85(pEST1414) cells on phenol-minimal plates. One Phe+ colony was isolated from each plate on day 2, and 32 of these preexisting mutants were further characterized. The spectrum of mutations in preexisting and postplating Phe+ mutants is compared in Table 1. Indeed, although one particular C → A transversion in the potential −10 element was a mutation hot spot for postplating mutants isolated from two separate experiments (76% and 72% from all events studied, respectively), the preexisting mutations appeared to be mainly different deletions (25% C → A transversion versus 59% deletions from all events studied). These deletions, like those characterized for postplating mutants described above, optimized the distance between the potential −10 and −35 elements of promoters (Fig. 1).
Note that Tn4652 transposition-generated Phe+ mutants appear only after a time lag: no insertions of Tn4652 were observed among the preexisting Phe+ mutants. To evaluate the frequency of Tn4652-linked mutants among all Phe+ mutants accumulated under selective conditions, ≈30–40 Phe+ colonies arising each day on phenol plates (see Fig. 2A) were analyzed by PCR for insertion of transposon Tn4652. The insertions of the transposon first appeared on day 3 after plating and accounted for 10% of the promoter-creating mutations arising in cells plated from stationary-phase culture. At the same time, we did not detect any transposon insertions in cells plated from exponentially growing cultures. On day 4, transposon insertions accounted for one-third of all mutations from both platings and then remained constant for a week. These data show that the initially higher rate of accumulation of Phe+ mutants described for stationary-phase cells compared with that for exponentially growing cells (Fig. 2A) cannot be ascribed to Tn4652 insertion mutations only. The delay in the appearance of Tn4652-linked Phe+ mutants suggests that Tn4652, like many transposons, tunes its transposition frequency to the physiological state of the host (40).
Effect of Starvation on Accumulation of the Phe+ Mutants Without Phenol Selection.
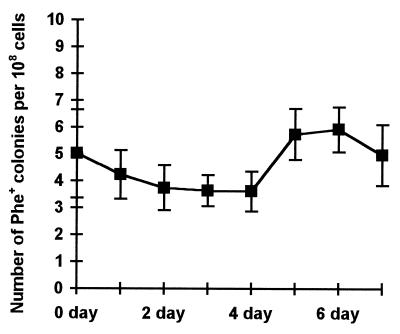
The effect of starvation on accumulation of the Phe+ mutants without selection for growth on phenol was studied in M9 liquid cultures. P. putida strain PaW85(pEST1414) was grown to saturation in glucose-minimal medium, pelleted, and resuspended in M9 minimal medium without any carbon source. Samples were taken on each day during 1 week, washed with M9, and plated on phenol-minimal plates. The Phe+ colonies emerging on plates were counted on day 2. Every day, the number of viable cells in the starving culture was also determined. Our data showed that the cells did not die in the starving culture; after 1 day of starvation, the number of viable cells doubled and remained constant thereafter for a week. According to our experiments, there was no accumulation of Phe+ mutants in the starving culture without the presence of phenol (Fig. 3).
Figure 3.
The effect of starvation on accumulation of Phe+ mutants without phenol selection.
Study of Survival of the Phe+ Mutants in the Starving Mixed Culture.
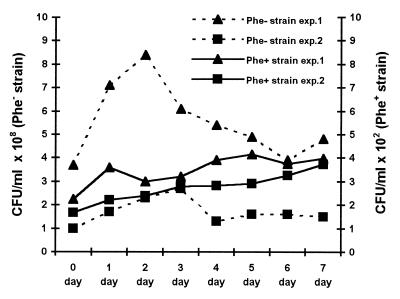
To control for the possibility that mutants constitutively expressing the pheBA genes die or revert rapidly in the absence of phenol selection, we mixed cultures of P. putida PaW85 strain carrying Phe+ mutant plasmid pPPM-CA (the common C → A substitution, see above) and strain containing the same plasmid without the pheBA genes. The mixed culture was aerated in M9 liquid medium without any carbon source for 1 week. On each day, samples were taken to measure the number of viable cells in the culture and to determine the number of the Phe+ colonies per 108 cells. Data presented in Fig. 4 indicate that the Phe+ mutants are not eliminated in a starving culture.
Figure 4.
Study of survival of Phe+ mutants in starving mixed culture. One milliliter of mixed culture contained 102 cells of P. putida PaW85 Phe+ strain carrying mutant plasmid pPPM-CA and 108 cells of P. putida PaW85 Phe− strain, which carried either wild-type plasmid pEST1414 (exp. 1) or vector plasmid pAYC32 (exp. 2). The Phe+ colonies were counted on day 2.
DISCUSSION
A favorite system for the study of mutational processes in starving bacteria has been E. coli strain FC40 (5), which has a deletion of the chromosomal lacZ gene and carries on F′lac plasmid an engineered lacI–lacZ fusion gene with +1 frameshift. Lac+ revertants arise on lactose-selective plates by mechanisms that require recombination functions (18) and plasmid transfer functions (21, 22, 23). The evidence that proofreading-defective DNA polymerase II increases frequency of adaptive mutations in E. coli FC40 has been reported (41). Additionally, a mismatch-repair deficiency was found to be responsible for the unusual revertant spectrum observed among Lac+ revertants that arose under selective conditions (24). Thus, a mechanism of mutability has been described for reversion of lacI–lacZ frameshift that differs from that in growing cells. Investigation of reversion of Salmonella typhimurium chromosomal lacZ mutants suggests that adaptive mutability is a more general phenomenon (42).
Herein, we used a promoterless pheBA gene cluster as a reporter to isolate and characterize mutations in P. putida cells that generate promoters for the transcription of the pheBA genes. We observed that the Phe+ mutants accumulate in the starving culture of P. putida in the presence of phenol but not without it (compare Figs. 2 and 3). Do our results reflect a novel mechanism of mutability, or are there some known possibilities based on bacterial physiology that could explain this phenomenon? Below we discuss different alternatives.
One explanation for the accumulation of certain mutants only after plating on the selective agent is based on selection against these mutants before plating (9, 10). If the mutant phenotype is deleterious in the absence of selective agent, the mutants may die or revert. It has also been argued that mutants grow more slowly than wild-type cells before the exposure to selective agent. Our results presented in Fig. 4 show no elimination of the Phe+ mutants from the mixed population under starvation conditions. We could also confirm that the Phe+ colonies were well detected on phenol-minimal medium 2 days after plating the cells from the mixed cultures.
We need to consider the possibility that the selective agent itself could behave as a mutagen. We did not detect a mutagenic effect of phenol in P. putida (data not shown). Similarly, phenol failed to increase mutation rates in Salmonella (43) and in Streptomyces (44).
Residual growth of bacteria on selective plates enables DNA replication. Slow utilization of phenol provides plasmid replication, even in no-dividing cells, and this replication may allow the accumulation of the mutants observed by us. A similar hypothesis has been postulated to account for adaptive reversion of the lacI–lacZ frameshift on F plasmid (22). Experiments with strain FC40 have shown that Lac+ reversion occurs only in the presence of lactose and the needed auxotrophic supplements (5, 22). The hypothesis that residual lactose metabolism is a requirement for reversion to Lac+ in FC40 has been raised (22, 42). The ability to metabolize lactose weakly was required for reversion to Lac+ under selective conditions also in the case of chromosomal lacZ mutations in S. typhimurium (42). Using residual growth rate and the nonselective reversion rate, the expected number of late revertants was calculated by the authors. For several leaky mutants, the authors found that the observed revertant number exceeded the expected number, suggesting that this excess arises from a adaptive mutability.
Our measurements of the number of viable cells on phenol-selective plates shown in Fig. 2B suggests that these cells did not divide remarkably on the plates. A slight increase in the number of viable cells on selective plates during the first days after plating was observed for the cells plated from exponentially growing culture, but viable cells remained constant for the cells plated from stationary-phase culture. At the same time, the initial accumulation rate of the Phe+ mutants on phenol-containing selective plates was found to be higher for bacteria plated from stationary-phase culture than that for cells from exponentially growing cultures (Fig. 2A). These results are unexplainable by the assumption that Phe+ mutations are associated with higher energy resources in logarithmic-phase cells and/or with growth of these bacteria. Our results shown in Fig. 2 indicate that some process predominating in stationary-phase cells facilitates generation and/or fixation of such mutations. We have not ruled out that some leakiness of the pheBA genes, sufficient for plasmid replication, is required for the generation of mutations.
One explanation for data presented in Fig. 2 is based on the possibility that the plasmid copy number is different in logarithmic- and stationary-phase cells, providing different numbers of targets for mutation. Plasmids used in our study are derivatives of the wide host range plasmid RSF1010. In E. coli, RSF1010 is present at a copy number of 12 per cell (45). Replication and copy number control of the plasmid RSF1010 is determined by plasmid-encoded proteins (46), and there are no data indicating that the physiological state of bacterium and host range influence copy number of this plasmid. Moreover, we have carried out quantitative PCR analysis using specific primers to amplify a segment of plasmid pEST1414 to observe differences in a plasmid copy number between logarithmic- and stationary-phase cells. Serial dilutions from logarithmic- and stationary-phase cultures were made, and the detection levels of the PCR products were found to be similar, ≈10 cells per reaction. Thus, our results do not support the idea that the higher accumulation rate of the Phe+ mutants on selective plates for cells plated from stationary-phase culture is caused by an exaggerated copy number of the plasmid in stationary-phase cells. In addition, mutations appearing in starving cells during selection were mainly C → A transversions, whereas mutations that arose during nonselective growth included mainly different deletions (Fig. 1 and Table 1). This also indicates that the mutation–generation process in starving cells is different from that in growing bacteria.
Several molecular models have been proposed to describe mechanisms in which randomly arising genetic variants proliferate differentially as a consequence of their fitness effects. Cairns et al. (1) proposed a “trial-and-error” hypothesis that involves the production of variant RNA messages or DNA copies, allowing “… individual cells to subject a subset of their informational macromolecules to the forces of natural selection.” Stahl’s model (47, 48) predicts that DNA mismatch repair is impaired in starving cells. In stationary phase, cells are sporadically replicating or repairing their DNA and the DNA replication could be more error-prone. A trial-and-error scenario presumes that variant DNA strands are under selection. If a coding strand in a starving cell contains a spontaneous mutation that can be transcribed and translated and the resulting protein allows the cell to grow and replicate its DNA, then one of the daughter cells could possess a mutation that is fixed in both DNA strands. If the mutation does not allow the cell to grow, the useless change will be eventually eradicated (11, 47–50). The hypothesis that mismatch repair is repressed during adaptive mutation is supported by the finding that the growth-dependent Lac+ reversion spectrum can be made indistinguishable from the adaptive mutation spectrum by eliminating mismatch repair during growth (24).
Initiation of transcription by E. coli RNA polymerase from a promoter sequence containing a mismatch bubble has been shown in vitro (51). σ70 holoenzyme can recognize the −10 consensus sequence as a single-stranded nontemplate strand in the melted transcription bubble (52). Drawing parallels with the model discussed above, we suggest that transcription of the pheBA operon can be initiated from a DNA sequence containing mismatch(es) within the potential −10 element, giving a cell a chance to accumulate energy and multiply, thereby fixing the mutation in the progeny.
DNA synthesis past an oxidatively damaged form of guanine, 7,8-dihydro-8-oxoguanine (8-oxoG), can result in misincorporation of adenine opposite the damaged guanine (53). To reduce this effect, bacteria have developed several enzymes, including MutY, that remove adenine from A/8-oxoG mispair (54). The error avoidance pathway, preventing mutations by 8-oxoG in DNA, plays an important role in maintaining replication fidelity: a mutMmutY double mutant has a mutation rate that is of the same order of magnitude as the mutation rate observed when the polymerase III proofreading function is disabled, and it is about an order of magnitude faster than the mutation rate of a strain lacking the mismatch repair system (54). Introduction of mutY deficiency into the tyrA14 ochre strain increased the appearance of tyrosine-independent mutants of bacteria incubated on plates lacking the required amino acid (55). Bridges proposed that the apparently directed nature of the Tyr+ phenotype could be explained by 8-oxoG mispairing events: miscoding during transcription would result in a transient Tyr+ phenotype, which could allow DNA replication and fixation of the mutation by a DNA A/8-oxyG mispairing.
Perhaps the mutY background would affect the nature and frequency of appearance of Phe+ mutants. The G:C → T:A transversion has been the most common mutation in our studies when the Phe+ mutants arose in starving cell populations under the selective conditions, but not in actively growing cells, where a variety of deletions created most of the functional promoters. The C → A substitution has occurred in the nontemplate DNA strand of a promoter. It is possible that 8-oxoG DNA lesions in a template strand cause mutation by A/8-oxyG mispairing during low-level plasmid DNA synthesis in starving bacteria. If such a mispair is not rapidly corrected, incorporation of an adenine into the nontemplate strand could allow RNA polymerase to recognize the promoter DNA and to initiate transcription from it.
Possibly the trial-and-error mechanism also acts in the case of Tn4652 transposition-generated fusion promoters. A potential −35 hexamer TTGCCT is located inside of the transposon. It could provide a recognition signal for RNA polymerase, if the flanking DNA contained a functional −10 element. The transposition process involves DNA synthesis at staggered ends of the element and target DNA junctions. As a result of this synthesis, the DNA element is flanked by two direct repeats of the target DNA. In three out of six fusion promoters identified by us (33), additional point mutations had occurred in the potential −10 elements, which made them even more similar to the E. coli σ70 promoter consensus sequence. We emphasize that all three of the Tn4652-generated fusion promoters exhibited changes that disrupted the symmetry—mutations occurred only in the target repeats, which served as −10 sequences for the fusion promoter (33).
The transient DNA sequence variants postulated by trial-and-error mechanisms have not been demonstrated. Our results show that the rate of accumulation of phenol-using mutants on selective plates depends on the physiological state of the culture before plating: the rate of accumulation of the mutants is higher in the case of stationary-phase cells. One possible explanation for this phenomenon is that there are more DNA sequence alterations serving as material for fixation of useful mutation in starving and nondividing cells than in a growing culture.
Acknowledgments
We thank R. Villems, T. Alamäe, F. Stahl, and two anonymous referees for helpful comments on the manuscript. This work was supported by grants from Estonian Science Foundation, Grant LCO 000 from the International Science Foundation, Grant No. LKH100 from the Joint Program of the Government of Estonia and the International Science Foundation, and grant from International Foundation for Science (Salen Foundation).
ABBREVIATION
- 8-oxoG
7,8-dihydro-8-oxoguanine
Footnotes
References
- 1.Cairns J, Overbaugh J, Miller S. Nature (London) 1988;335:142–145. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- 2.Hall B G. Genetics. 1988;120:887–897. doi: 10.1093/genetics/120.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall B G. Genetics. 1990;126:5–16. doi: 10.1093/genetics/126.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson S A, DeCloux A M, Munro J. Genetics. 1991;129:647–658. doi: 10.1093/genetics/129.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cairns J, Foster P L. Genetics. 1991;128:695–701. doi: 10.1093/genetics/128.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foster P L, Cairns J. Genetics. 1992;131:783–789. doi: 10.1093/genetics/131.4.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas A W, Lewington J, Hope S, Topping A W, Weightman A J, Slater J H. Arch Microbiol. 1992;158:176–182. doi: 10.1007/BF00290813. [DOI] [PubMed] [Google Scholar]
- 8.Hall B G. Proc Natl Acad Sci USA. 1992;89:4300–4303. doi: 10.1073/pnas.89.10.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lenski R E, Mittler J E. Science. 1993;259:188–194. doi: 10.1126/science.7678468. [DOI] [PubMed] [Google Scholar]
- 10.Lenski R E, Sniegowski P D. Science. 1995;269:285. doi: 10.1126/science.7618089. [DOI] [PubMed] [Google Scholar]
- 11.Lenski R E, Sniegowski P D. Curr Biol. 1995;5:97–99. doi: 10.1016/s0960-9822(95)00023-6. [DOI] [PubMed] [Google Scholar]
- 12.Foster P L. Annu Rev Microbiol. 1993;47:467–504. doi: 10.1146/annurev.mi.47.100193.002343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shapiro J A. Science. 1995;268:373–374. doi: 10.1126/science.7716540. [DOI] [PubMed] [Google Scholar]
- 14.Hall B G. FEMS Microbiol Lett. 1994;117:237–242. [Google Scholar]
- 15.Hall B G. Proc Natl Acad Sci USA. 1995;92:5669–5673. doi: 10.1073/pnas.92.12.5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brenner S. Curr Biol. 1992;2:167–168. doi: 10.1016/0960-9822(92)90496-w. [DOI] [PubMed] [Google Scholar]
- 17.Maynard Smith J. The Theory of Evolution. Cambridge, U.K.: Cambridge Univ. Press; 1993. pp. 2–3. [Google Scholar]
- 18.Harris R S, Longerich S, Rosenberg S M. Science. 1994;264:258–260. doi: 10.1126/science.8146657. [DOI] [PubMed] [Google Scholar]
- 19.Rosenberg S M, Longerich S, Gee P, Harris R S. Science. 1994;265:405–407. doi: 10.1126/science.8023163. [DOI] [PubMed] [Google Scholar]
- 20.Foster P L, Trimarchi J M. Science. 1994;265:407–409. doi: 10.1126/science.8023164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radicella J P, Park P U, Fox M S. Science. 1995;268:418–420. doi: 10.1126/science.7716545. [DOI] [PubMed] [Google Scholar]
- 22.Galitski T, Roth J R. Science. 1995;268:421–423. doi: 10.1126/science.7716546. [DOI] [PubMed] [Google Scholar]
- 23.Foster P L, Trimarchi J M. Proc Natl Acad Sci USA. 1995;92:5487–5490. doi: 10.1073/pnas.92.12.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Longerich S, Galloway A M, Harris R S, Wong C, Rosenberg S M. Proc Natl Acad Sci USA. 1995;92:12017–12020. doi: 10.1073/pnas.92.26.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyer H W, Roulland-Dussoix D. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 26.Bayley S A, Duggleby C J, Worsey M J, Williams P A, Hardy K G, Broda P. Mol Gen Genet. 1977;154:203–204. doi: 10.1007/BF00330838. [DOI] [PubMed] [Google Scholar]
- 27.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. pp. 352–355. [Google Scholar]
- 28.Adams M H. Bacteriophages. New York: Wiley Interscience; 1959. pp. 445–447. [Google Scholar]
- 29.Kivisaar M, Kasak L, Nurk A. Gene. 1991;98:15–20. doi: 10.1016/0378-1119(91)90098-v. [DOI] [PubMed] [Google Scholar]
- 30.Nurk A, Kasak L, Kivisaar M. Gene. 1991;102:13–18. doi: 10.1016/0378-1119(91)90531-f. [DOI] [PubMed] [Google Scholar]
- 31.Hanahan D. J Mol Biol. 1983;166:577–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 32.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nurk A, Tamm A, Hõrak R, Kivisaar M. Gene. 1993;127:23–29. doi: 10.1016/0378-1119(93)90612-7. [DOI] [PubMed] [Google Scholar]
- 34.Blomberg P, Wagner G E, Nordström K. EMBO J. 1990;9:2331–2340. doi: 10.1002/j.1460-2075.1990.tb07405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stefano J E, Gralla J D. Proc Natl Acad Sci USA. 1982;79:1069–1072. doi: 10.1073/pnas.79.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao I, Gussin G N. J Bacteriol. 1991;173:394–397. doi: 10.1128/jb.173.1.394-397.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gragerov A I, Chenchik A A, Aivasashvilli A, Beabealashvilli R S, Nikiforov V. Mol Gen Genet. 1984;195:511–515. doi: 10.1007/BF00341455. [DOI] [PubMed] [Google Scholar]
- 38.Luria S E, Delbrück M. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prival M J, Cebula T A. Genetics. 1992;132:303–310. doi: 10.1093/genetics/132.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higgins N P. Trends Biochem Sci. 1992;17:207–211. doi: 10.1016/0968-0004(92)90376-k. [DOI] [PubMed] [Google Scholar]
- 41.Foster P L, Gudmunsson G, Trimarchi J M, Cai H, Goodman M F. Proc Natl Acad Sci USA. 1995;92:7951–7955. doi: 10.1073/pnas.92.17.7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galitski T, Roth J R. Genetics. 1996;143:645–659. doi: 10.1093/genetics/143.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeMarini D M, Brooks H G, Parkes D G., Jr Environ Mol Mutagen. 1990;15:1–9. doi: 10.1002/em.2850150102. [DOI] [PubMed] [Google Scholar]
- 44.Buchholz S E, Omer C A, Viitanen P V, Sariaslani F S, Stahl R G., Jr Appl Biochem Biotechnol. 1992;32:149–158. doi: 10.1007/BF02922155. [DOI] [PubMed] [Google Scholar]
- 45.Bagdasarian M M, Scholz P, Frey J, Bagdasarian M. Evolution and Environmental Spread of Antibiotic Resistance Genes. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. pp. 209–223. [Google Scholar]
- 46.Scherzinger E, Haring V, Lurz R, Otto S. Nucleic Acids Res. 1991;19:1203–1211. doi: 10.1093/nar/19.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stahl F W. Nature (London) 1988;335:112–113. doi: 10.1038/335112a0. [DOI] [PubMed] [Google Scholar]
- 48.Stahl F W. Genetics. 1992;132:865–867. doi: 10.1093/genetics/132.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boe L. Mol Microbiol. 1990;4:597–601. doi: 10.1111/j.1365-2958.1990.tb00628.x. [DOI] [PubMed] [Google Scholar]
- 50.Sniegowski P D, Lenski R E. Annu Rev Ecol Syst. 1995;26:553–578. [Google Scholar]
- 51.Aiyar S E, Helmann J D, deHaseth P L. J Biol Chem. 1994;269:13179–13184. [PubMed] [Google Scholar]
- 52.Roberts C W, Roberts J W. Cell. 1996;86:495–501. doi: 10.1016/s0092-8674(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 53.Michaels M L, Miller J H. J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Michaels M L, Cruz C, Grollman A, Miller J H. Proc Natl Acad Sci USA. 1992;89:7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bridges B A. Nature (London) 1995;375:741. doi: 10.1038/375741a0. [DOI] [PubMed] [Google Scholar]