Abstract
Src-homology 3 (SH3) domains recognize PXXP core motif preceded or followed by positively charged residue(s). Whether SH3 domains recognize motifs other than proline-based sequences is unclear. In this study, we report SH3 domain binding to a novel proline-independent motif in immune cell adaptor SKAP55, which is comprised of two N-terminal lysine and arginine residues followed by two tyrosines (i.e. RKxxYxxY). Domains capable of binding to class I proline motifs bound to the motif, while the class II domains failed to bind. Peptide precipitation, alanine scanning and in vivo co-expression studies demonstrated a requirement for the arginine, lysine and tandem tyrosines of the motif. Two-dimensional NMR analysis of the peptide bound FYN-SH3 domain showed overlap with the binding site of a proline-rich peptide on the charged surface of the SH3 domain, while resonance signals for other residues (W119, W120, Y137) were not perturbed by the RKGDYASY based peptide. Expression of the RKGDYASY peptide potently inhibited TcRζ/CD3-mediated NF-AT transcription in T cells. Our findings extend the repertoire of SH3 domain binding motifs to include a tyrosine-based motif and demonstrate a regulatory role for this motif in receptor signaling.
Keywords: protein–protein binding/SH3 domain/SKAP55/tyrosine
Introduction
Src-homology 3 (SH3) domains mediate protein–protein interactions involved in the subcellular localization of proteins, cytoskeletal organization and signal transduction (Mayer and Baltimore, 1993; Pawson and Scott, 1997). Since the initial description of Abelson (ABL) SH3 domain recognition of the 3BP1 protein (Cicchetti et al., 1992; Ren et al., 1993), numerous SH3 domain-mediated interactions have been documented. These include SH3-mediated interactions between the adaptor GRB-2 (growth factor receptor-bound protein-2) and Son-of-Sevenless (SOS), Src kinase SH3 domain binding to the p85 subunit of phosphatidylinositol 3-kinase, Crk SH3 domain binding to C3G and many others (Mayer and Gupta, 1998; Musacchio et al., 1994b). SH3 domains consist of two anti-parallel β sheets packed at right angles to one other (Musacchio et al., 1994b). Two SH3 variable loops, the RT and n-Src loops, flank a ligand-binding region formed by the contribution of residues that are well conserved in the SH3 family and this region binds peptides with the consensus sequence Pro-X-X-Pro (PXXP) (Musacchio et al., 1992, 1994a; Yu et al., 1992, 1994; Feng, 1994; Goudreau et al., 1994; Lim et al., 1994). The proline-based motif adopts a pseudosymmetrical polyproline class II (PPII) helix that is stabilized primarily by hydrophobic interactions and, in some cases, by additional electrostatic interactions. It has been proposed that the requirement for prolines is related to that fact that this residue is the only natural N-substituted amino acid (Nguyen et al., 1999).
The detailed requirements of SH3 domain binding to ligand have been defined by the combined use of chemical and phage display combinatorial peptide libraries, nuclear magnetic resonance (NMR) and crystal structure analysis. From this, two classes of interactions, sometimes termed class I (R/KxxPxxP) and class II (PxxPxR), have been defined (Mayer and Gupta, 1998). Due to the pseudosymmetrical nature of the PP II helix, SH3 domains can bind ligands in either a N- to C-terminal, or C- to N-terminal orientation (Yu et al., 1992; Feng et al., 1994; Lim et al., 1994; Musacchio et al., 1994a). Directionality is conferred by the interaction of the arginine or lysine residues with the charged outer face on the SH3 domain, while the tandem prolines bind to two hydrophobic pockets. In all reported cases, at least one of the hydrophobic residues of the core peptide has been found to be a proline residue, usually as PxxP or, as shown recently, PxxDY (Mongiovi et al., 1999).
SH3 domains are found in Src-related kinases and in several of the newly discovered adaptor proteins of immune cells (Rudd, 1999; Samelson, 1999). This family includes linker for activation of T cells (LAT), SH2-domain-containing leukocyte protein of 76 kDa (SLP-76), FYN T-binding protein/SLP-76-associated protein (FYB/SLAP) and Src kinase-associated protein of 55 kDa (SKAP55) (Jackman et al., 1995; da Silva et al., 1997; Marie-Cardine et al., 1997; Musci et al., 1997; Liu et al., 1998). LAT and SLP-76 are substrates for the tyrosine kinase ZAP-70 (Wardenburg et al., 1996; Raab et al., 1997; Samelson, 1999) and are needed for T-cell antigen receptor (TcR) and pre-TcR signaling in T cells and thymocytes, respectively (Clements et al., 1998; Pivniouk et al., 1998; Yablonski et al., 1998). FYB and SKAP55 are unique proteins with C-terminal SH3 domains (da Silva et al., 1997; Musci et al., 1997; Liu et al., 1998; Marie-Cardine et al., 1998; Veale et al., 1999). Each is preferentially phosphorylated by the Src kinase FYN-T (Liu et al., 1998; Marie-Cardine et al., 1998; Raab et al., 1999). The SH3 domain of SKAP55 can bind to a proline-rich region in FYB (Marie-Cardine et al., 1998), while the target of the FYB SH3 domain has yet to be elucidated. While the expression of FYB/SLAP alone in T cells can either weakly potentiate or inhibit anti-CD3-mediated interleukin 2 (IL-2) production (da Silva et al., 1997; Musci et al., 1997), co-expression with FYN and SLP-76 cooperatively up-regulates IL-2 transcription in transfection assays (Geng et al., 1999; Raab et al., 1999; Veale et al., 1999).
Although SH3 domains are well established in their recognition of proline-rich motifs, little is known about their ability to recognize other motifs in the formation of protein–protein complexes. In this study, we report the identification of a SH3 domain binding motif on SKAP55, which is comprised of adjacent arginine and lysine residues followed by tandem tyrosines (i.e. RKxxYxxY). SH3 domains capable of binding to class I motifs also bound to the RKxxYxxY motif, while class II SH3 domains of GRB-2 failed to bind. The FYB SH3 domain bound with some 10-fold higher affinity than observed for the domains of FYN and LCK. Mutational analysis demonstrated a requirement for the arginine, lysine and tandem tyrosine residues, while NMR spectroscopy showed that the peptide binding site overlaps with a negatively charged patch that interacts with proline-based sequences. Over-expression of the RKGDYASYY peptide inhibited TcR-mediated nuclear factor of activated T cells (NF-AT) transcription in T cells. Overall, our observations extend the repertoire of SH3 domain binding sequences to include sequences comprised of arginine and lysine residues linked to tandem tyrosines.
Results
SH3 domain binding to a novel tyrosine-based RKxxYxxY motif
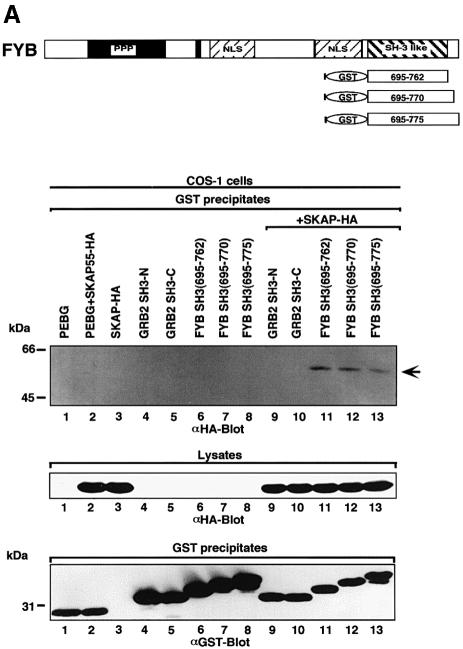
FYB and SKAP55 interact with each other, in part by virtue of SKAP55 SH3 domain binding to FYB (Liu et al., 1998; Marie-Cardine et al., 1998). To determine whether the SH3 domain of FYB was also able to bind to a site on SKAP55, the FYB SH3 domain tagged with glutathione S-transferase (GST) and SKAP55 tagged with hemagluttinin (HA) were co-transfected in COS cells and assessed for binding by co-precipitation (Figure 1A). Three constructs of the FYB SH3 domain of slightly different sizes (residues 695–762, 695–770 and 695–775) were used. Glutathione beads were used to precipitate FYB, while the presence of co-precipitated SKAP55 was detected by immunoblotting with antibody against HA. Under these conditions, each SH3 domain precipitated a 55 kDa protein that corresponded to SKAP55 (Figure 1A, upper panel, lanes 11–13). Specificity in binding was shown by the inability of the GRB-2 N- and C-terminal SH3 domains to precipitate the protein (Figure 1A, lanes 9 and 10, respectively).
Fig. 1. FYB SH3 domain binding to the SK4 region of SKAP55. (A) The scheme shows mouse full-length FYB, sub-regions and GST–SH3 domain constructs. (Upper panel of gel) FYB SH3 domain binds full-length SKAP55 in COS-1 cells. COS-1 cells were transfected with full-length SKAP55 alone, or with the GRB-2 or various FYB SH3 domain(s) and assessed for complex formation. Glutathione beads were used to precipitate the GST fusion proteins. Lane 1, pEBG; lane 2, pEBG and SKAP55-HA; lane 3, SKAP55-HA; lane 4, GRB-2 N-terminal SH3 domain; lane 5, GRB-2 C-terminal SH3 domain; lane 6, FYB SH3(695–762); lane 7, FYB SH3(695–770); lane 8; FYB SH3(695–775); lane 9, GRB-2 N-terminal SH3 domain and SKAP55-HA; lane 10, GRB-2 C-terminal SH3 domain and SKAP55-HA; lane 11, FYB SH3(695–762) and SKAP55-HA; lane 12, FYB SH3(695–770) and SKAP55-HA; lane 13, FYB SH3(695–775) and SKAP55-HA. The precipitates were separated on a 10% SDS–polyacrylamide gel followed by anti-HA blotting. (Middle panel) Levels of SKAP55 protein expression. As in upper panel, except cell lysate was blotted with anti-HA. (Lower panel) Levels of GST fusion protein expression. As in upper panel, except that precipitates were blotted with anti-GST. (B) Schematic representation of SKAP55 and various sub-regions. Various DNA fragments encoding the SKAP55 were generated by PCR and inserted into the pSRα at the BamHI and KpnI sites with an in-frame HA tag. (Upper panel of gel) In vivo association of FYB SH3 and SKAP55 at SK4 region. COS-1 cells were co-transfected with different GST–SKAP55 subdomains and FYB SH3 domain and assessed for complex formation. Glutathione–Sepharose beads were used to precipitate the GST fusion proteins. Lane 1, pSRαHa plus FYB SH3; lane 2, pSRαHa-SKAP55(1–106) plus FYB SH3; lane 3, pSRαHa SKAP55(105–208) plus FYB SH3; lane 4, pSRαHa-SKAP55(204–299) plus FYB SH3; lane 5, pSRα Ha-SKAP55(300–369) plus FYB SH3. The precipitates were separated on a 10% SDS–polyacrylamide gel and immunoblotted with anti-HA monoclonal antibody. (Middle panel) Levels of FYB protein expression. As in upper panel except that the cell lysate was blotted with anti-HA. (Lower panel) Levels of GST fusion protein expression. As in upper panel except that precipitates were blotted with anti-GST. (C) (Above) In vitro association of FYB SH3 and SKAP55 at SK4 region. The different truncated GST–SKAP55 fusion proteins used in the far-western assay. Lane 1, pGEX-SKAP55(1–106); lane 2, pGEX-SKAP55(105–208); lane 3, pGEX-SKAP55(204–299); lane 4, pGEX-SKAP55(300–369); lane 5, pGEX-SK1(204–238); lane 6, pGEX-SK2(239–255); lane 7, pGEX-SK3(256–276); lane 8, pGEX-SK4(277–298). The different truncated GST–SKAP55 fusion proteins were separated on a 10% SDS–polyacrylamide gel, transferred to nitrocellulose membranes and probed with Flag-tagged FYB SH3 protein and immunoblotted with anti-Flag monoclonal antibody. The sequence of the motifs contained within the GST fusion proteins is as follows: SK1, 204QISFLLKDLSSLTIPYEEDEEEEEKEETYDDIDGF238; SK2, 239DSPSCGSQCRPTILPGS255; SK3, 256VGIKEPTEEKEEEDIYEVLPD276; SK4, 277EEHDLEEDESGTRRKGDYASYY298. (Lower panel) FYB SH3 domain recognizes the SK4 region of SKAP55. The various GST–SH2 and –SH3 fusion proteins used in the far-western assay. Lane 1, pGEX-SK4; lane 2, CRKL SH3; lane 3, LCK SH3; lane 4, p85 SH3; lane 5, Src SH3; lane 6, ABL SH2; lane 7, GRB-2 N SH2; lane 8, FYN, SH2; lane 9, LCK SH2; lane 10, PLCγ SH2-N.
To identify the binding region in SKAP55, HA-tagged sub-regions [the N-terminus (residues 1–106), the PH (pleckstrin homology) domain (residues 105–204), an intervening region termed SK (residues 204–299) and the SH3 domain (residues 300–369) of SKAP55] were generated (Figure 1B). Each of the subregions was co-expressed with the FYB SH3 domain in COS cells followed by precipitation with glutathione beads and immunoblotting with anti-HA. Under these conditions, the FYB SH3 domain selectively precipitated the SK fragment (Figure 1B, lane 4). No binding to other regions was noted (lanes 2, 3 and 5). This finding was confirmed by protein–protein blotting using GST-tagged SKAP sub-regions and Flag-tagged FYB SH3 domains (Figure 1C, middle panel). Again, the FYB SH3 domain bound specifically to the SK sub-region (compare lane 3 with lanes 1, 2 and 4).
To dissect further the binding region within the SK region of SKAP55, GST-linked sub-regions within the SK segment were generated that correspond to residues 204–238 (SK1), 239–255 (SK2), 256–276 (SK3) and 277–298 (SK4) (Figure 1C, upper panel). Protein–protein blotting showed that the FYB SH3 domain bound exclusively to the SK4 sub-region (middle panel, compare lane 8 with lanes 5–7). As a negative control, no binding of the FYB SH3 domain was evident against a panel of control proteins including the CRKL (CRK-like), LCK, p85 and SRC SH3 domains, as well as the ABL, GRB-2, FYN, LCK and PLCγ Src-homology 2 (SH2) domains (Figure 1C, lower panel, compare lane 1 with lanes 2–10). Taken together, these findings demonstrate that the FYB SH3 domain can bind to a specific region in the SKAP55 adaptor protein.
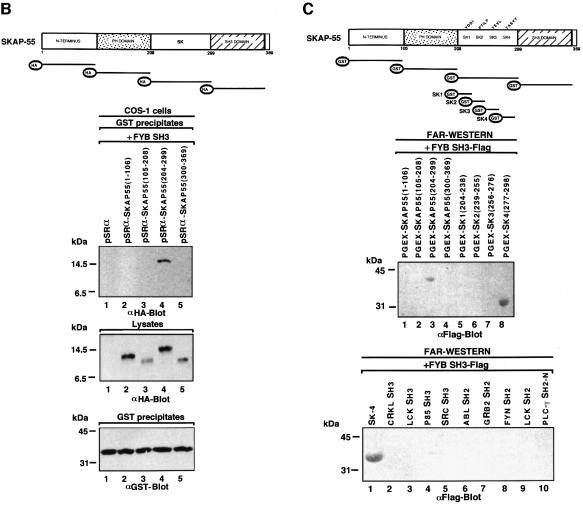
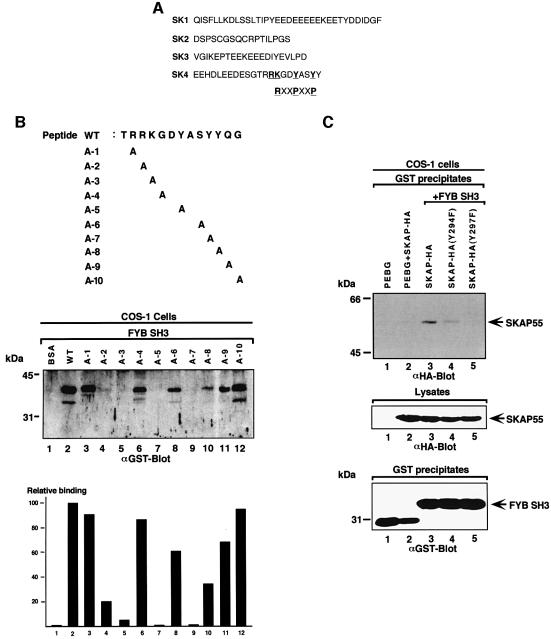
An analysis of the amino acid sequence of the SK4 peptide (EEHDLEEDESGTRRKGDYASYY) showed the absence of prolines residues that could form class I R/KxxPxxP or class II PxxPxR motifs (Figure 2A). The closest approximation to an R/KxxPxxP motif was found in the sequence R289RKGDYASYY298. This sequence is reminiscent of a class I motif where the N-terminal charged RK residues are followed by a YxxY motif that could mimic the proline residues of a classic PXXP motif. An assessment of whether this motif can bind to the SH3 domain was carried out by coupling peptide to Affigel beads. These peptide-bound beads were then used to precipitate the GST–FYB SH3 domain from lysates derived from transfected cells (Figure 2B). Under the conditions, the TRRKGDYASYYQG peptide readily precipitated the FYB SH3 domain (lane 2). Alanine scanning was then conducted to assess the importance of individual residues. Substitution of residues R290, K291, Y294 and Y297 reduced binding by 85–99% as detected by densiometry (Figure 2B, middle panel, lanes 4, 5, 7 and 9, respectively; see also lower histogram panel). In contrast, substitution of the intervening amino acids G292 or S296 or the terminal Q299 and G300 residues had no effect on binding (lanes 6, 8, 11 and 12). Substitution of the most N-terminal R289 also had no effect on binding. Substitution of the third tyrosine (Y298), which is C-terminal to the YxxY motif, had a partial effect (Figure 2B, middle and lower panels, lane 10). These findings indicate that certain key residues, R290, K291, Y294 and Y297, are required for binding. While the requirement for the charged R290 and K291 residues and the YxxY motif suggests similarities to a class I R/KxxPxxP motif, the requirement for both the arginine and lysine residues (instead of just one) in binding suggested differences in the molecular basis of SH3 domain binding to this motif.
Fig. 2. Key residues are required for RKGDTASYY motif binding to the FYB SH3 domain. (A) Amino acid sequence of peptides corresponding to SK1–SK4. The SK4 region, carrying the RKGDYASY motif, is compared with a classic class I RxxPxxP. (B, upper panel) The SKAP55 peptide (TRRKGDYASYYQG; residues 288–300) used in precipitation studies. Various sequential mutants were synthesized in which alanine was used to substitute for amino acids. (Middle panel) AminoLink-Plus coupled to various peptides was used to precipitate the GST-tagged FYB SH3 domain from lysates from COS-1 cells. GST–FYB SH3 domain was detected by anti-GST immunoblotting. Lane 1, bovine serum albumin; lane 2, wild-type TRAKGDYASYYQG peptide; lane 3, TARKGDYASYYQG A-1 peptide; lane 4, TRAKGDYASYYQG A-2 peptide; lane 5, TRRAGDYASYYQG A-3 peptide;; lane 6, TRRKADYASYYQG A-4 peptide; lane 7, TRRKGDAASYYQG A-5 peptide; lane 8, TRRKGDYAAYYQG A-6 peptide; lane 9, TRRKGDYASAYQG A-7 peptide; lane 10, TRRKGDYASYAQG A-8 peptide; lane 11, TRRKGDYASYYAG A-9 peptide; lane 12, TRRKGDYASYYQA A-10 peptide. (Lower panel) Densitometric profile of SKAP55 in GST–FYB SH3 precipitates using a Scantjet laser scanner (Hewlett-Packard). (C) (Upper panel) Mutation of tyrosine residues Y294F/Y297F attenuates in vivo SKAP55 binding to the FYB SH3 domain. COS-1 cells were transfected with SKAP55, SKAP55(Y294F), SKAP55(Y297F) and FYB SH3 domain and assessed for complex formation. Glutathione–Sepharose beads were used to precipitate the GST-tagged FYB SH3 domain. Co-precipitated HA-tagged SKAP55 was detected by anti-HA immunoblotting. Lane 1, pEBG; lane 2, pEBG plus SKAP55; lane 3, SKAP55 plus FYB SH3; lane 4, SKAP55 (Y294F) plus FYB SH3; lane 5, SKAP55 (Y294F) plus FYB SH3 domain. The precipitates were separated on a 10% SDS–polyacrylamide gel and subjected to anti-HA blotting. (Middle panel) Levels of SKAP-HA protein expression. Cell lysates were separated by SDS–PAGE, transferred to nitrocellulose and subjected to blotting with anti-HA. (Lower panel) Levels of GST fusion protein expression. As in middle panel, except that lysates were blotted with anti-GST.
To test further the importance of the individual tyrosine residues in an in vivo context, Y294 and Y297 mutants of SKAP55 were co-expressed with the GST–FYB SH3 domain in COS cells (Figure 2C). Substitution of Y297 caused a complete loss of binding, while loss of Y294 resulted in a partial loss of binding (Figure 2C, upper panel, compare lanes 4 and 5 with lane 3). Equivalent levels of SKAP55 and the GST–FYB SH3 domain were expressed in the different transfectants (Figure 2C, middle and lower panels, respectively). Taken together, these findings from in vitro and in vivo studies demonstrate that specific residues in the R289RKGDYASYY298 motif are needed for an interaction with the SH3 domain.
Other SH3 domains recognize the RKxxYxxY motif
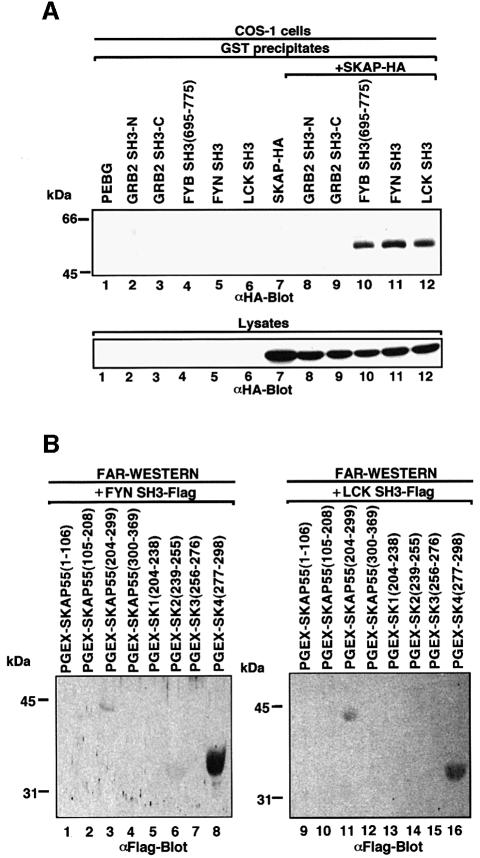
Given the similarity of the RKxxYxxY motif to class I proline-based motifs, we next assessed whether the motif could be recognized by other SH3 domains (Figure 3). In vivo co-expression of either GST-tagged FYN or LCK SH3 domains and HA-tagged SKAP55 resulted in binding as detected by co-precipitation (Figure 3A, lanes 11 and 12). In contrast, neither the N-terminal nor the C-terminal SH3 domain of GRB-2 precipitated the protein (lanes 8 and 9). The SH3 domains of GRB-2 recognize the oppositely oriented PxxPxR motif (Lim et al., 1994). Protein–protein blotting confirmed that the FYN and LCK SH3 domains recognized the SK subregion (Figure 3B, lanes 3 and 11, respectively). Furthermore, within this region, they recognized the SK4 peptide (Figure 3B, lanes 8 and 16, respectively). As previously seen with the FYB SH3 domain, specificity was observed where the FYN and LCK SH3 domains failed to recognize the other SK regions (Figure 3B, lanes 13–15 and 5–7). These findings indicate that the recognition of the RKxxYxxY motif can be extended to other SH3 domains with an ability to recognize proline-based class I motifs.
Fig. 3. Other SH3 domains capable of recognizing proline-based class I motifs also recognize tyrosine-based motifs. (A; upper panel) Co-expressed FYN and LCK SH3 domains also interact with HA-tagged SKAP55. COS-1 cells were transfected with SKAP55, GRB-2 SH3, FYB SH3, FYN SH3 and LCK SH3 in pEBG and assessed for complex formation. Glutathione beads were used to precipitate the GST fusion proteins, while co-precipitated HA-tagged SKAP55 was detected by anti-HA blotting. Lane 1, pEBG; lane 2, GRB-2 N-terminal SH3; lane 3, GRB-2 C-terminal SH3; lane 4, FYB SH3; lane 5, FYN SH3; lane 6, LCK SH3; lane 7, SKAP55-HA; lane 8, GRB-2 N-terminal SH3 and SKAP55-HA; lane 9, GRB-2 C-terminal SH3 and SKAP55-HA; lane 10, FYB SH3 and SKAP55-HA; lane 11, FYN SH3 and SKAP55-HA; lane 12, LCK SH3 and SKAP55-HA. The precipitates were separated on a 10% SDS–polyacrylamide gel, transferred to nitrocellulose and subjected to anti-HA blotting. (Lower panel) Expression of SKAP55. As in upper panel except that cell lysate was run on gel and subjected to anti-HA blotting. (B) In vitro binding of FYN SH3 and LCK SH3 with the SK and SK4 regions of SKAP55. SKAP55 constructs were expressed in T cells and subjected to protein–protein blotting with FYN SH3 (lanes 1–8) and LCK SH3 (lanes 9–16). Detection was conducted using anti-Flag antibody coupled to alkaline phosphatase. Lane 1, pGEX-SKAP55(1–106); lane 2, pGEX-SKAP55(105–208); lane 3, pGEX-SKAP55(204–299); lane 4, pGEX-SKAP55(300–369); lane 5, pGEX-SK1(204–238); lane 6, pGEX-SK2(239–255); lane 7, pGEX-SK3(256–276); lane 8, pGEX-SK4(277–298); lane 9, pGEX-SKAP55(1–106); lane 10, pGEX-SKAP55(105–208); lane 11, pGEX-SKAP55(204–299); lane 12, pGEX-SKAP55(300–369); lane 13, pGEX-SK1(204–238); lane 14, pGEX-SK2(239–255); lane 15, pGEX-SK3(256–276); lane 16, pGEX-SK4(277–298).
Surface plasmon resonance measurements of binding to the non-phosphorylated RKxxYxxY-containing peptide
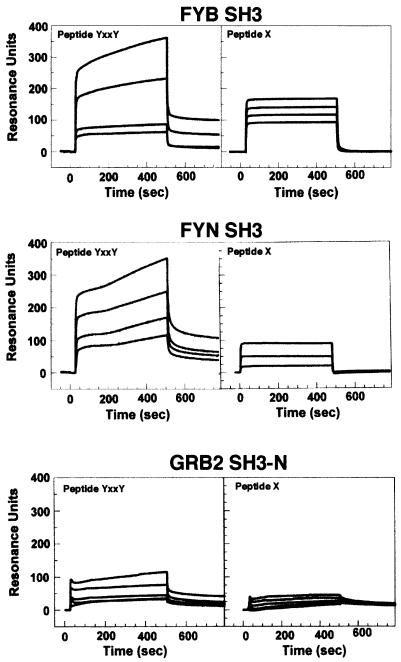
Given the unorthodox nature of RKxxYxxY recognition, we extended our study by measuring the interaction by surface plasmon resonance. SH3 domains bind proline-rich peptide motifs with affinities in the 1–200 µM range (Mayer and Gupta, 1998). Significantly, the FYB, LCK and FYN SH3 domains bound to the RKGDYASYY motif with sufficient affinity to be detected in in vitro peptide precipitation and in vivo co-expression studies (Figures 1–3). The interaction is relatively resistant to exposure and washes in detergent (Figures 1–3). To obtain further data on the affinity of binding, surface plasmon resonance measurements were carried out using immobilized SH3 domains incubated with a range of peptide concentrations. SH3 domains were immobilized on biosensor chips and exposed to increasing concentrations of peptide. Over multiple experiments, the FYB SH3 domain showed specific binding to the TRRKGDYASYYQG peptide with an affinity in the micromolar range (Kd 2–6 × 10–5 M) (Figure 4, upper panel). The SEM was <7.5% in every instance. This affinity is of the same order as that observed for other SH3 domain interactions (i.e. 1–100 µM) (Chen et al., 1993; Feng et al., 1994; Yu et al., 1994). The FYN SH3 domain was also exposed to peptide and assessed for binding; the peptide bound specifically, but with ∼10-fold lower affinity (Figure 4, middle panel). In contrast, as a negative control, GRB-2 N-terminal SH3 domain did not bind (lower panel). The same purified GRB-2 domain bound to the PII peptide of SOS with a Kd in the nanomolar range (data not shown). An additional negative control, an unrelated peptide, TTGFVKMPPTE, failed to bind (data not shown).
Fig. 4. Surface plasmon resonance confirms FYB and FYN SH3 domain binding to TRAKGDYASYYQG peptide. Binding of the TRAKGDYASYYQG peptide to immobilized FYB SH3 domain (upper panel), FYN SH3 domain (middle panel) and GRB2 N-terminal SH3 domain (lower panel). Surface plasmon resonance was conducted as described in Materials and methods. Peptide TTGVFVKMPPTE was used as an irrelevant control. Peptides were injected at 35, 70, 175 or 350 µM. Binding was measured in resonance units (RU), where 1 RU corresponds to the binding of 1 ng/mm2 of flow cell surface.
Two-dimensional NMR spectroscopy confirms the specificity of the interaction
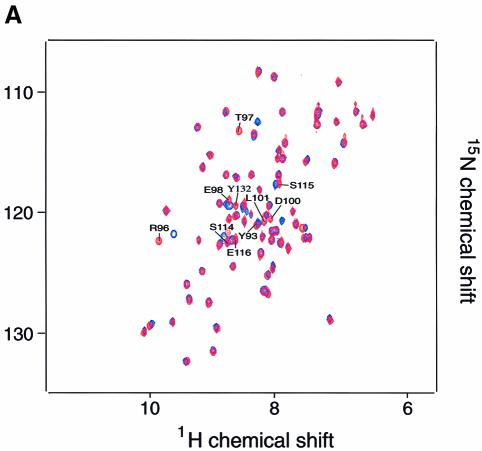
We next investigated the binding of the TRRKGD YASYYQG SKAP55 peptide to an SH3 domain by two-dimensional NMR spectroscopy. The FYN SH3 domain was used since it was more readily purified and NMR assignments could be based on previously published resonance shifts (Mal et al., 1998). Equal amounts of this peptide were added to a 0.3 mM solution of the 15N-labeled, purified FYN domain and chemical shift differences of individual NH groups within the protein were examined. Figure 5A shows a comparison of the isolated (red) and complexed (blue) FYN domain 15N–1H correlation spectra. A number of peaks that are labeled in Figure 5A were significantly shifted upon addition of the peptide ligand. The 10 residues that displayed the largest chemical shift changes upon peptide binding are located in the RT loop (Y93, R96, T97, E98, D100 and L101), the n-Src loop (S114, S115 and E116) and the β-strand (Y132). Since a ligand-induced conformational change is unlikely to have caused the observed resonance shifts of the individual NH groups, it is most likely that these residues represent the sites of interaction with the peptide ligand. Mapping these residues on the surface of the three-dimensional solution structure of the FYN domain (Noble et al., 1993; Morton et al., 1996) shows that they form a contiguous area (Figure 5B). This area maps to a region of the SH3 domain that is known to be involved in binding to proline-rich consensus ligands. A plot of the combined chemical shift change indices for all individual backbone NH groups of the FYN domain upon binding is shown in Figure 5C. In a control experiment we used the peptide PPRPLPVAPGSSKT from phosphatidyl inositol 3-kinase (PI3 kinase), which we previously showed can bind to the FYN SH3 domain (Prasad et al., 1993; Kapeller et al., 1994). Figure 5D shows the combined chemical shift change index for a 0.3 mM sample of the FYN domain upon addition of a stoichiometric amount of the PI3 kinase peptide. A comparison of the data in Figure 5C and D reveals that the chemical shift changes upon addition of the proline-rich peptide were much larger than those for the tyrosine-based SKAP55 peptide. This indicates the higher binding affinity for the proline-rich peptide, where most of the protein is in the bound state at the concentrations used. However, the pattern of the chemical shift changes upon ligand binding is similar for these two peptides. The most prominent chemical shift changes are observed for residues within the RT loop in both cases; it is well established that this loop plays a pivotal role in the binding of SH3 domains to its cognate sequences (Musacchio et al., 1994b). The overlap of the binding site of the FYN domain for the tyrosine-based peptide with the established proline-rich sequence recognition site argues for a weak but specific interaction of the SKAP55 peptide with the FYN domain. A hydrophobic control peptide, TTGVFVKMPPTE, did not induce any chemical shift perturbations upon addition to the 15N-labeled FYN domain (data not shown).
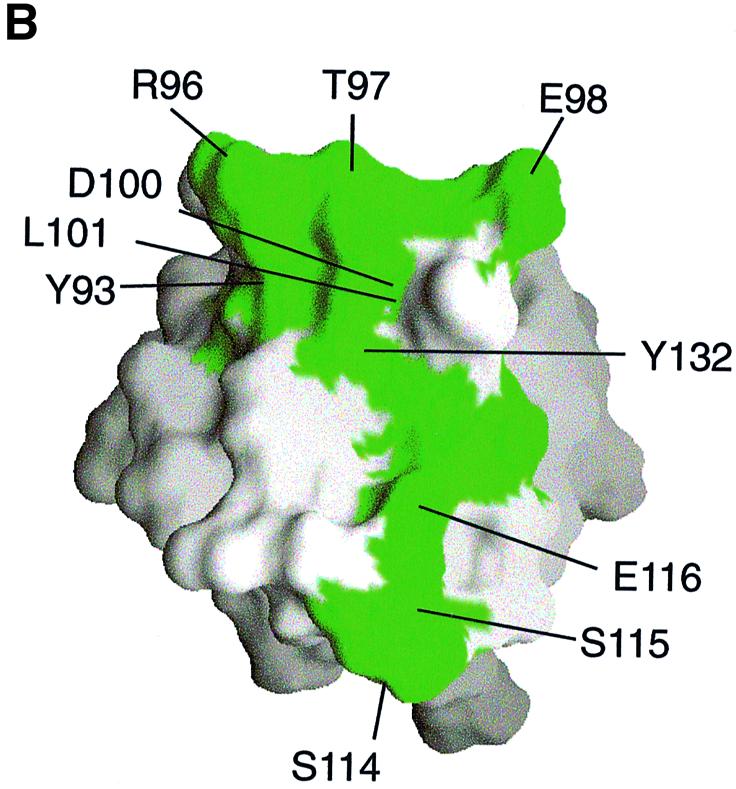
Fig. 5. Two-dimensional NMR spectra of TRAKGDYASYYQG peptide binding to the FYN SH3 domain. (A) 15N–1H NMR correlation spectra of the isolated and complexed FYN SH3 domain. 15N–1H resonances that become significantly shifted upon addition of equimolar amounts of the SK4 peptide ligand are denoted by amino acid type and number within the full-length FYN kinase. The 10 residues that display the largest chemical shift changes upon peptide binding are located in the RT loop (Y93, R96, T97, E98, D100 and L101), the n-Src loop (S114, S115 and E116) and the β-strand (Y132). (B) GRASP (Nicholls et al., 1991) surface representations of the FYN SH3 domain (Morton, 1996). The 10 residues with the largest combined chemical shift index [as in (A)] are depicted in green and are labeled according to residue type and amino acid number within the full-length kinase. (C and D) Combined chemical shift change indices for individual backbone NH groups of the FYN SH3 domain at 0.3 mM upon addition of equimolar amounts of the peptide TRRKGDYASYYQG (C) or PPRPLPVAPGSSKT (D).
Interestingly, there are a few distinct differences in the chemical shift change indices of the tyrosine-based compared with the proline-rich peptide (Figure 5C and D). Residues W119, W120 and Y137 are shifted upon addition of the proline-rich, but not by the tyrosine-based ligand. W119 and Y137 have been shown to contact the proline-rich peptide ligand directly in NOE-based NMR experiments (Morton, 1996). Based on the absence of chemical shift perturbations, it is unlikely that W119 and Y137 contact the tyrosine-based ligand. In the FYB domain, W119 is substituted with another residue (possibly a lysine or tyrosine, depending on the alignment). It is possible that this change renders a higher affinity interaction of the FYB domain for the tyrosine-based peptide. The binding experiments performed show that the peptide ligand is a fast exchange with off rates faster than ∼103 s−1, since the chemical shifts of the FYN domain HSQC (heteronuclear single quantum coherence) resonances change in a manner dependent on the concentration of the ligand (unpublished results). Only a fraction of the FYN domain is in the bound conformation under the conditions used here. NMR titration experiments indicate a Kd in the high micromolar range, a value that is in accord with the BIAcore measurements. Due to the low solubility of the FYB SH3 domain, NMR studies on this higher affinity interaction were not possible (data not shown).
RKxxYxxYY binding mediates TcR regulation of IL-2 transcription
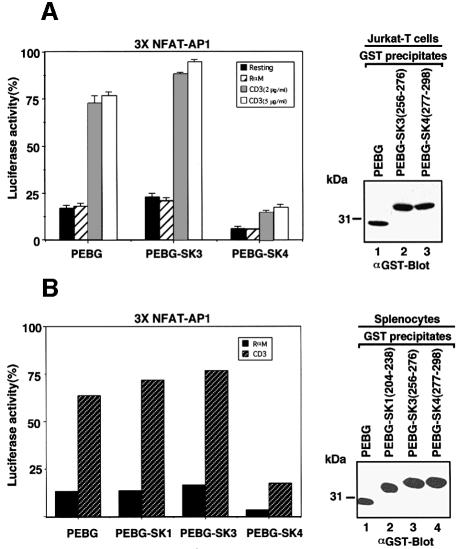
If the interaction of the RKxxYxxY motif with the SH3 domain were of biological importance, it might be expected to interfere with receptor-mediated signaling in cells. A GST-tagged version of the TRRKGDYASYYQG peptide was expressed in Jurkat T cells and murine splenocytes to determine whether it interfered with activation of NF-AT transcription of the IL-2 promoter by engagement of the T-cell receptor–CD3 complex (Figure 6A). Cells were co-transfected with the NF-AT reporter and a second construct (pRL-TK) for normalization of transfection efficiencies. Untransfected and GST-transfected cells showed a 3- to 4-fold increase in NF-AT transcriptional activity in response to anti-CD3 (2 and 5 µg/ml). Expression of GST–SK4 caused a consistent 50–80% inhibition of transcription in seven experiments. Importantly, expression of an irrelevant GST–SK3 peptide did not inhibit transcription. Immunoblotting showed that each of the transfected proteins was expressed at equivalent levels (upper right panel). These data indicate that the RKxxYxxY motif plays a central role in TcR-mediated IL-2 transcription in Jurkat T cells.
Fig. 6. SK4 peptide attentuates TcR up-regulation of IL-2 transcription. (A, left panel) Jurkat T cells were subjected to electroporation using 5 µg NF-AT of the IL-2 promoter luciferase reporter plasmid and 0.2 µg pRL-TK plasmid together with either 20 µg pEBG vector, pEBG-SK3 or pEBG-SK4 constructs. Cells were unstimulated (black bars) or exposed to rabbit anti-mouse (hatched bars), anti-CD3 (OKT3, 2 µg/ml, grey bars) or anti-CD3 (OKT3, 5 µg/ml, open white bars) for 6 h and assayed for luciferase activity. Luciferase units of the experimental vector were normalized to the level of the control vector in each sample. The data are representative of seven independent experiments. Results are the means and standard errors from three replicate experiments. (Right panel) Levels of GST fusion protein expression. Cell lysates from Jurkat T cells that had been transfected with GST–SK4 were subjected to immunoblotting with an anti-GST antibody. Lane 1, pEBG; lane 2, pEBG-SK3; lane 3, pEBG-SK4. (B, left panel) ConA-activated splenocytes 48 h after activation were subjected to electroporation using 5 µg NF-AT of the IL-2 promoter luciferase reporter plasmid and 0.2 µg pRL-TK plasmid together with either 20 µg pEBG vector, pEBG-SK1, pEBG-SK3 or pEBG-SK4 constructs. Cells were exposed to rabbit anti-mouse (black bars) or anti-CD3 (2C11, 2 µg/ml, hatched bars) for 6 h and assayed for luciferase activity. Luciferase activity was measured in spleen cells treated as described in Materials and methods. (Right panel) Levels of GST fusion protein expression. Cell lysates from spleen cells that had been transfected with GST–SK4 were subjected to immunoblotting with an anti-GST antibody. Lane 1, pEBG; lane 2, pEBG-SK1; lane 3, pEBG-SK3; lane 4, pEBG-SK4.
To apply this finding to normal peripheral T cells, murine splenocytes were activated by concanavalin A (Con A) for 48 h prior to transfection with various constructs and the NF-AT promoter construct (Figure 6B). We routinely observe a transfection efficiency of 10–15% of the total population of cells; of those cells that are able to take up DNA, the majority (80%) take up both the transfected DNA and the IL-2 construct (data not shown). Under these conditions, co-expression of the GST SK4 peptide inhibited IL-2 transcription by 70–80%. Importantly, neither the SK1 nor SK3 peptide had an effect on TcR activation. Immunoblotting showed that the transfected proteins were expressed at equivalent levels (lower right panel). Taken together, these data are consistent with the notion that SK4 motif recognition is needed for TcR signaling in normal blood T lymphocytes.
Discussion
SH3 domains are well established protein modules that recognize extended proline-rich peptide sequences (class I, RxxPxxP; class II, PxxPxR) (Mayer and Gupta, 1998). A modification of this theme has recently been reported with Eps8 SH3 domain recognition of the sequence, PxxDY, where the aspartic acid (D) can substitute for the first proline (Mongiovi et al., 1999). In this study, we extend the diversity of SH3 domain binding motifs by demonstrating binding to a tyrosine-based motif composed of arginine and lysine residues adjacent to spaced tyrosines (RKxxYxxY). The combined approaches of in vivo co-expression, protein–protein blotting, peptide precipitation, alanine scanning mutagenesis, surface plasmon resonance and two-dimensional NMR spectroscopy confirmed the interaction. Although lacking proline residues, this novel motif could be viewed as retaining an overall signature of class I proline-based motifs in which tyrosines might substitute for proline residues. In this case, positively charged N-teminal arginine and lysine residues are followed by adjacent tyrosine residues separated by two intervening amino acids. Alanine substitution and peptide precipitation analysis confirmed the need for the arginine and lysine residues (R290 and K291) as well as the tyrosine residues in FYB SH3 domain binding (Figure 2B). Other intervening residues had little effect on binding. Consistent with this, SH3 domains capable of recognizing class I proline motifs, such as those in the Src kinases FYN and LCK, also recognize the RKxxYxxYY sequence, while GRB-2 class II domains failed to bind. Lastly, the functional importance of peptide recognition was shown by the ability of SK4 peptide to block TcR up-regulation of IL-2 transcription (Figure 6). Taken together, our findings identify a novel mode of SH3 domain interaction that may eventually be found to operate in other protein–protein interactions.
Although two-dimensional NMR spectroscopy of the FYB SH3 domain binding was not possible because of technical problems in the purification of the protein, the specificity of the interaction was confirmed with the FYN SH3 domain. This demonstrated that the ligand-binding site is similar to the site for the binding to proline-based peptides with a chemical shift perturbation of residues Y93, R96, T97, E98, D100, L101, S114, S115, E116 and Y132 (Figure 5A). These residues form a contiguous surface area with a predominantly negative surface potential (Figure 5B). The importance of this region correlates well with the skewed importance of the RK residues (i.e. each of the charged residues is needed) in binding, as observed with alanine scanning and peptide precipitation analysis (Figure 2B). The RK residues of the SKAP peptide complement the negative surface of the SH3 domain in the recognition event. Whether the tyrosine residues of the peptide involve van der Waals interactions and hydrogen bonding to backbone atoms similar to those seen for proline–SH3 domain interactions remains to be determined. In either case, the tyrosine-based peptide is unlikely to form a pseudosymmetric PPII helix as observed for proline-based motfis. This suggests that the two tyrosines are not likely to be aligned in their interaction with the surface of the SH3 domain, as has been observed for PxxP binding (Musacchio et al., 1992; Yu et al., 1992, 1994; Feng et al., 1994; Goudreau et al., 1994; Lim et al., 1994). Indeed, the absence of a shift perturbation for the NH groups of W119 (backbone and side-chain NH) and Y137 indicates that these residues do not contribute to the binding of the tyrosine-based ligand. In contrast, large chemical shift differences are seen for these NH groups upon binding of the proline-rich ligand. We therefore expect the interaction of the tyrosine-based peptide to differ from the consensus rules established for the binding of proline-rich ligands to SH3 domains. The smaller contact surface for the tyrosine-based ligand may account for the lower affinity observed for SKAP55 peptide binding.
Surface plasma resonance measurements indicated that the FYB SH3 domain bound with a higher affinity to the SKAP55 peptide than the FYN and LCK SH3 domains (Figure 4). Given the higher affinity, binding to peptide is expected to involve additional interactions not observed for the FYN SH3 domain to the peptide. In this regard, the FYB SH3 domain differs from other SH3 domains in the absence of key tryptophans (W119 in FYN) that are needed in binding to proline motifs (da Silva et al., 1997). Correspondingly, as mentioned, these same residues failed to undergo a shift perturbation with TRRKGDYASYYQG peptide binding to the FYN SH3 domain (Figure 5). This substitution in FYB may confer a higher affinity interaction of the domain for the tyrosine-based peptide. The FYB SH3 domain also has an extended RT loop that may make additional contacts not encountered in FYN SH3 domain binding. RT loop interactions can increase the affinity of binding as observed in binding between the FYN SH3 domain and the Nef protein of HIV-1 (Arg96–IIe) (Lee et al., 1995, 1996; Saksela et al., 1995). Further studies will be needed to determine the molecular basis of the FYB SH3 domain binding to the TRRKGD YASYYQG motif and the manner by which it differs from the FYN SH3 domain.
A BLAST search identified a limited number of sequences with similarity to the SKAP55 tyrosine-based motif. These sequences include the human TRABID protein (gi6634459), the 26S proteasome subunit (gi6624601), flavocytochrome c fumarate reductase (gi6573311) and nuclear FMRP interacting protein 1 (gi6525071). Preliminary data indicate that the RKxxYxxF motif of SKAP-55 related (SKAP55R) protein (Liu et al., 1998) also binds to the FYB and FYN-T SH3 domains (data not shown). Although the RKxxYxxY motif is limited in expression, further studies will be needed to determine the overall prevalence of SH3 domain binding to tyrosine-based motifs in different biological systems. These interactions could act alone in the mediation of protein–protein interactions, or operate in conjunction with other SH3 domain interactions to increase the overall avidity of binding. For example, while the FYB SH3 domain can recognize tyrosine-based sequences in SKAP55, the SKAP55 SH3 domain can bind to proline-rich sequences in FYB (Liu et al., 1998; Marie-Cardine et al., 1998).
Lastly, the functional importance of the RKxxYxxYY motif in signal transduction was observed with SK4 peptide blockage of TcR up-regulation of IL-2 transcription (Figure 6). Although we cannot attribute this effect specifically to an interaction with SH3 domains, it suggests an importance of the peptide in signaling pathways leading to IL-2 production. Neither of the other SK peptides, such as SK1 and SK3, had an effect on transcription. Inhibition was observed with only a 2- to 3-fold increase in expression of the peptide relative to endogenous SKAP55 expression (data not shown). In addition to Jurkat cells, inhibition was observed in peripheral blood T cells, indicating that the peptide sequence is of importance in signaling by normal T cells. This is one of the first examples of blockage of IL-2 transcription by a single intracellular peptide. A recent report documented inhibition via a calcineurin binding peptide (Aramburu et al., 1999). TRRKGDYASYYQG-based peptides may be useful for suppressing the immune response, which is presently achieved with cyclosporin A, FK506 and related compounds.
Materials and methods
Construction of expression plasmids
Construction of plasmids FYB has been described elsewhere. A portion of the FYB SH3 gene was amplified by PCR. The 5′ primer was 5′-GAAAAAAGGATCCTACGACGGTGAAATTCG-3′ and the 3′ primers were 5′-GCGGCGGCCGCTTAGATTTCTCCCATCATTG-3′ (for FYB SH3 domain, amino acids 695–762), 5′-GTCAGCGGCCGC CTAGCAGCCATCAGCAATATC-3′ (for FYB SH3 domain, amino acids 695–770) and 5′-GAACGCGGCCGCGTGCTAGTCATTGTC ATAGAT-3′ (for FYB SH3 domain, amino acids 695–775). The PCR fragments were then cloned into plasmid pEBG (pEF-BOS-GST; kind gift of Dr B.Mayer, Children’s Hospital, Boston, MA) and pGEX 4T-2 (Phamacia) expression vector at the BamHI and NotI sites. All restriction endonucleases and DNA modification enzymes used for plasmid construction were purchased from New England Biolabs. The full-length SKAP55-HA expression plasmid was made by PCR using the synthetic oligonucleotide primer 5′-GCCGAGCGTCGACCAGGCCGCCGCCCT CCCTGAG-3′ and 5′-GAATATGGTACCTGGGTTTCATCTTTCTT CCAC-3′. The product was digested with SalI and KpnI and then inserted into the SalI and KpnI sites of pSRα-HA expression vector (gift of Dr M.Streuli, Dana Farber Cancer Institute, Boston, MA). The GRB-2 N- and C-terminal SH3 sequences were generated by PCR with the primers 5′-GCGCTCGGATCCGAAGCCATCGCCAAA-3′, 5′-GCCAAAAAC CAGGTACCTCATTTCATTTC-3′, 5′-GCAGGGATCCTACGTCCAG GCCCT-3′ and 5′-GACTCTTAGACGGTACCTTACACGGGGGTG-3′ using full-length GRB-2. The PCR product was digested with SalI and KpnI and cloned into pGEX 4T-2 expression vector. GST–T7 tagged expression vector was constructed by annealing two oligonucleotides, 5′-GAATTGCCACCATGGGAATGGCTAGCATGACTGGTGGACAGC AAATGGGTTCTGAATTCGAGCTC-3′ and 5′-GAGCTCGAATTCA GAACCCATTTGCTGTCCACCAGTCATGCTAGCCATTCCCATGG TGGCAATTC-3′, incubation with Klenow and nucleotides to fill in, and ligation into pGEX 4T-2 expression vector.
PCR-based mutagenesis
Site-directed PCR mutagenesis was used to generate SKAP55 with substitution at Y294F and Y297F using the 5′ primer 5′-GCC GAGCGTCGACCAGGCCGCCGCCCTCCCTGAG and the 3′ primer GGAGTAGACTTTGCCAGTTACTACCAGGGCCTA (for SKAP55, Y294F) or GGAGTAGACTATGCCAGTTTCTACCAGGGCCTA (for SKAP55, Y297F). PCR fragments were digested with SalI and AccI and full-length SKAP55 digested with AccI and KpnI, and the resulting fragments were ligated into the SalI and KpnI sites of pSRα-HA expression vector. The PCR-derived fragment was confirmed by DNA sequence analysis.
Expression and purification of GST fusion proteins
Plasmids were transformed into the DH5α strain of Escherichia coli and induced with isopropyl-β-d-thiogalactopyranoside (IPTG) to produce GST fusion proteins. The bacteria were collected by centrifugation and resuspended in E.coli lysis buffer [40 mM Tris–HCl pH 7.5, 5 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), diisopropyl fluorophosphate and 1% Triton X-100]. Vigorous sonication was performed before centrifugation at 100 000 g for 30 min. The resulting supernatants were saved as crude extracts containing GST fusion proteins. For thrombin cleavage, washed gluthathione–Sepharose beads bound to GST fusion protein were incubated with an empirically determined amount of human thrombin (Sigma) in phosphate-buffered saline (PBS) supplemented with 2.5 mM CaCl2 and the eluted cleaved Flag-tagged FYB proteins were separated from beads by pouring into a disposable column. Na2-EDTA was added to 2.5 mM, and PMSF was added to 1 mM to inactivate thrombin. Purified proteins were stored at –70°C.
Cell culture and DNA transfection
COS-1, Jurkat T cells and mouse spleen cells were maintained in RPMI 1640 medium supplemented with 10% (v/v) fetal bovine serum (FBS; Intergen), 1% (w/v) penicillin and streptomycin (Gibco) and 1% (v/v) l-glutamine (Gibco). COS-1 cells were transfected with cDNAs inserted into the various expression vectors. COS-1 cell transfection was conducted according to standard protocols (Raab et al., 1999).
Immunoprecipitation and immunoblotting
Immunoprecipitation and immunoblotting were performed as described previously (da Silva et al., 1997). Briefly, 2 × 105 COS-1 cells were transfected with cDNA using DEAE–dextran. After 2 days, cells were harvested and lysed with 200 µl lysis buffer [20 mM Tris–HCl pH 8.0, 150 mM NaCl, 1% (v/v) Triton X-100, 1 mM PMSF, 1 mM Na3VO4, 1 mM NaF, 1 mM leupeptin, 1 mM pepstatin and 1% aprotinin]. Immunoprecipitation was carried out by incubation of the lysate with the antibody for 1 h at 4°C, followed by incubation with 50 µl of glutathione–Sepharose beads (50% w/v) for 1 h at 4°C. Immunoprecipitates were washed three times with ice-cold lysis buffer and subjected to SDS–PAGE. For immunoblotting, the immunoprecipitates were separated by SDS–PAGE and transferred on to nitrocellulose filters (Schleicher and Schuell). Filters were blocked with 5% (w/v) skimmed milk for 1 h in Tris-buffered saline (TBS) pH 8.0 and then probed with the indicated antibody. Bound antibody was revealed with horseradish peroxidase-conjugated rabbit anti-mouse antibody using enhanced chemiluminescence (ECL, Amersham). Anti-GST monoclonal antibody was purchased from Santa Cruz Biotechnology, and anti-HA monoclonal antibody from Boehringer Mannheim Biochemicals. Anti-phosphotyrosine monoclonal antibody 4G10 was kindly provided by Dr Tom Roberts (Dana-Farber Cancer Institute, Boston, MA). Anti-FYN-T, LCK and ZAP70 monoclonal antibodies were purchased from Transduction Laboratories.
Peptide binding analysis
Peptides were synthesized and HPLC-purified by the Molecular Biology Core Facility (Dana-Farber Cancer Institute). The sequences of the peptides used were as follows: unphosphorylated peptide TRRKGDYASYYQG and irrelevant peptide TTGVFVKMPPTE. For the binding analysis, peptides were coupled to AminoLink-Plus gel beads (Pierce). The beads were added to the lysate and incubated for 2 h at 4°C. The precipitates were then washed three times with ice-cold lysis buffer and subjected to SDS–PAGE and immunoblotting.
Measurement of peptide binding using surface plasmon resonance
All interactions were determined using a fully upgraded BIAcore 1000 instrument. Various SH3 fusion proteins (10 µg/ml in 100 mM sodium acetate pH 4.5) were immobilized on CM5 Biosensor chips (BIAcore) through cross-linking of free amine groups to the NHS (N-hydroxysuccinimide)/EDC (1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride) activated flow cell surface, followed by blocking of free succinimide ester with 1 M ethanolamine. After extensive washing of the surface with 150 mM NaCl, peptide binding was assessed by injecting the indicated concentrations in 150 mM NaCl over the flow cell surface at 5 µl/min. When required, the surface was regenerated using 1 M NaCl containing 0.05% P20 detergent (BIAcore). The data were analyzed using the BIAevaluation v3.0 software and fitted to a 1:1 Langmuir binding model with separate kd and ka determination. The association constant (Ka) was determined as ka/kd, and the dissociation constant (Kd) as 1/Ka.
Two-dimensional NMR spectroscopy
The FYN domain used for the NMR studies comprises amino acid residues 83–152 of the full-length protein. The expression vector pBAT was used for expression of the recombinant protein in E.coli BL21 (DE3) (Musacchio et al., 1996). The purification of the protein was achieved with a single-step purification on DEAE–Sepharose. The buffer used for the backbone assignment was 50 mM Na2HPO4, 50 mM NaCl pH 6.8. Backbone assignments were achieved using the HNCA (amide proton-to-nitrogen-to-α carbon correlation) experiment (Kay et al., 1990); comparison with the published HSQC spectra of the 15N-labeled FYN domain (Morton, 1996; Mal et al., 1998) helped in initial assignments.
Peptide titration experiments were performed in 20 mM MOPS and 150 mM NaCl. Increasing amounts of the respective peptides were added and HSQC spectra (Sklenar et al., 1993; Mori et al., 1995) were taken after each addition. The combined chemical shift index at an equimolar ratio of 0.5 mM FYN SH3 domain and SK4 peptide was determined as: [(D1H_cs)2 + (D15N_cs)2]1/2, where D1H is in units of 0.1 p.p.m. and D15N in units of 0.5 p.p.m.
Luciferase assay
Jurkat T cells and murine splenocytes (2 × 107) were co-transfected with 20 µg cDNA alone or in combinations plus 3× NFAT-LUC reporter plasmid (kindly provided by Dr Burakoff, Dana-Farber Cancer Institute) and 0.2 µg of pRL-TK (Promega) for normalization of transfection efficiency. Cells were pulsed using a BTX Gene Pulser at 250 mV, 800 µF in 10% FCS. Cells (1 × 106) were aliquoted into a 12 well plate 16 h after transfection and cultured in a final volume of 1 ml RPMI growth medium. Cells were unstimulated or stimulated at 37°C with anti-CD3 (OKT3: 1 µg/ml and 2 µg/ml; 2C11: 2 µg/ml). After 6 h, stimulation cells were lysed in 100 µl lysis buffer (Promega kit) followed by Stop and Go reaction to measure the control reporter plasmid (Dual luciferase system kit; Promega). Luciferase units of the experimental vector were normalized to the level of the control vector in each sample.
References
- Aramburu J., Yaffe,M.B., Lopez-Rodriquez,C., Cantley,L.C., Hogan,P.G. and Rao,A. (1999) Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science, 285, 2129–2133. [DOI] [PubMed] [Google Scholar]
- Chen J.K., Lane,W.S., Brauer,A.W., Tanaka,A. and Schreiber,S.L. (1993) Biased combinatorial libraries: novel ligands for the SH3 domain of phosphatidylinositol 3-kinase. J. Am. Chem. Soc., 115, 12591–12592. [Google Scholar]
- Cicchetti P., Mayer,B.J., Thiel,G. and Baltimore,D. (1992) Identification of a protein that binds to the SH3 region of Abl and is similar to Bcr and GAPρ. Science, 257, 803–806. [DOI] [PubMed] [Google Scholar]
- Clements J.L., Yang,B., Ross-Barta,S.E., Eliason,S.L., Hirstka,R.F., Williamson,R.A. and Koretzky,G.A. (1998) Requirement for the leucocyte-specific adapter protein SLP-76 for normal T cell development. Science, 281, 416–419. [DOI] [PubMed] [Google Scholar]
- da Silva A.J., Li,Z., de Vera,C., Canto,E., Findell,P. and Rudd,C.E. (1997) Cloning of a novel T-cell protein FYB that binds FYN and SH2-domain-containing leukocyte protein 76 and modulates interleukin 2 production. Proc. Natl Acad. Sci. USA, 94, 7493–7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Chen,J.K., Yu,H., Simon,J.A. and Schreiber,S.L. (1994) Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3–ligand interaction. Science, 266, 1241–1247. [DOI] [PubMed] [Google Scholar]
- Geng L., Raab,M. and Rudd,C.E. (1999) Cutting edge: SLP-76 cooperativity with FYB/FYN-T in the up-regulation of TCR-driven IL-2 transcription requires SLP-76 binding to FYB at Tyr595 and Tyr651. J. Immunol., 163, 5753–5757. [PubMed] [Google Scholar]
- Goudreau N., Cornille,F., Duchesne,M., Parker,F., Tocque,B., Garbay,C. and Roques,B.P. (1994) NMR structure of the N-terminal SH3 domain of GRB2 and its complex with a proline rich peptide from Sos. Nature Struct. Biol., 1, 898–907. [DOI] [PubMed] [Google Scholar]
- Jackman J.K., Motto,D.G., Sun,Q., Tanemoto,M., Turck,C.W., Peltz,G.A., Koretzky,G.A. and Findell,P.R. (1995) Molecular cloning of SLP-76, a 76-kDa tyrosine phosphoprotein associated with Grb2 in T cells. J. Biol. Chem., 270, 7029–7032. [DOI] [PubMed] [Google Scholar]
- Kapeller R., Prasad,K.V., Janssen,O., Hou,W., Schaffhausen,B.S., Rudd,C.E. and Cantley,L.C. (1994) Identification of two SH3-binding motifs in the regulatory subunit of phosphatidylinositol 3-kinase. J. Biol. Chem., 269, 1927–1933. [PubMed] [Google Scholar]
- Kay L.E., Ikura,M., Tschudin,R. and Bax,A. (1990) Three-dimensional triple resonance NMR spectroscopy of isotopically enriched proteins. J. Magn. Reson., 89, 496–514. [DOI] [PubMed] [Google Scholar]
- Lee C.-H., Leung,B., Lemmon,M.A., Sheng,J., Cowbrun,D., Kuriyan,J. and Saksela,K. (1995) A single amino acid in the SH3 domain of HcK determines the high afffinity and specifcity of binding to the HIV Nef protein. EMBO J., 14, 5006–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-H., Saksela,K., Mirza,U.A., Chait,B. and Kuriyan,J. (1996) Crystal structure of the conserved core of the HIV-1 Nef complexed with a Src family SH3 domain. Cell, 85, 931–942. [DOI] [PubMed] [Google Scholar]
- Lim W.A., Richards,F.M. and Fox,R.O. (1994) Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature, 372, 375–379. [DOI] [PubMed] [Google Scholar]
- Liu J., Kang,H., Raab,M., da Silva,A., Kraeft,S.-K. and Rudd,C.E. (1998) FYB (FYN binding protein) serves as a binding partner for lymphoid protein and FYN kinase substrate SKAP55 and a SKAP55-related protein in T cells. Proc. Natl Acad. Sci. USA, 95, 8779–8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mal T.K., Matthews,S.J., Kovacs,H., Campbell,I.D. and Boyd,J. (1998) Some NMR experiments and a structure determination employing a [15N,2H] enriched protein. J. Biomol. NMR, 12, 259–276. [DOI] [PubMed] [Google Scholar]
- Marie-Cardine A., Bruyns,E., Eckerskorn,C., Kirchgessner,H., Meuer,S. and Schraven,B. (1997) Molecular cloning of SKAP55, a novel protein that associates with the protein tyrosine kinase p59fyn in human T-lymphocytes. J. Biol. Chem., 272, 16077–16080. [DOI] [PubMed] [Google Scholar]
- Marie-Cardine A., Hendricks-Taylor,L.R., Boerth,N.J., Zhao,H., Schraven,B. and Koretzky,G.A. (1998) Molecular interaction between the Fyn-associated protein SKAP55 and the SLP-76-associated phosphoprotein SLAP-130. J. Biol. Chem., 273, 25789–25795. [DOI] [PubMed] [Google Scholar]
- Mayer B. and Baltimore,D. (1993) Signalling through SH2 and SH3 domains. Trends Cell Biol., 3, 8–13. [DOI] [PubMed] [Google Scholar]
- Mayer B.J. and Gupta,R. (1998) Functions of SH2 and SH3 domains. Curr. Top. Microbiol. Immunol., 228, 1–22. [DOI] [PubMed] [Google Scholar]
- Mongiovi A.M., Romano,P.R., Panni,S., Mendoza,M., Wong,W.T., Musacchio,A., Cesareni,G. and Di Fiore,P.P. (1999) A novel peptide–SH3 interaction. EMBO J., 18, 5300–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S., Abeygunawardara,J.M.O. and van Zijl,P.C. (1995) Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J. Magn. Reson. B, 108, 94–98. [DOI] [PubMed] [Google Scholar]
- Morton C.J. Pugh,D.J., Brown,E.L., Kahmann,J.D., Rezoni, D.A. and Campbell,I.D. (1996) Solution structure and peptide binding of the SH3 domain from human Fyn. Structure, 4, 705–714. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Noble,M., Pauptit,R., Wierenga,R. and Saraste,M. (1992) Crystal structure of a src-homology 3 (SH3) domain. Nature, 359, 851–855. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Saraste,M. and Wilmanns,M. (1994a) High resolution crystal structure of tyrosine kinase SH3 domains complexed with proline-rich peptides. Nature Struct. Biol., 1, 546–551. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Wilmanns,M. and Saraste,M. (1994b) Structure and function of the SH3 domain. Prog. Biophys. Mol. Biol., 61, 283–297. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Cantley,L.C. and Harrison,S.C. (1996) Crystal structure of the breakpoint cluster region-homology domain from phosphoinositide 3-kinase p85 α subunit. Proc. Natl Acad. Sci. USA, 93, 14373–14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musci M.A., Hendricks-Taylor,L.R., Motto,D.G., Paskind,M., Kamens,J., Turck,C.W. and Koretzky,G.A. (1997) Molecular cloning of SLAP-130, an SLP-76-associated substrate of the T cell antigen receptor-stimulated protein tyrosine kinases. J. Biol. Chem., 272, 11674–11677. [DOI] [PubMed] [Google Scholar]
- Nguyen J.T., Turck,C.W., Cohen,F.E., Zuckermann,R.N. and Lim,W.A. (1999) Exploiting the basis of proline recogntion by CH3 and WW domains: design of N-substituted inhibitors. Science, 282, 2088–2092. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 282–293. [DOI] [PubMed] [Google Scholar]
- Noble M.E., Musacchio,A., Saraste,M., Courtneidge,S.A. and Wierenga,R.K. (1993) Crystal structure of the SH3 domain in human Fyn; comparison of the three-dimensional structures of SH3 domains in tyrosine kinases and spectrin. EMBO J., 12, 2617–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T. and Scott,J.D. (1997) Signaling through scaffold, anchoring, and adaptor proteins. Science, 278, 2075–2080. [DOI] [PubMed] [Google Scholar]
- Pivniouk V., Tsitsikov,E., Swinton,P., Rathbun,G., Alt,F.W. and Geha,R.S. (1998) Impaired viability and profound block in thymocyte development in mice lacking the adaptor protein SLP-76. Cell, 94, 229–238. [DOI] [PubMed] [Google Scholar]
- Prasad K.V., Janssen,O., Kapeller,R., Raab,M. Cantley,L.C. and Rudd,C.E. (1993) Src-homology 3 domain of protein kinase p59fyn mediates binding to phosphatidylinositol 3-kinase in T cells. Proc. Natl Acad. Sci. USA, 90, 7366–7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M., da Silva,A.J., Findell,P.R. and Rudd,C.E. (1997) Regulation of Vav-SLP-76 binding by ZAP-70 and its relevance to TcRζ/CD3 induction of interleukin 2. Immunity, 6, 1–11. [DOI] [PubMed] [Google Scholar]
- Raab M., Kang,H., da Silva,A., Zhu,X. and Rudd,C.E. (1999) FYN–FYB–SLP-76 interactions define a T-cell receptor/CD3-mediated tyrosine phosphorylation pathway that up-regulates interleukin 2 transcription in T-cells. J. Biol. Chem., 274, 21170–21179. [DOI] [PubMed] [Google Scholar]
- Ren R., Mayer,B.J., Cicchetti,P. and Baltimore,D. (1993) Identification of a ten-amino acid proline-rich SH3 binding site. Science, 259, 1157–1161. [DOI] [PubMed] [Google Scholar]
- Rudd C.E. (1999) Adaptors and molecular scaffolds in immune cell signaling. Cell, 96, 1–20. [DOI] [PubMed] [Google Scholar]
- Saksela K., Cheng,G. and Baltimore,D. (1995) Proline-rich (PxxP) motifs in HIV-1 Nef bind to SH3 domains of a subset of Src kinases and are required for the enhanced growth of Nef+ viruses but not for down-regulation of CD4. EMBO J., 14, 484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samelson L.E. (1999) Adaptor proteins and T-cell antigen receptor signaling. Prog. Biophys. Mol. Biol., 71, 393–403. [DOI] [PubMed] [Google Scholar]
- Sklenar V., Piotto,M., Leppik,R. and Saudek,V. (1993) Gradient-tailored water suppression for 1H–15N HSQC experiments optimized to retain full sensitivity. J. Magn. Reson., 102, 241–245. [Google Scholar]
- Veale M., Raab,M., Li,Z., da Silva,A., Kraeft,S.-K., Weremowicz,S., Morton,C.C. and Rudd,C.E. (1999) Novel isoform of lymphoid adaptor FYN-T-binding protein (FYB-130) interacts with SLP-76 and up-regulates interleukin 2 production. J. Biol. Chem., 274, 28427–28435. [DOI] [PubMed] [Google Scholar]
- Wardenburg J.B. et al. (1996) Phosphorylation of SLP-76 by the ZAP-70 protein-tyrosine kinase is required for T-cell receptor function. J. Biol. Chem., 271, 19641–19644. [DOI] [PubMed] [Google Scholar]
- Yablonski D., Kuhne,M.R., Kadlecek,T. and Weiss,A. (1998) Uncoupling of nonreceptor tyrosine kinases from PLC-g1 in a SLP-76-deficient T cell. Science, 218, 413–416. [DOI] [PubMed] [Google Scholar]
- Yu H., Rosen,M.K., Shin,T.B., Seidel-Dugan,C., Brugge,J.S. and Schreiber,S.L. (1992) Solution structure of the SH3 domain of src and identification of its ligand-binding site. Science, 258, 1665–1668. [DOI] [PubMed] [Google Scholar]
- Yu H.S., Chen,J.K., Feng,S., Dalgarno,D.C., Brauer,A.W. and Schreiber,S.L. (1994) Structural basis for the binding of proline-rich peptides to SH3 domains. Cell, 76, 933–945. [DOI] [PubMed] [Google Scholar]