Abstract
The propagation of herpesviruses has long been viewed as a temporally regulated sequential process that results from the consecutive expression of specific viral transactivators. As a key step in this process, lytic viral DNA replication is considered as a checkpoint that controls the expression of the late structural viral genes. In a novel genetic approach, we show that both hypotheses do not hold true for the Epstein–Barr virus (EBV). The study of viral mutants of EBV in which the early genes BZLF1 and BRLF1 are deleted allowed a precise assignment of the function of these proteins. Both transactivators were absolutely essential for viral DNA replication. Both BZLF1 and BRLF1 were required for full expression of the EBV proteins expressed during the lytic program, although the respective influence of these molecules on the expression of various viral target genes varied greatly. In replication-defective viral mutants, neither early gene expression nor DNA replication was a prerequisite for late gene expression. This work shows that BRLF1 and BZLF1 harbor distinct but complementary functions that influence all stages of viral production.
Keywords: Epstein–Barr virus/immediate early genes/lytic DNA replication
Introduction
Herpesviruses encode between 60 and 100 genes, and productive lytic infection is characterized by a temporally regulated cascade in which three distinct kinetic classes of transcripts are expressed in a sequential manner (Honess and Roizman, 1974; Honess and Roizman, 1975; for a review, see Roizman and Sears, 1996). Immediate early (α) gene products are expressed under conditions of lytic infection in the absence of de novo protein synthesis and orchestrate the ensuing transcriptional cascade. The early (β) genes and their products are involved primarily in viral DNA replication and comprise the essential virus-encoded DNA replication machinery and a number of enzymes that contribute to the biosynthesis of deoxynucleoside triphosphates. The late (γ) genes are subdivided into two classes based on their apparent requirement for DNA replication (Roizman and Sears, 1996). The expression of true late genes, also called γ2 genes, seems to be strictly dependent on the onset or completion of lytic DNA replication since their expression is not observed in the presence of inhibitory concentrations of drugs that block viral DNA synthesis or with herpesviruses carrying conditional mutations in early genes (Honess and Roizman, 1974; Powell et al., 1975; Ward and Stevens, 1975; Holland et al., 1980; Conley et al., 1981; Mavromara-Nazos and Roizman, 1987, 1989).
In contrast to the majority of herpesviruses, infection with members of the γ-herpesvirus family rarely leads to the production of viral particles (for a review, see Rickinson and Kieff, 1996). Instead, a state of latency is readily established in which the viral DNA is transmitted to the daughter cells. Nevertheless, reactivation from this state of equilibrium to the productive, lytic phase can occur. As a consequence, the host cell is destroyed and viral particles are assembled and released. In the case of Epstein–Barr virus (EBV), the physiological conditions that lead to a reactivation of the latent virus are not clearly defined, but expression of the viral gene BZLF1 is sufficient for lytic production in permissive cells (Countryman and Miller, 1985; Takada et al., 1986). BZLF1 is a transactivator that shows homologies to members of the fos family of proteins (Farrell et al., 1989; Urier et al., 1989) and has been implicated in the transcription of human and viral genes (for a review, see Speck et al., 1997). Several studies have identified a second viral transactivator, BRLF1, as having the capacity to induce the EBV lytic program (Hardwick et al., 1988; Zalani et al., 1996; Ragoczy et al., 1998). These observations suggested that both proteins have redundant properties. An additional level of complexity stems from the observation that the BZLF1 protein activates the promoters of BZLF1 and BRLF1 and, vice versa, the BRLF1 gene product up-regulates the expression of the BZLF1 gene (Sinclair et al., 1991; Ragoczy et al., 1998).
According to the model initially based on the study of the herpes simplex virus, the expression of the γ2 late genes of herpesviruses is strictly dependent on the completion of lytic viral DNA replication (see above). An identical model has been proposed for EBV on the basis of experiments using inhibitors of the viral DNA polymerase, such as phosphonoacetic acid (PAA) (Summers and Klein, 1976; Yajima et al., 1976; Datta and Hood, 1981). Incubation of EBV-infected cells in the presence of PAA blocks not only the viral lytic DNA replication, but also the expression of EBV late genes (Serio et al., 1997). This assumption is supported by the findings with the Raji cell line, which carries a mutant EBV. The Raji EBV genome contains a deletion in the virally encoded single-stranded DNA-binding protein BALF2 and is defective for lytic viral DNA replication and late gene expression (Baer et al., 1984; Decaussin et al., 1995).
Since the experimental link between herpesvirus DNA replication and progression through the different phases of the lytic viral program was only indirect, we wished to address this question using a genetic approach. Recently, we have succeeded in cloning the complete EBV genome in Escherichia coli on a single plasmid (Delecluse et al., 1998). After DNA transfection into 293 cells, this EBV genome is fully functional in that the latent phase of EBV’s life cycle is maintained and production of infectious virion particles can be induced at will. In addition, any mutation can be introduced into the EBV genome in E.coli such that even genetic alterations that affect essential genetic elements can be studied (Delecluse et al., 1999). In order to examine the genetic requirement for induction of the lytic phase of EBV as well as its full execution, we introduced two non-functional mutations in the EBV genome. By insertional mutagenesis, the BZLF1 and BRLF1 genes were disrupted, rendering the mutant genomes non-functional in terms of virion synthesis. The mutant virus genomes were termed BZLF1-KO and BRLF1-KO, respectively. They were transferred via DNA transfection into 293 cells in order to obtain latently infected cell lines (Delecluse et al., 1998). The ability of the BZLF1 and BRLF1 immediate early genes to induce early and late gene expression and DNA replication was analyzed in these cell lines.
We demonstrate here that both transactivators are required for early and late gene expression as well as for viral DNA replication. It appears that (i) both BZLF1 and BRLF1 are mandatory at all stages of virion production and (ii) have non-redundant functions. Unexpectedly, lytic DNA replication is not required to allow expression of some late genes of this herpesvirus.
Results
Construction of the EBV mutant viruses
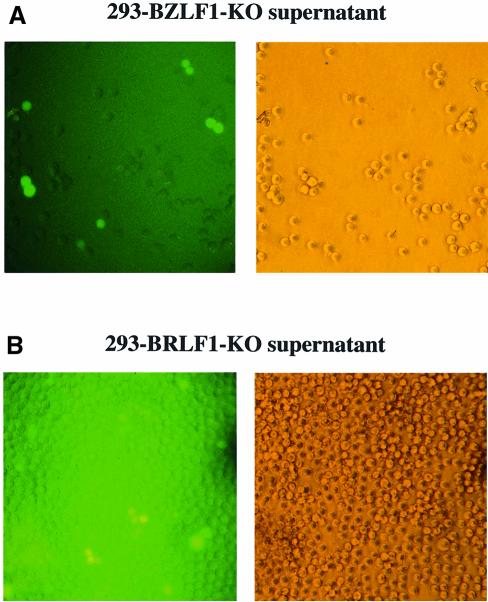
The complete genome of the B95.8 strain of EBV cloned onto an F-factor replicon, termed EBV/F-factor, was moved into a recA-positive, recBC-negative E.coli strain (BJ5183) (Delecluse et al., 1998). This strain allows further genetic manipulation with linear DNA fragments by homologous recombination (Hanahan, 1983; Delecluse et al., 1999). The BZLF1 gene and the BRLF1 gene were disrupted by insertional mutagenesis with the aid of the kanamycin or tetracycline resistance gene using the method of targeted allelic exchange as described in Materials and methods. In order to confirm the successful disruption of the target gene, DNA plasmids derived from bacterial clones that carry individual recombinant EBV/F-factor molecules were digested with restriction enzymes (Figure 1). DNAs were analyzed further by Southern blot analysis with specific probes. The results confirmed the mutations of the two loci (data not shown). The modified genomic EBV DNAs (BZLF1-KO and BRLF1-KO) were used to stably transfect 293 cells that have been shown to support the EBV lytic program (Delecluse et al., 1998). After transfection of the BZLF1-KO DNA and hygromycin selection, two cell clones grew out. The clones were green fluorescent protein (GFP) positive (the F-factor replicon also carries the gene for GFP) and encompassed the rearranged BZLF1 locus by Southern blot analysis (data not shown). An identical approach gave rise to the generation of ten 293 cell clones that carry a viral strain in which the BRLF1 gene was deleted (data not shown). For convenience, these cells are called 293-BZLF1-KO and 293-BRLF1-KO throughout the manuscript. To confirm the integrity of the BZLF1-KO mutant, 293-BZLF1-KO cells were transfected with a BZLF1 expression plasmid and cell-free supernatants were used to infect Raji cells. The supernatants proved to contain infectious particles as indicated by GFP expression (Figure 2). The viral concentration in these supernatants was ∼103 viral particles/ml as estimated by counting the proportion of GFP-positive Raji cells. Similar results were obtained by transfection of a BRLF1 expression plasmid into the 293-BRLF1-KO cells (Figure 2). These experiments show that the phenotype introduced by disruption of the BZLF1 or BRLF1 gene can be functionally complemented in trans by the introduction of the missing proteins.
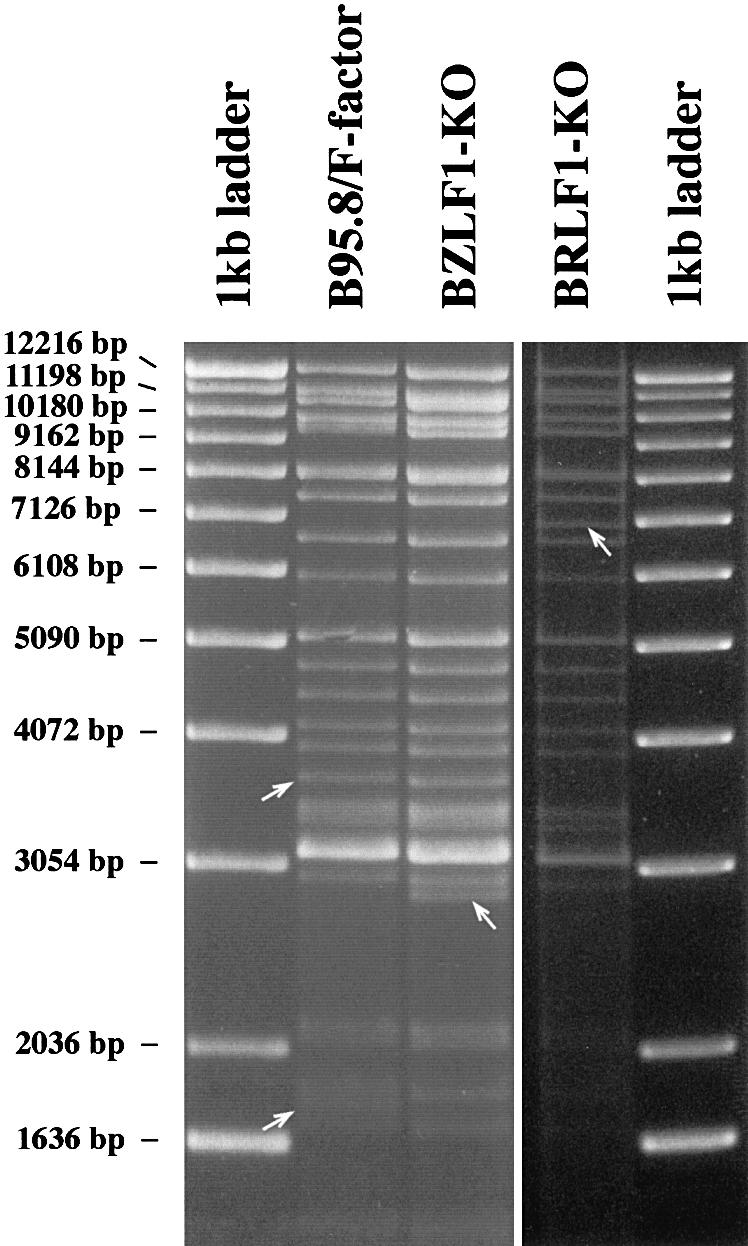
Fig. 1. Restriction fragment analysis of both BZLF1-KO and BRLF1-KO mutant DNAs in comparison with the wild-type B95.8/F-factor DNA. The BZLF1 gene was exchanged against the kanamycin resistance gene via homologous recombination. DNA was purified from a chloramphenicol- and kanamycin-resistant E.coli clone and digested with the BamHI restriction enzyme. The restriction pattern of the mutant BZLF1-KO DNA was compared with that obtained with wild-type B95.8/F-factor DNA. Both restriction fragment patterns were identical, with two exceptions. A new fragment was generated by replacement of the BamHI fragment that carries the BZLF1 gene (disappearance of the 1.8 kb fragment in B95.8/F-factor) with the one that carries the kanamycin resistance gene (appearance of a 2.8 kb fragment in BZLF1-KO DNA) (see also Figure 6A). Both rearranged fragments are indicated by an arrow. The same procedure was applied to the BRLF1 gene. In this case, the 3.6 kb BamHI fragment that carries the BRLF1 gene was exchanged with a 6.9 kb fragment that carries the tetracycline resistance gene.
Fig. 2. Complementation of the BRLF1 and BZLF1 mutants. GFP expression in Raji cells incubated with supernatants from 293-BZLF1-KO cells transfected with an expression plasmid encoding BZLF1 (A) or from 293-BRLF1-KO cells transfected with BRLF1 (B). A 1 ml aliquot of supernatant was mixed with 104 Raji cells. GFP fluorescence was investigated 48 h after infection (left panel); corresponding phase contrast light microscopy (right panels) (magnification ×200). After incubation with supernatants from 293-BZLF1-KO cells, ∼10% of the Raji cells were GFP positive, indicating a virus titer of at least 103 infectious viruses/ml. In the case of 293-BRLF1-KO cells, the virus titer could also be estimated as being at least 103 infectious viruses/ml.
BZLF1 and BRLF1 mutually induce their expression
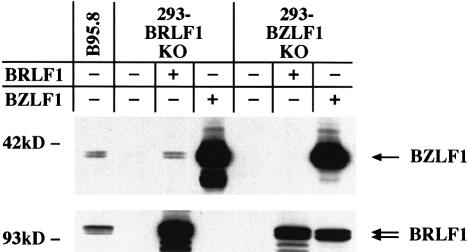
We first wished to test the ability of both immediate early genes to activate their expression mutually. As expected, transfection of BZLF1 into the 293-BZLF1-KO cell line resulted in expression of the BRLF1 protein (Figure 3, lower panel). Conversely, the transfection of the BRLF1 expression plasmid into the 293-BRLF1-KO cell line led to the synthesis of the BZLF1 protein, as shown by western blots with monoclonal antibodies (Figure 3, upper panel).
Fig. 3. Reciprocal induction of BZLF1 and BRLF1. Western blot analysis of 293-BZLF1-KO or 293-BRLF1-KO cells transfected with an expression vector encoding either BRLF1 or BZLF1. Upper panel: BZLF1 expression pattern. Lower panel: BRLF1 expression pattern. The B95.8 cell line that spontaneously produces EBV virions was used as a positive control. Complementation of 293-BRLF1-KO with a BRLF1 expression plasmid or of 293-BZLF1-KO with a BZLF1 expression plasmid leads to the expression of both transactivators, showing that BZLF1 induces the expression of BRLF1, and vice versa. In 293-BZLF1-KO cells transfected with a BRLF1 expression plasmid or in 293-BRLF1-KO cells transfected with a BZLF1 expression plasmid, the expression pattern was limited to the expression of the transfected gene. Untransfected 293-BZLF1-KO or 293-BRLF1-KO cells provided additional negative controls.
Characterization of the 293-BZLF1-KO cell line
Using the 293-BZLF1-KO cell line, we investigated the role of BRLF1 in the viral lytic program, the contribution of BZLF1 to B-cell immortalization and the molecular basis for the ability of 12-O-tetradecanoyl-13-acetate (TPA) and butyrate to activate the lytic program in permissive cells.
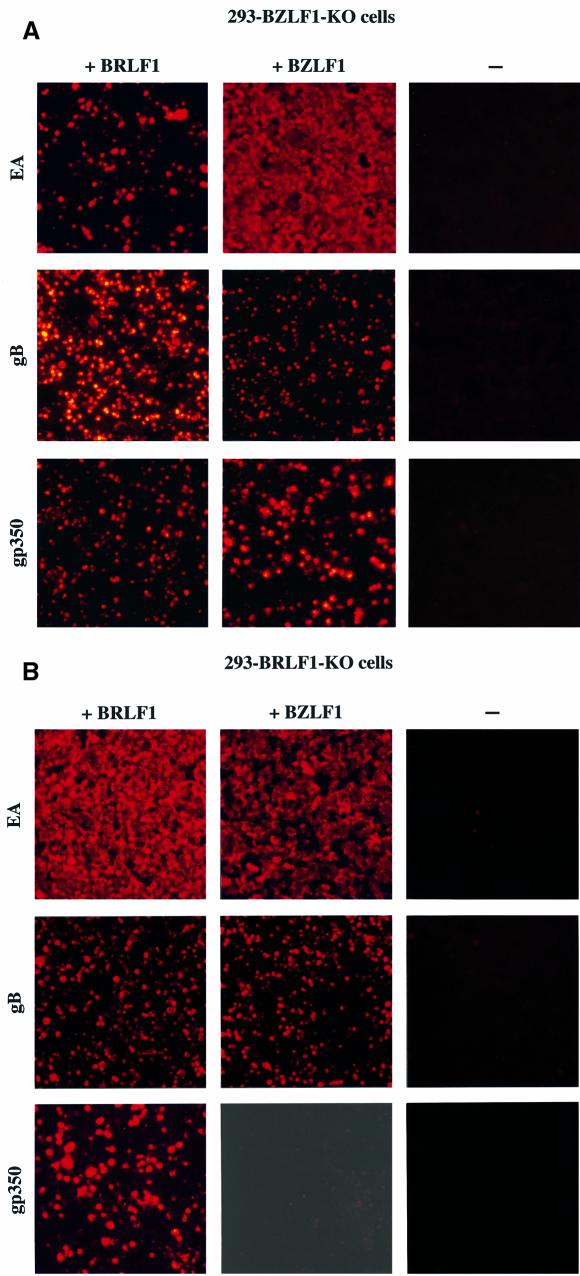
Lytic DNA replication was readily restored after transfection of the BZLF1 gene (Hammerschmidt and Sugden, 1988) into the 293-BZLF1-KO cells, as expected (Figure 4). Similarly, all early and late viral proteins tested could be detected after transfection with BZLF1 (Table I; Figure 5A). In contrast, lytic DNA replication did not take place nor was virus released when 293-BZLF1-KO cells were transfected with BRLF1 alone (Table I; Figure 4). Furthermore, only limited expression of the early antigen EA-D (BMRF1) could be observed in these settings (Table I). Both findings could be related since EA-D antigens include proteins that are mandatory for the formation of a functional DNA replication complex (Fixman et al., 1992, 1995). The expression of the three viral late proteins gB (or gp125 encoded by the BALF4 gene), p160 [or major viral capsid protein (m-VCA) encoded by the BcLF1 gene] and the viral envelope glycoprotein gp350 (encoded by the BLLF1 gene) was analyzed further in the cell line that carries the BZLF1-KO mutant virus genome, by immunostaining techniques (Figure 5A; Table I). The results of these experiments are summarized in Table I. They show that gB-positive and gp350-positive cells could be detected in 293-BZLF1-KO cells after transfection with the BRLF1 gene. In contrast, staining for the m-VCA antigen stayed negative. Transfection with a control vector consisting of the empty expression plasmid backbone alone did not give rise to any viral gene expression (data not shown). After introduction of BRLF1 or BZLF1 into 293-BZLF1-KO cells, the proportion of positive cells for the late viral proteins gp350 and gB was comparable (Table I). However, the intensity of staining for gp350 was slightly stronger after transfection with BZLF1 than after transfection with BRLF1. The percentage of gB-positive cells corresponded directly to the percentage of BRLF1- positive cells (Table I).
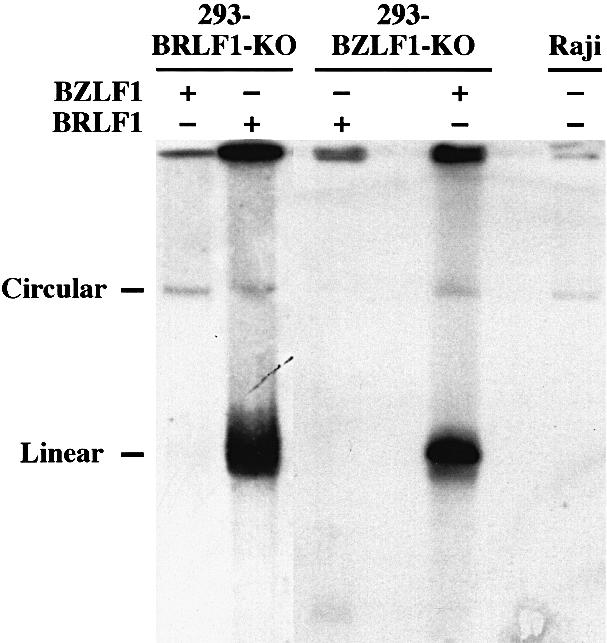
Fig. 4. Analysis of DNA replication in BZLF1-KO and BRLF1-KO viral mutants after transfection of expression plasmids that carry the BZLF1 or BRLF1 genes. In cells latently infected with EBV, the viral DNA is present as an episome. In contrast, during lytic replication, newly synthesized viral DNA is linear. Both circular and linear forms can be distinguished readily after separation on a Gardella agarose gel electrophoresis, followed by Southern blot hybridization with an EBV-specific probe. Lytic replication was clearly identified after transfection of BZLF1 into the 293-BZLF1-KO cell line or of BRLF1 into the 293-BRLF1-KO cell line. In contrast, neither transfection of BRLF1 into the 293-BZLF1-KO cell line nor transfection of BZLF1 into the 293-BRLF1-KO cell line could give rise to any detectable DNA replication. The cell line Raji is defective for the lytic program and was used as a negative control.
Table I. Viral protein expression pattern.
| Cells | Plasmids | Anti-BRLF1 | Anti-BZLF1 | Anti-EAa | Anti-gB | Anti-m-VCA | Anti-gp350 | Raji cell infectionb |
|---|---|---|---|---|---|---|---|---|
| 293-BRLF1-KO | BRLF1 | +++ | +++ | +++ | +++ | +++ | +++ | yes |
| BZLF1 | – | +++ | +++c | + | – | – | no | |
| 293-BZLF1-KO | BRLF1 | +++ | – | + | +++c | – | ++ | no |
| BZLF1 | +++ | +++ | +++ | +++ | +++ | +++ | yes |
+, <2% of cells are positive; ++, 2–15% of cells are positive; +++, >15% of cells are positive.
aOwing to the diffuse character of the staining, a precise evaluation of the number of positive cells is difficult. +++ indicates that the majority of the cells are positive, + indicates that <10% are positive.
b10% of the Raji cells are positive for GFP after incubation with supernatants from the indicated cell line after transfection with the indicated plasmids.
cStaining less intense than that obtained with the wild-type virus.
Fig. 5. Production of the EBV lytic proteins EA-D, gB and gp350 after transfection of a BRLF1 or BZLF1 expression plasmid into 293-BZLF1-KO cells (A) or 293-BRLF1-KO cells (B). In each panel, a negative control consisting of untransfected cells is included. Fixed cells were incubated with a specific monoclonal antibody and a second anti-mouse antibody coupled to the Cy5 fluorochrome. Stained cells were visualized under UV light (magnification ×100).
As EBV predominantly infects B cells, we extended our experiments to include these cells. Three immortalized B-cell lines generated with the BZLF1-KO mutant (see the next paragraph) were transfected with BZLF1 and BRLF1. In the immortalized B-cell lines, due to the low transfection efficiency, only a few cells (<1 out of 1000) showed expression of both gB and gp350 after transfection with BZLF1 (data not shown). Transfection with BRLF1 alone led to infrequent expression of BRLF1, gB and gp350 (<1 cell out of 1000 was positive for these proteins, data not shown). In general, these results are in agreement with those obtained in 293 cells. In conclusion, the BRLF1 protein alone leads to synthesis of some but not all EBV late genes in lytic replication-defective EBV mutants, in both epithelial and lymphoid cells, but the viral lytic program is not fully complete in the absence of BZLF1.
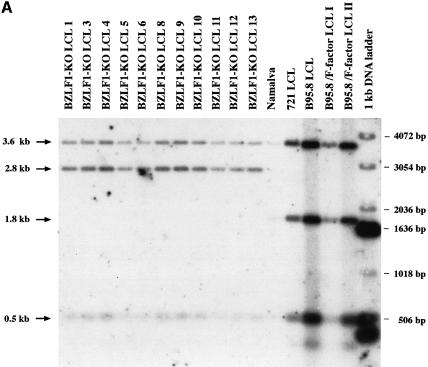
To determine whether BZLF1 contributes to the immortalizing potential of EBV, virus stocks were produced after transfection of the BZLF1 gene into 293-BZLF1-KO cells and used to infect primary B lymphocytes from the peripheral blood of a healthy donor. Four weeks after infecting 107 primary B cells, 20 cell clones grew up that expressed GFP. B cells from the same donor were also infected with supernatants from the B95.8 cell line. No difference in cell proliferation was noticed between the established cell lines generated with the BZLF1-KO virus or with B95.8 virus (data not shown). In order to exclude the possibility that the virus that led to B-cell immortalization was the result of recombination between the genome of the BZLF1-KO virus and the transfected BZLF1 expression plasmid, Southern blot analysis was performed. None of the B-cell lines obtained after infection with BZLF1-KO virus showed any evidence of such recombination events (Figure 6A). Similarly, when one of the immortalized B-cell lines was induced with TPA and butyrate for 3 days and tested for BZLF1 expression by western blot analysis, no expression of BZLF1 could be detected (Figure 6B). Therefore, it appears that immortalized B-cell lines infected with either wild-type EBV or the BZLF1-KO mutant virus do not differ with regard to B-cell proliferation.

Fig. 6. Immortalized B cells generated by infection of primary lymphoid cells with the BZLF1-KO EBV mutant do not carry the BZLF1 gene. (A) Southern blot analysis of cell lines infected either with the BZLF1-KO mutant or with wild-type EBV. Blots were hybridized with the construct used to generate the BZLF1-KO mutant that consists of the kanamycin resistance gene flanked by the first two-thirds of the first BZLF1 exon and sequences 5′ and 3′ from this gene. Whereas the wild-type EBVs carry the intact BZLF1 gene (1.8 kb fragment), only recombined fragments could be detected in the cell population infected by the viral mutant (2.8 kb fragment) (see also Figure 1). (B) Western blot analysis of cell lines infected either with the BZLF1-KO mutant or with wild-type EBV. After TPA and butyrate induction, cells that carry the wild-type EBV DNA (B95.8 and B95.8 LCL), but not those that carry the EBV viral mutant (BZLF1-KO LCL9), show clear production of the BZLF1 protein. Proteins extracted from the above-described lymphoid cell lines (20 µg for the BZLF1-KO mutant, 4 µg for the control cells) were separated by SDS–PAGE, blotted onto a nitrocellulose membrane and incubated with an antibody specific to the BZLF1 protein (Z130).
Co-culturing of 293 cells that carry the complete EBV genome with primary human B lymphocytes leads to the generation of immortalized B-cell lines (Sugden, 1984; Delecluse et al., 1998). In contrast, co-culturing of primary B lymphocytes with 293-BZLF1-KO cells did not give rise to any immortalized B cells even after incubation for >6 weeks on feeder cells. In a very similar experiment, treatment of 293-BZLF1-KO cells with TPA and butyrate, two drugs known to activate EBV’s lytic program in latently infected cells (zur Hausen et al., 1978), followed by extensive washings and co-incubation with primary B lymphocytes, did not give rise to immortalized cells. In line with these observations, no GFP-positive cells could be detected after incubation of Raji cells with culture supernatants from 293-BZLF1-KO cells induced with TPA and butyrate. In conclusion, the BZLF1-KO mutant definitely requires BZLF1 in trans for virion production.
Characterization of the 293-BRLF1-KO cell line
The 293-BRLF1-KO cell line allows an evaluation of the functions of BZLF1 in the absence of BRLF1. Transfection of a BZLF1 expression plasmid into the 293-BRLF1-KO cell line was followed by strong expression of the EA-D antigen (Figure 5B). However, the level of EA-D expression was less intense than that obtained after introduction of BRLF1 into the 293-BRLF1-KO cells (Figure 5B; Table I). No DNA replication could be observed after transfection of 293-BRLF1-KO cells with BZLF1, using a Gardella gel analysis, indicating that BRLF1 is absolutely required for viral lytic DNA replication (Figure 4). At this stage, we cannot distinguish between a direct contribution of BRLF1 itself to the lytic replication, and an indirect mechanism in which BRLF1 activates viral proteins required for DNA replication. Finally, the pattern of expression of the late EBV proteins was investigated in 293-BRLF1-KO cells transfected with BZLF1. Whereas no expression of the gp350 protein or the m-VCA antigen could be detected, the gB protein was found to be expressed, although the level of expression was lower than that observed after transfection with BRLF1 (Figure 5B; Table I).
The BRLF1 gene product induces the expression of the gp350 gene in EBV-negative cells
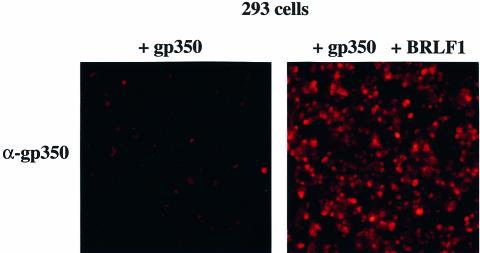
In order to dissect further the observed effect of BRLF1 on the expression of the EBV late genes, we cloned the gp350 gene and its promoter region onto a simple prokaryotic vector plasmid. In a transient transfection assay, this plasmid was introduced alone and in combination with the BRLF1 expression plasmid into EBV-negative 293 cells. The gp350 gene was expressed at a low level when transfected alone, but strong induction was observed when both BRLF1 and gp350 were introduced concomitantly into these cells (Figure 7). This result indicates that BRLF1 alone is able to activate the expression of this EBV late gene even in the absence of the viral genome.
Fig. 7. The BRLF1 immediate early protein induces the expression of gp350 in EBV-negative cells. Expression of the EBV late protein gp350 after transfection of the gp350 gene that codes for gp350 (left panel, magnification ×100) and after co-transfection of the gp350 gene and BRLF1 into 293 cells (right panel, magnification ×100).
To ensure that the observed activation of gp350 gene expression by the gp350 protein reflects the situation during the lytic program of EBV, the 5′ terminus of the gp350 mRNA was analyzed using primer extension. In this assay, a radiolabeled oligonucleotide specific for gp350 mRNA was used as a primer for reverse transcription of total RNA extracted from B95.8 cells, 293-BZLF1-KO cells transfected with a BRLF1 expression plasmid and 293 cells co-transfected transiently with the gp350 gene and a BRLF1 expression plasmid. The result of this experiment is shown in Figure 8. This experiment confirmed the ability of BRLF1 to activate the expression of gp350, even in an EBV-negative cell. The level of expression of the gp350 gene in 293-BZLF1-KO induced with the expression vector for BRLF1 was comparable in intensity to that observed in B95.8 cells as a positive control. In B95.8 cells, this analysis revealed two slightly divergent gp350 mRNAs. In all cases, the 5′ terminus of the generated gp350 mRNA was nearly identical, suggesting that BRLF1 induces the gp350 gene from the identified promoter (Beisel et al., 1985).
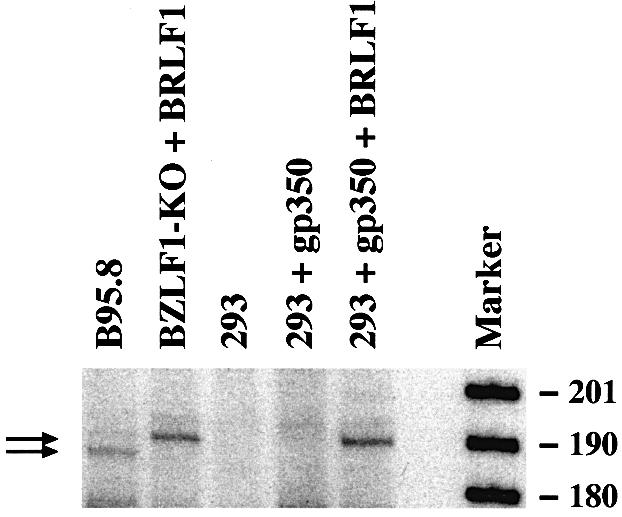
Fig. 8. The BRLF1 protein activates the expression of the gp350 gene from its established promoter. The 5′ end of the gp350 mRNA from 293 or 293-BZLF1-KO cells transfected with an expression plasmid encoding BRLF1 was determined using primer extension. B95.8 cells and untransfected 293 cells provided the positive and negative controls, respectively. Introduction of a BRLF1 expression plasmid into 293 or 293-BZLF1-KO cells gave rise to the synthesis of a gp350-specific mRNA, similar in size to the gp350 mRNA found in the B95.8 cells. The BRLF1-mediated activation of the gp350 mRNA therefore reflects the situation during the EBV lytic program in B95.8 cells. However, an additional gp350 mRNA transcript, slightly shorter in size, could also be detected in B95.8. At this stage, it is not clear whether this reflects the fact that the B95.8 cell line is derived from marmoset cells, or the contribution of additional viral factors apart from BRLF1 during viral lytic replication.
Discussion
As a member of the herpesvirus family, EBV codes for a complex protein network, and the switch between the latent and the lytic gene expression program illustrates this complexity. The observation that only a minority of cell lines infected with the same viral strain will support the EBV lytic cycle spontaneously suggests that cellular conditions interfere with various stages of virus synthesis. Therefore, the appropriate experimental setting for the dissection of the viral molecular mechanisms that lead to the production of infectious particles is EBV-positive cell lines that support the lytic program, in which the viral genes that govern virus production can be induced or suppressed at will. Similarly, a genetic approach with knock-out mutants of the different genes and cis-elements is critical to dissect essential aspects of this program. In this study, we analyze two viral mutants that lack either the BZLF1 gene or the BRLF1 gene. These mutations, which both block the completion of the lytic phase of EBV’s life cycle at different stages, allowed us (i) to illustrate the functions of the immediate early genes BRLF1 and BZLF1 in an independent manner and (ii) to analyze the link between lytic DNA replication and late gene expression. As a technical prerequisite, cloning of the complete EBV genome in a prokaryotic host turned out to be an important step towards the generation of loss-of-function viral mutations.
The first information we gained from the analysis of these EBV mutants is that both BRLF1 and BZLF1 genes can reactivate the viral genome from its latent state with comparable efficiencies. In the context of a natural infection, it is unclear which of the two proteins is activated first. Our data argue against a scenario that puts the BZLF1 protein upstream of the BRLF1 protein in the lytic cascade. This observation does not rule out cellular factors in some cell types further modulating the interaction between both transactivators, and therefore accounts for apparently contradictory reports in the literature regarding the ability of BRLF1 to induce the lytic program in B cells.
A step-by-step analysis of the viral synthesis showed that both BRLF1 and BZLF1 are required to complete the lytic program (Figures 2–5; Table I). The production of the EA-D viral antigen is mainly the result of BZLF1 expression, although BRLF1 can induce this antigen at a low level. Both BZLF1 and BRLF1 were found to be necessary for lytic DNA replication. At present, it is not clear whether BRLF1 interacts directly with the DNA molecule during viral DNA replication, as has been proven for BZLF1, or whether its contribution to replication is limited to the induction of proteins that act in trans on viral DNA replication. 293 cells that carry the BZLF1-KO mutant genome in a latent form do not release virus spontaneously or after stimulation with TPA and/or butyrate. This mutant genome can be induced to undergo virus production only after transfecting an expression plasmid encoding BZLF1. This result confirms previous observations that TPA and butyrate activate the EBV lytic program indirectly by induction of BZLF1 (Flemington and Speck, 1990; Speck et al., 1997 and references therein). Extending our observations to the late viral proteins, we show here that expression of the gp350 protein absolutely requires BRLF1 but does not seem to be influenced directly by BZLF1. The ability of BRLF1 to induce the synthesis of the gp350 transcript could be confirmed using primer extension. The gp350 mRNA produced after introduction of a BRLF1 expression plasmid into the 293-BZLF1-KO cells was very close in size to that identified in the B95.8 cell line, which supports the EBV lytic program spontaneously, indicating nearly identical promoter usage. This provides additional support for a physiological role for BRLF1 in inducing the expression of the gp350 gene. BRLF1 remains less potent than BZLF1 in inducing the expression of gp350 in 293-BZLF1-KO cells, which means that both transactivators together are more effective than BRLF1 alone (Table I). Therefore, BZLF1 must contribute indirectly to the expression of gp350. Other genes, such as gB, seem to be induced by both immediate early proteins, but again wild-type level of expression could be obtained only with a combination of BRLF1 and BZLF1. Finally, the induction of the p160 component of the VCA complex could be observed only when both transactivators were present, whereas the single expression of one of them was ineffective (Table I). These results indicate that both transactivators are required at all steps of the lytic program, mainly in a co-operative manner, although the contributions of both immediate early proteins varies greatly with the target gene. Using the Raji cell line, Ragoczy and Miller (1999) partially reached our conclusions with regard to the viral protein expression pattern. These authors found that in Raji cells, BZLF1 has a negative effect on the ability of BRLF1 to induce late gene expression. We did not find any evidence for such an effect in our system, but in contrast showed that BZLF1 itself induces the expression of the gB protein and clearly contributes indirectly to the expression of gp350 and m-VCA. The fact that BRLF1 cannot induce the expression of BZLF1 in Raji cells shows that this cell line has encountered additional modifications that render a precise analysis of the function of both transactivators difficult to interpret.
The co-transfection of BRLF1 and gp350 in EBV-negative 293 cells led to a strong expression of the late protein, indicating that gp350 expression is independent of other viral proteins and/or ongoing DNA replication (Figure 7). This observation does not rule out other viral proteins positively or negatively modulating the expression of gp350 or other late genes. As no typical BRLF1-specific binding site could be detected in the promoters of the genes for m-VCA and the gp350, no direct mechanism for this induction currently can be proposed (Gruffat et al., 1990). Using primer extension, we could confirm the ability of BRLF1 to activate the expression of the gp350 gene in an EBV-negative cellular context. This experiment also showed that the gp350 mRNA starts from the genuine promoter region, reinforcing the idea that the observed transactivating potential of BRLF1 was specific.
The analysis of three immortalized B-cell lines generated with the viral BZLF1-KO mutant showed the same results with regard to the ability of BRLF1 to induce the expression of the late genes, although the number of gB- and BRLF1-positive cells was much lower, reflecting the technical difficulties in transfecting newly established immortalized B-cell lines.
The viral particles that were produced after transfection of BZLF1 in 293-BZLF1-KO cells were infectious and retained their ability to immortalize B cells, indicating that the BZLF1 gene is not required in cis for the maintenance of B-cell immortalization. This result does not rule out a contribution of the BZLF1 gene product at the level of initiation of B-cell immortalization since the BZLF1 protein might be incorporated into the virion particle, as has been discussed before (Serio et al., 1996), similarly to the herpes simplex virus transactivator VP16 (Vmw65) (Post et al., 1981; Preston et al., 1984; Campbell and Villarreal, 1988).
In both 293-BZLF1-KO and 293-BRLF1-KO cells, late gene expression could be achieved even in the absence of DNA replication (Figures 4 and 5). At first sight, this observation contradicts previous work that showed a strict dependency of viral late gene expression on viral DNA replication (for a review see Roizman and Sears, 1996). In theory, viral replication can influence gene expression either by providing a multitude of viral templates available for transcription of late genes, or by giving rise to unmodified, e.g. unmethylated, or, as is the case for herpesviruses, even histone-free DNA progeny. Our results clearly show that the amplification of the viral DNA template or ongoing lytic DNA replication is not required for late gene expression. One theoretical possibility would be that both transactivators induce a modification of the viral DNA, e.g. demethylation, independently of viral replication per se. The transfection of BZLF1 into the 293-BZLF1-KO cell line did not influence the viral methylation status (our unpublished data), but additional DNA modifications cannot be excluded. Serio et al. reported data on the basis of transient reporter assays in which synthesis of the viral late proteins followed activation of the EBV lytic program, but in which a bona fide replication of the DNA template to be transcribed was not required (Serio et al., 1997, 1998). This type of experiment has been debated controversially in the field of herpes simplex virus (Roizman and Sears, 1996). One of the central arguments that favors the existence of a link between EBV lytic DNA replication and viral gene expression is the indirect inhibition of late genes in lytically induced cells by compounds such as PAA or others that interfere with viral DNA replication (see Introduction). Since, to our knowledge, no universal genetic system to study the lytic phase of herpesvirus replication has been available previously, the possibility that PAA directly influences the expression of late genes has not been tested. In line with this assertion, we found that PAA reduces, but does not completely inhibit, the transactivating effects of BRLF1 on the gp350 gene in EBV-negative 293 cells (our unpublished observations).
In conclusion, the analysis of these two EBV mutants unequivocally assigned different but co-operative roles to the BRLF1 and BZLF1 viral genes at each step of the viral lytic synthesis. Our study underscores the potential of genetic analysis to address essential lytic functions of herpesviruses.
Materials and methods
Cells
The 293 line is a transformed epithelial cell line whose differentiation stage is not fully characterized (Graham et al., 1977). Raji and Namalwa are EBV-positive human Burkitt’s cell lines (Pulvertaft, 1964; Nyormoi et al., 1973). B95.8 is a lymphoblastoid cell line obtained by infection of marmoset monkey peripheral blood leukocytes with EBV (Miller et al., 1972). B lymphocytes from peripheral blood buffy coats, adenoids and tonsils were purified on a Ficoll cushion after T-cell rosetting using sheep erythrocytes as described (Zeidler et al., 1996). All cell lines were grown in RPMI 1640, 10% fetal calf serum (Life Technologies, Eggenstein, Germany). Induction of the EBV lytic program was achieved by incubation of the cell lines with a combination of the phorbol ester TPA (100 ng/ml final concentration) and sodium butyrate (3 mM final concentration) in standard medium for 3 days.
Recombinant plasmids
pMBO131 is an F-factor-based prokaryotic replicon that carries the F-factor origin of replication, the chloramphenicol resistance gene and partitioning genes (O’Connor et al., 1989). To construct the BZLF1-KO mutant, the XbaI–SalI fragment (EBV coordinates 98 398–105 301) that spans the BZLF1 locus in the EBV genome was cloned onto the plasmid pBluescript IIsk(–) to yield p2084. The XbaI fragment from the pCP15 plasmid (Cherepanov and Wackernagel, 1995) that carries the kanamycin resistance gene was ligated to the NheI-cleaved p2084 plasmid to produce p2088. This cloning step resulted in the deletion of the second exon, both introns and the last third of the first exon of the BZLF1 gene (EBV coordinates 102 389–103 388), as well as in a frameshift of the last BZLF1 exon by the insertion of the kanamycin resistance gene. The p2451 plasmid consists of the p2084 plasmid in which the SfiI–EcoNI fragment that spans the BRLF1 locus was replaced by the PvuII fragment from pCP16, which carries the tetracycline resistance gene. After linearization, DNA fragments from p2088 or from p2451 were introduced into the BJ5183 bacterial strain as described (Delecluse et al., 1999). The expression plasmid for BZLF1 has been described by Hammerschmidt and Sugden (1988). The HindIII fragment from the EBV genome that contains the BRLF1 gene was cloned onto the pKRV expression plasmid (p2130). The p2303 plasmid clone consists of the gp350 gene cloned onto the pBluescript IIsk(+) vector. Plasmids were propagated in E.coli strain DH5α, with the exception of the F-factor-based constructs, which were amplified in E.coli strain DH10B.
DNA transfections
DNA transfections into 293 cells were performed using lipofectamine in Optimem minimal medium (Life Technologies, Eggenstein, Germany). For electroporation, lymphoid cells (107 cells) were washed in RPMI 1640 without fetal calf serum, resuspended in 250 µl of the same medium and placed with the plasmid DNA to be transfected in 0.4 cm wide electroporation cuvettes. Cells were transfected using an electroporator (Bio-Rad Laboratories, Munich, Germany) at 250 V and 960 µF.
Infections
Raji cells (1 × 104) were infected overnight with filtered (0.45 µm pore size) supernatants (1 ml) from 293-BZLF1-KO or 293-BRLF1-KO cells in which the lytic program had been induced by transfecting an expression plasmid encoding BZLF1 or BRLF1 (Hammerschmidt and Sugden, 1988). Primary B cells (1 × 107) were infected with 5 ml of BZLF1-KO-containing supernatants as described (Hammerschmidt and Sugden, 1989). B cells were then plated in 96-well cluster plates (2 × 105/well) and fed once a week. Infection of the same primary B cells with the wild-type EBV (B95.8) was performed in parallel, as well as mock infections.
Southern blot analysis
DNA preparation and hybridization were performed as described (Delecluse et al., 1998).
Immunostaining
Detection of the diffuse early antigen (EA-D or BMRF-1) (Chemicon International, Temecula, CA), the gB and the p160 components of the viral capsid antigen complex encoded by BALF4 and BcLF1, respectively (Chemicon International, Temecula, CA; Kishishita et al., 1984), and of the gp350 antigen (Hoffman et al., 1980; hybridoma HB168 obtained from ATCC) in lytically induced 293-BZLF1-KO or 293-BRLF1-KO cells was carried out as described previously (Delecluse et al., 1998). Immunostaining with the antibody specific to the BRLF1 gene product (clone 5A9, 1:500 dilution) was performed after fixation of the cells in acetone/methanol (1/1 v/v) for 20 min.
Western blot analysis
Proteins were extracted by incubating the cells in a standard buffer (1% NP-40, 0.1% Tween-20, 1 mM phenylmethylsulfonyl fluoride) for 15 min followed by sonication for 5 s. After centrifugation, aliquots of the supernatants were kept at –80°C in 20% glycerol. Up to 20 µg of proteins denatured in Laemmli buffer for 5 min at 95°C were separated on a 10 or 12.5% SDS–polyacrylamide gel and electroblotted onto a nitrocellulose membrane (Hybond C, Amersham). After pre-incubation of the blot in phosphate-buffered saline (PBS) 1% milk dry powder, a monoclonal antibody against the BZLF1 protein (Z130) or the BRLF1 protein (clone 5A9) was added overnight at 4°C (Mikaelian et al., 1993). After extensive washings in PBS, the blot was incubated for 1 h with a goat anti-mouse secondary antibody coupled to horseradish peroxidase (Immunotech, Marseille, France). Bound antibodies were revealed using the ECL detection reagent (Amersham, UK).
Primer extension
An oligonucleotide (TTGATGCTTTTTGCCCCCGACATCG; EBV coordinates 91 974–91 998) specific for the gp350 gene was 5′ radiolabeled using T4 polynucleotide kinase in the presence of [γ-32P]ATP (Amersham, UK). The labeled oligonucleotide was purified on a 10% acrylamide gel and annealed with mRNAs from the B95.8 cell line, from 293 cells transfected with an expression plasmid coding for BRLF1 or from 293-BRLF1-KO cells transfected with an expression plasmid coding for BRLF1. After reverse transcription with Superscript™II Reverse Transcriptase (Life Technologies, Eggenstein, Germany) and addition of RNase A, the samples were separated on an 8% denaturing acrylamide gel. Gels were vacuum dried for 1 h at 80°C and exposed for 3 days at –80°C using intensifying screens. MspI-digested pBR322 DNA (New England Biolabs, MA) was radiolabeled and used as a DNA molecular weight reference.
Acknowledgments
Acknowledgements
We thank B.Neuhierl for helpful discussions and critical reading of the manuscript. This work was supported by Public Health Service grant CA70723, grant Ha 1354/3 and SFB 455 by the Deutsche Forschungsgemeinschaft, and grant 10-2016-Ze from the Deutsche Krebshilfe to W.H.
References
- Baer R. et al. (1984) DNA sequence and expression of the B95-8 Epstein–Barr virus genome. Nature, 310, 207–211. [DOI] [PubMed] [Google Scholar]
- Beisel C., Tanner,J., Matsuo,T., Thorley-Lawson,D., Kezdy,F. and Kieff,E. (1985) Two major outer envelope glycoproteins of Epstein–Barr virus are encoded by the same gene. J. Virol., 54, 665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell B.A. and Villarreal,L.P. (1988) Functional analysis of the individual enhancer core sequences of polyomavirus: cell-specific uncoupling of DNA replication from transcription. Mol. Cell. Biol., 8, 1993–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P.P. and Wackernagel,W. (1995) Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene, 158, 9–14. [DOI] [PubMed] [Google Scholar]
- Conley A.J., Knipe,D.M., Jones,P.C. and Roizman,B. (1981) Molecular genetics of herpes simplex virus. VII. Characterization of a temperature-sensitive mutant produced by in vitro mutagenesis and defective in DNA synthesis and accumulation of γ polypeptides. J. Virol., 37, 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Countryman J. and Miller,G. (1985) Activation of expression of latent Epstein–Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc. Natl Acad. Sci. USA, 82, 4085–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A.K. and Hood,R.E. (1981) Mechanism of inhibition of Epstein–Barr virus replication by phosphonoformic acid. Virology, 114, 52–59. [DOI] [PubMed] [Google Scholar]
- Decaussin G., Leclerc,V. and Ooka,T. (1995) The lytic cycle of Epstein–Barr virus in the nonproducer Raji line can be rescued by the expression of a 135-kilodalton protein encoded by the BALF2 open reading frame. J. Virol., 69, 7309–7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delecluse H.J., Hilsendegen,T., Pich,D., Zeidler,R. and Hammerschmidt,W. (1998) Propagation and recovery of intact, infectious Epstein–Barr virus from prokaryotic to human cells. Proc. Natl Acad. Sci. USA, 95, 8245–8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delecluse H.J., Pich,D., Hilsendegen,T., Baum,C. and Hammerschmidt,W. (1999) A first-generation packaging cell line for Epstein–Barr virus-derived vectors. Proc. Natl Acad. Sci. USA, 96, 5188–5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell P.J., Rowe,D.T., Rooney,C.M. and Kouzarides,T. (1989) Epstein–Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J., 8, 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fixman E.D., Hayward,G.S. and Hayward,S.D. (1992) trans-acting requirements for replication of Epstein–Barr virus ori-Lyt. J. Virol., 66, 5030–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fixman E.D., Hayward,G.S. and Hayward,S.D. (1995) Replication of Epstein–Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol., 69, 2998–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemington E. and Speck,S.H. (1990) Identification of phorbol ester response elements in the promoter of Epstein–Barr virus putative lytic switch gene BZLF1. J. Virol., 64, 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F.L., Smiley,J., Russell,W.C. and Nairn,R. (1977) Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol., 36, 59–74. [DOI] [PubMed] [Google Scholar]
- Gruffat H., Manet,E., Rigolet,A. and Sergeant,A. (1990) The enhancer factor R of Epstein–Barr virus (EBV) is a sequence-specific DNA binding protein. Nucleic Acids Res., 18, 6835–6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt W. and Sugden,B. (1988) Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein–Barr virus. Cell, 55, 427–433. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt W. and Sugden,B. (1989) Genetic analysis of immortalizing functions of Epstein–Barr virus in human B lymphocytes. Nature, 340, 393–397. [DOI] [PubMed] [Google Scholar]
- Hanahan D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol., 166, 557–580. [DOI] [PubMed] [Google Scholar]
- Hardwick J.M., Lieberman,P.M. and Hayward,S.D. (1988) A new Epstein–Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J. Virol., 62, 2274–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman G.J., Lazarowitz,S.G. and Hayward,S.D. (1980) Monoclonal antibody against a 250,000-dalton glycoprotein of Epstein–Barr virus identifies a membrane antigen and a neutralizing antigen. Proc. Natl Acad. Sci. USA, 77, 2979–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland L.E., Anderson,K.P., Shipman,C.,Jr and Wagner,E.K. (1980) Viral DNA synthesis is required for the efficient expression of specific herpes simplex virus type 1 mRNA species. Virology, 101, 10–24. [DOI] [PubMed] [Google Scholar]
- Honess R.W. and Roizman,B. (1974) Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol., 14, 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honess R.W. and Roizman,B. (1975) Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc. Natl Acad. Sci. USA, 72, 1276–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishishita M., Luka,J., Vroman,B., Poduslo,J.F. and Pearson,G.R. (1984) Production of monoclonal antibody to a late intracellular Epstein–Barr virus-induced antigen. Virology, 133, 363–375. [DOI] [PubMed] [Google Scholar]
- Mavromara-Nazos P. and Roizman,B. (1987) Activation of herpes simplex virus 1 γ 2 genes by viral DNA replication. Virology, 161, 593–598. [DOI] [PubMed] [Google Scholar]
- Mavromara-Nazos P. and Roizman,B. (1989) Delineation of regulatory domains of early (β) and late (γ2) genes by construction of chimeric genes expressed in herpes simplex virus 1 genomes. Proc. Natl Acad. Sci. USA, 86, 4071–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikaelian I., Drouet,E., Marechal,V., Denoyel,G., Nicolas,J.C. and Sergeant,A. (1993) The DNA-binding domain of two bZIP transcription factors, the Epstein–Barr virus switch gene product EB1 and Jun, is a bipartite nuclear targeting sequence. J. Virol., 67, 734–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Shope,T., Hermann,L., Stitt,D. and Lipman,M. (1972) Epstein–Barr virus: transformation, cytopathic changes and viral antigens in squirrel monkey and marmoset leucocytes. Proc. Natl Acad. Sci. USA, 69, 383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyormoi O., Klein,G., Adams,A. and Dombos,L. (1973) Sensitivity to EBV superinfection and IUdR inducibility of hybrid cells formed between a sensitive and a relatively resistant Burkitt lymphoma cell line. Int. J. Cancer, 12, 396–408. [DOI] [PubMed] [Google Scholar]
- O’Connor M., Peifer,M. and Bender,W. (1989) Construction of large DNA segments in Escherichia coli. Science, 244, 1307–1312. [DOI] [PubMed] [Google Scholar]
- Post L.E., Mackem,S. and Roizman,B. (1981) Regulation of α genes of herpes simplex virus: expression of chimeric genes produced by fusion of thymidine kinase with α gene promoters. Cell, 24, 555–565. [DOI] [PubMed] [Google Scholar]
- Powell K.L., Purifoy,D.J. and Courtney,R.J. (1975) The synthesis of herpes simplex virus proteins in the absence of virus DNA synthesis. Biochem. Biophys. Res. Commun., 66, 262–271. [DOI] [PubMed] [Google Scholar]
- Preston C.M., Cordingley,M.G. and Stow,N.D. (1984) Analysis of DNA sequences which regulate the transcription of a herpes simplex virus immediate early gene. J. Virol., 50, 708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulvertaft R.J.V. (1964) Cytology of Burkitt’s tumour (African lymphoma). Lancet, i, 238–240. [DOI] [PubMed] [Google Scholar]
- Ragoczy T. and Miller,G. (1999) Role of the Epstein–Barr virus RTA protein in activation of distinct classes of viral lytic cycle genes. J. Virol., 73, 9858–9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragoczy T., Heston,L. and Miller,G. (1998) The Epstein–Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol., 72, 7978–7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickinson A.B. and Kieff,E. (1996) Epstein–Barr virus. In Fields,B.N., Knipe,D.M., Howley,P.M., Chanock,R.M., Melnick,J.L., Monath,T., Roizman,B. and Straus,S.E. (eds), Field’s Virology. Lippincott-Raven, Philadelphia, PA, Vol. 2, pp. 2397–2446. [Google Scholar]
- Roizman B. and Sears,A.E. (1996) Herpes simplex viruses and their replication. In Fields,B.N., Knipe,D.M., Howley,P.M., Chanock,R.M., Melnick,J.L., Monath,T., Roizman,B. and Straus,S.E. (eds), Field’s Virology. Lippincott-Raven, Philadelphia, PA, Vol. 2, pp. 2231–2295. [Google Scholar]
- Serio T.R., Angeloni,A., Kolman,J.L., Gradoville,L., Sun,R., Katz,D.A., Van Grunsven,W., Middeldorp,J. and Miller,G. (1996) Two 21-kilodalton components of the Epstein–Barr virus capsid antigen complex and their relationship to ZEBRA-associated protein p21 (ZAP21). J. Virol., 70, 8047–8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio T.R., Kolman,J.L. and Miller,G. (1997) Late gene expression from the Epstein–Barr virus BcLF1 and BFRF3 promoters does not require DNA replication in cis. J. Virol., 71, 8726–8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio T.R., Cahill,N., Prout,M.E. and Miller,G. (1998) A functionally distinct TATA box required for late progression through the Epstein–Barr virus life cycle. J. Virol., 72, 8338–8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair A.J., Brimmell,M., Shanahan,F. and Farrell,P.J. (1991) Pathways of activation of the Epstein–Barr virus productive cycle. J. Virol., 65, 2237–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck S.H., Chatila,T. and Flemington,E. (1997) Reactivation of Epstein–Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol., 5, 399–405. [DOI] [PubMed] [Google Scholar]
- Sugden B. (1984) Expression of virus-associated functions in cells transformed in vitro by Epstein–Barr virus: Epstein–Barr virus cell surface antigen and virus-release from transformed cells. In Purtilo,D.T. (ed.), Immune Deficiency and Cancer. Plenum Press, New York, pp. 165–177. [Google Scholar]
- Summers W.C. and Klein,G. (1976) Inhibition of Epstein–Barr virus DNA synthesis and late gene expression by phosphonoacetic acid. J. Virol., 18, 151–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada K., Shimizu,N., Sakuma,S. and Ono,Y. (1986) Trans activation of the latent Epstein–Barr virus (EBV) genome after transfection of the EBV DNA fragment. J. Virol., 57, 1016–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urier G., Buisson,M., Chambard,P. and Sergeant,A. (1989) The Epstein–Barr virus early protein EB1 activates transcription from different responsive elements including AP-1 binding sites. EMBO J., 8, 1447–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward R.L. and Stevens,J.G. (1975) Effect of cytosine arabinoside on viral-specific protein synthesis in cells infected with herpes simplex virus. J. Virol., 15, 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima Y., Tanaka,A. and Nonoyama,M. (1976) Inhibition of productive replication of Epstein–Barr virus DNA by phosphonoacetic acid. Virology, 71, 352–354. [DOI] [PubMed] [Google Scholar]
- Zalani S., Holley-Guthrie,E. and Kenney,S. (1996) Epstein–Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl Acad. Sci. USA, 93, 9194–9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler R., Meißner,P., Eißner,G., Lazis,S. and Hammerschmidt,W. (1996) Rapid proliferation of B-cells from adenoids in response to Epstein–Barr virus infection. Cancer Res., 56, 5610–5614. [PubMed] [Google Scholar]
- zur Hausen H., O’Neill,F.J. and Freese,U.K. (1978) Persisting oncogenic herpesvirus induced by tumour promoter TPA. Nature, 272, 373–375. [DOI] [PubMed] [Google Scholar]