Abstract
Sex combs reduced (SCR) is a Drosophila Hox protein that determines the identity of the labial and prothoracic segments. In search of factors that might associate with SCR to control its activity and/or specificity, we performed a yeast two-hybrid screen. A Drosophila homologue of the regulatory subunit (B′/PR61) of serine-threonine protein phosphatase 2A (dPP2A,B′) specifically interacted with the SCR homeodomain. The N-terminal arm within the SCR homeodomain was shown to be a target of phosphorylation/dephosphorylation by cAMP-dependent protein kinase A and protein phosphatase 2A, respectively. In vivo analyses revealed that mutant forms of SCR mimicking constitutively dephosphorylated or phosphorylated states of the homeodomain were active or inactive, respectively. Inactivity of the phosphorylated mimic form was attributed to impaired DNA binding. Specific ablation of dPP2A,B′ gene activity by double-stranded RNA-mediated genetic interference resulted in embryos without salivary glands, an SCR null phenotype. Our data demonstrate an essential role for Drosophila PP2A,B′ in positively modulating SCR function.
Keywords: dephosphorylation/double-stranded RNA-mediated ablation/HOX function/protein phosphatase 2A/Sex combs reduced
Introduction
Hox genes encode transcription factors that contain a highly conserved homeodomain of 60 amino acids that is capable of binding to DNA in a sequence-specific manner. The homeodomain consists of three α-helices that establish specific contacts with the DNA, and an N-terminal arm that is disordered flexibly in solution but contacts the minor groove upon binding to the DNA (Gehring et al., 1994a,b). It is believed that Hox genes exert their function by regulating the expression of a specific array of downstream target genes (Graba et al., 1997). In vitro, the homeodomain-containing proteins recognize similar DNA sequences. In fact, the analysis of transgenic flies expressing chimeric Antennapedia (ANTP)–Sex combs reduced (SCR) proteins has demonstrated that the specific identity of body segments is determined by the N-terminal arm within the homeodomain (Furukubo-Tokunaga et al., 1993; Zeng et al., 1993).
How the N-terminal flexible arm of the homeodomain brings about subtle differences in DNA binding is unclear at present. Specificity could be achieved directly due to differences in critical amino acids, or by the association with particular cofactor(s) that themselves confer target gene specificity. Alternatively, putative cofactors might modify the N-terminal arm post-translationally, in this way modulating its DNA-binding specificity and/or activity. To identify and isolate proteins important for conferring functional specificity to SCR, we employed a yeast two-hybrid screen (Bartel and Fields, 1995). We report the isolation of a regulatory subunit (dPP2A,B′) of the serine-threonine protein phosphatase 2A (PP2A) and demonstrate specific interaction between SCR and dPP2A,B′. 32P-labelling experiments revealed that the N-terminal arm of the SCR homeodomain can be phosphorylated and dephosphorylated by cAMP-dependent protein kinase A (PKA) and PP2A, respectively. We also show that SCR-AA that mimics the constitutively dephosphorylated state of the N-terminal arm of the homeodomain is functionally active in fly embryos, while an equivalent transgene encoding SCR-DD that mimics the constitutively phosphorylated state is inactive. Fly embryos with a genomic deletion encompassing the dPP2A,B′ locus, and embryos with a functional knockout of the dPP2A,B′ gene by double-stranded RNA (dsRNA)-mediated genetic interference exhibit an SCR null-like phenotype. Our results demonstrate that dPP2A,B′ is essential for SCR function, strongly suggesting that the activity of SCR, and possibly other homeobox-containing transcription factors, is controlled by PP2A-mediated dephosphorylation of a conserved consensus site within the homeodomain.
Results
The SCR homeodomain interacts with a regulatory subunit of serine-threonine PP2A
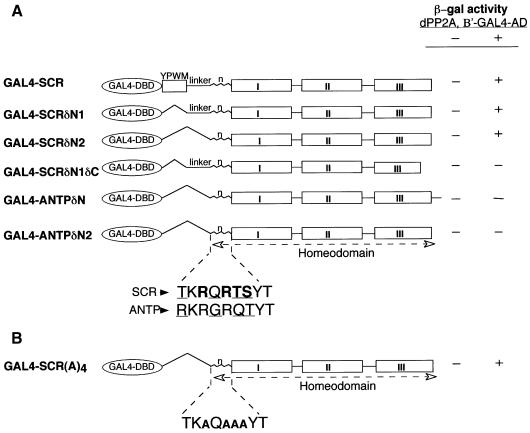
To isolate Drosophila cDNAs encoding proteins that interact with the SCR homeodomain, we have used the yeast two-hybrid system (Bartel and Fields, 1995). For the bait, we constructed a chimeric protein composed of the GAL4 DNA-binding domain (DBD) and amino acids 297–378 of SCR corresponding to the YPWM motif (Mann, 1995) and the homeodomain (N-terminal arm and the three helices), except its last five amino acids, which flank helix 3 at its C-terminal end (GAL4–SCR, Figure 1A). One of the partial cDNAs that interacted specifically with GAL4–SCR encoded a 425 amino acid polypeptide, which was identified in a database search as a Drosophila homologue of the regulatory subunit (B′/PR61) of serine-threonine PP2A (PP2A,B′) (Groves et al., 1999). Three full-length cDNAs corresponding to the Drosophila PP2A,B′ (dPP2A,B′) were subsequently isolated from a Drosophila embryo cDNA library, encoding a 670 amino acid protein (Figure 2). The fact that all three independent cDNA isolates were identical in sequence in the coding region suggests that Drosophila embryos might express only one isoform of dPP2A,B′. Moreover, in situ hybridization to polytene chromosomes with dPP2A,B′ cDNA gave a positive signal only at position 90E on chromosome III, indicating a single gene locus. Based on the degree of sequence conservation, the mammalian homologues appear to be the closest relatives of Drosophila dPP2A,B′ (data not shown). Whole-mount in situ hybridization on developing fly embryos revealed that dPP2A,B′ is present ubiquitously throughout embryogenesis (data not shown).
Fig. 1. (A) dPP2A,B′ interacts specifically with the SCR homeodomain in a yeast two-hybrid screen. Different GAL4 fusion constructs containing the GAL4 DNA-binding domain (amino acids 1–147) fused to distinct regions of SCR and ANTP are shown in the scheme. The amino acid co-ordinates of the regions of SCR and ANTP in the constructs are provided in Materials and methods. The GAL4–HOX protein chimeras were co-transformed into yeast, with either GAL4 AD alone (–) or the GAL4 AD fused to amino acids 245–670 of dPP2A,B′ (+). The differences in the amino acids within the N-terminal arm of SCR and ANTP homeodomains are underlined. Two consensus phosphorylation sites in the SCR homeodomain are depicted in bold. (B) Conservation of the phosphorylation sites within the N-terminal arm of the SCR homeodomain is not essential for the interaction with dPP2A,B′. The four amino acids (RRTS) of the consensus phosphorylation sites in the SCR homeodomain are substituted by alanines in GAL4–SCR(A)4.
Fig. 2. Full-length nucleotide and deduced amino acid sequence of dPP2A,B′ cDNA. The 5′- and 3′-untranslated sequences are presented in lower case. The open reading frame is shown in upper case. The numbers corresponding to the nucleotide and the amino acid sequence are indicated on either side of the respective sequences.
The homeodomain of SCR is sufficient for interaction with dPP2A,B′
To identify the minimum region of the SCR fragment used in the two-hybrid screen essential for the interaction with dPP2A,B′, several deletion mutants of SCR were assayed for the interaction with dPP2A,B′ in yeast. Our two-hybrid analysis showed that the YPWM motif and the linker region between this motif and the homeodomain were not required for interaction with dPP2A,B′, and that the isolated homeodomain of SCR(GAL4–SCRδN2) was sufficient (Figure 1A, compare the reporter gene expression by GAL4–SCRδN2, GAL4–SCR and GAL4–SCRδN1). On the other hand, the ANTP homeodomain lacking the equivalent N-terminal region (construct GAL4–ANTPδN) did not interact with dPP2A,B′, neither did GAL4–ANTPδN2 representing the ANTP homeodomain lacking the extra amino acids present at the C-terminus of the homeodomain. The only difference between constructs GAL4–SCRδN2 and GAL4–ANTPδN2 resides in four amino acids in the N-terminal arms of the respective homeodomains (see Figure 1A), strongly suggesting that the N-terminal arm of the SCR homeodomain is essential for the specific interaction with dPP2A,B′. To delineate further the region critical for interaction, an additional construct (GAL4–SCRδN1δC) that included the linker region at the N-terminal end but lacked helix 3 of the homeodomain from amino acid 45 onwards (numbered according to Gehring et al., 1994b) was tested. This bait failed to interact with dPP2A,B′ (Figure 1A), suggesting that additional contacts formed by helix 3 might be essential or, alternatively, that the absence of helix 3 might have given rise to a misfolded homeodomain. Based upon these data, we demonstrate that an intact SCR homeodomain is essential and sufficient for the interaction with dPP2A,B′.
Two consensus sites for phosphorylation by cAMP-dependent protein kinase (Kennelly and Krebs, 1991) were identified in the N-terminal arm of the SCR homeodomain (for the respective sequence, see Figure 1A). We substituted the amino acid residues at positions 3, 5, 6 and 7 of the homeodomain, which form the consensus phosphorylation sites, with alanines [GAL4–SCR(A)4], and found that this mutant protein still interacted with dPP2A,B′ (Figure 1B). We therefore conclude that the critical amino acid residues responsible for the interaction of the SCR homeodomain with dPP2A,B′ are not part of the consensus phosphorylation sites.
The N-terminal arm of the SCR homeodomain is a target of phosphorylation/dephosphorylation events
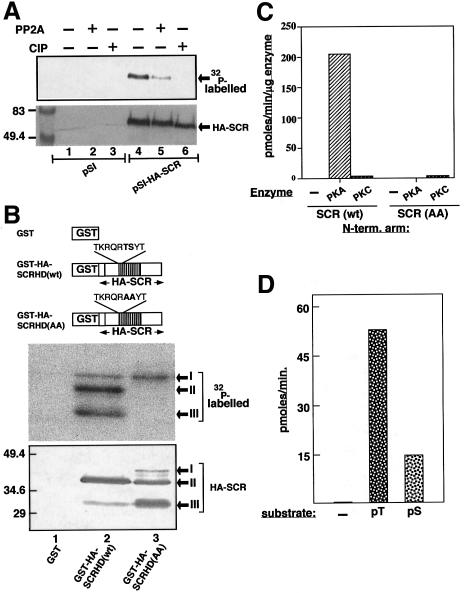
Although no information regarding the in vivo phosphorylation status of SCR could be found in the literature, careful examination of the amino acid sequence revealed several potential phosphorylation/dephosphorylation sites, including the two sites in the N-terminal arm of the homeodomain mentioned above (data not shown and Figure 1A). To obtain experimental evidence for this observation, we transfected an expression vector encoding haemagglutin-tagged SCR (HA-SCR), and empty vector as control, into COS-1 cells, a mammalian cell line known for its robust expression of recombinant proteins. Since SCR and its mammalian homologues, such as HOX A-5, are functionally conserved (Zhao et al., 1993), COS-1 cells were considered an adequate recipient cell line. Transfected cells were labelled with [32P]orthophosphate, and the immunoprecipitated HA-SCR was analysed by western blotting. No signal was detected in the lanes corresponding to empty vector (Figure 3A, lanes 1–3). In contrast, a specific band corresponding to 32P-labelled HA-SCR was observed by autoradiography and/or immunostaining (Figure 3A, lanes 4–6), thus demonstrating that SCR is phosphorylated in vivo. Furthermore, SCR could be dephosphorylated efficiently by PP2A or calf intestine alkaline phosphatase (Figure 3A, compare lanes 5 and 6 with lane 4).
Fig. 3. In vivo and in vitro phosphorylation and dephosphorylation of SCR. (A) 32P-labelled extracts of empty vector (pSI) or pSI-HA-SCR-transfected COS-1 cells were immunoprecipitated and subjected to dephosphorylation by the PP2A catalytic subunit (lanes 2 and 5) and by calf intestine alkaline phosphatase (CIP, lanes 3 and 6). Western blot was revealed by autoradiography (upper panel) and by alkaline phosphatase immunostaining to normalize the SCR content in each lane (lower panel). The position of HA-SCR is indicated with arrows. Molecular size markers in kDa are shown on the left. (B) Schematic representation of GST fusion constructs utilized in the present study. The depicted scale is arbitrary. The GST fusion constructs encode the GST protein alone (GST), GST–HA-SCRHD(wt) and GST–HA-SCRHD(AA). The homeodomain is represented by the shaded box. The wild-type and AA mutant sequences of the N-terminal arm in the homeodomain are also indicated (upper panel). The GST–SCRs were detected on western blot by alkaline phosphatase staining, to monitor the relative levels of protein products in each lane (lower panel), and subsequently by autoradiography (middle panel). Note that the GST–SCRs are visible in three different sizes corresponding to the full-length (band I) and its C-terminal truncations (bands II and III), which are indicated by arrows. (C) Comparison of phosphorylation of the wild-type and the AA mutant versions of the N-terminal arm of the SCR homeodomain in the absence and presence of kinases. Biotinylated peptides (200 µM) SCR(wt): TKRQRTSYT and SCR(AA): TKRQRAAYT were used as substrates. The specific activity of the [γ-32P]ATP per reaction was typically 2000 c.p.m./pmol. A 5 ng aliquot of PKA, PKC or no enzyme (–) was included in the assays. The enzyme reactions were carried out at 30°C for 2–5 min. The incorporation of [γ-32P]ATP into the peptides is depicted on the y-axis in pmol/min/µg enzyme. The values are an average of six and three independent experiments for PKA and PKC, respectively. (D) Dephosphorylation of the N-terminal arm of the SCR homeo-domain by PP2A: no peptide (–) or 200 µM of phosphopeptides pT: TKRQR(pT)SYT and pS: TKRQRT(pS)YT (x-axis) were used as substrates for dephosphorylation by PP2A. The assays were performed at 30°C for 15 min using a serine-threonine phosphatase (non-radioactive) assay kit (Promega) in the presence of 50 ng of the catalytic subunit of PP2A. The activity was monitored spectrophoto-metrically by measuring the release of pmoles of free phosphate per minute (y-axis). The depicted values are an average of four individual experiments.
In order to see whether the N-terminal arm of the SCR homeodomain could be phosphorylated, we produced unfused glutathione S-transferase (GST) and fused GST–SCRs [GST–HA-SCRHD(wt) and (AA)]. While GST–HA-SCRHD(wt) encodes the wild-type sequence of the SCR homeodomain, GST–HA-SCR(AA) encodes a homeodomain in which the threonine and serine at positions 6 and 7 are substituted by alanines (which cannot be phosphorylated) (Figure 3B, upper panel). Purified GST recombinant proteins were included as substrates in PKA reactions in the presence of [γ-32P]ATP. The western blots of kinase reaction products revealed two faster migrating major bands and one slower migrating minor band [barely visible in the GST–HA-SCRHD(wt) lane]. The minor band, i.e. of weak intensity, corresponds to the expected full-length chimeric protein, whereas the two predominant bands correspond to specific proteolytic products of the full-length protein (Figure 3B, middle and lower panels). Since we had re-purified the GST proteins on glutathione–Sepharose subsequent to the kinase reactions, all three products (bands I–III) must have retained an intact GST moiety at their N terminal end, implying that the proteolytic products (bands II and III) must be truncated at the C-terminus. The autoradiography of the blot showed that all the three forms of GST–HA-SCRHD(wt) are phosphorylated, whereas in the case of GST–HA-SCRHD(AA), only the full-length mutant protein is phosphorylated, albeit to a lower level than the wild-type version, and its truncated forms remain completely unphosphorylated (Figure 3B, compare lanes 2 and 3, middle and lower panels). Since the only difference between the ‘wt’ and ‘AA’ mutants of GST–HA-SCRHD proteins is at positions 6 and 7 of the homeodomain, our data prove that these residues in the N-terminal arm indeed become phosphorylated. Moreover, there may be additional phosphorylation site(s) in the C-terminal region outside the homeodomain, since the full-length forms of both wild-type and AA-substituted GST–SCR-HDs are 32P-labelled.
To test directly whether the potential PKA phosphorylation sites in the N-terminal arm of the homeodomain are targets of PKA, we performed PKA assays using biotinylated peptides as substrates comprising the wild-type (TS) or mutant (TS→AA) N-terminal arm of the SCR homeodomain. While cAMP-dependent PKA phosphorylated the wild-type N-terminal arm of the SCR homeodomain, mutant AA remained unlabelled. Since the threonine at position 6 of the homeodomain also fulfils the criterion of a consensus PKC phosphorylation site, we also measured PKC activity on the above peptides. Our in vitro kinase assays clearly show that the N-terminal arm of the homeodomain of SCR is phosphorylated specifically by PKA but not by PKC (Figure 3C). To determine whether the PKA phosphorylation sites in the N-terminal arm of the homeodomain are also targets of PP2A-mediated dephosphorylation, we performed PP2A assays on phosphopeptides that were phosphorylated either at threonine at position 6 (pT) or at serine at position 7 (pS) of the homeodomain. Spectrophotometric analysis of in vitro phosphatase reaction products revealed that both pT and pS phosphopeptides were dephosphorylated efficiently, albeit with different efficiencies (pT>pS; Figure 3D). Taken together, our studies clearly demonstrate that the N-terminal arm of the SCR homeodomain is indeed a target of PKA-mediated phosphorylation and PP2A-mediated dephosphorylation events.
Analysis of SCR transgenic flies—experimental outline
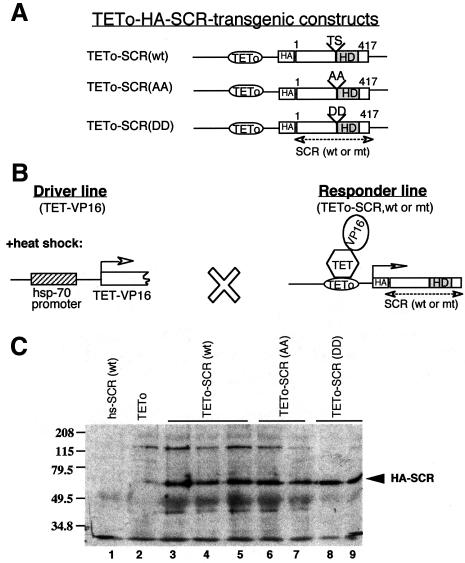
We next tested whether the functional activity of SCR was influenced by the phosphorylation status of the threonine (T) and serine (S) residues at amino acids 6 and 7 in the N-terminal arm of the SCR homeodomain, which we have shown to serve as targets of PKA- and PP2A-mediated phosphorylation/dephosphorylation, respectively (see above). These residues correspond to amino acids 329 and 330 of full-length SCR. When expressed in flies, the SCR(AA) and SCR(DD) mutants are expected to mimic constitutively dephosphorylated and constitutively phosphorylated states of the N-terminal arm of the homeodomain, respectively (from here on, referred to simply as the dephosphorylated and phosphorylated states of SCR).
The cDNAs encoding SCR(wt), SCR(AA) and SCR(DD) were placed under the control of the tetracycline operator (see Figure 4A). The constructs were microinjected into eggs, giving rise to several transgenic responder lines. The responder TETo-SCR lines (wild-type, AA or DD mutants) were crossed to a driver line (Tet-VP16), which, in our study, expresses Tet-VP16 under the control of the heat shock promoter (hsp-70). Heat-shocked embryos carrying the constructs express either HA-SCR(wt), HA-SCR(AA) or HA-SCR(DD). Transgene expression levels were determined by western blot analysis of embryonic cell extracts (Figure 4C).
Fig. 4. Expression of SCR transgenes in Drosophila embryos. (A) Schematic representation of the constructs used to generate transgenic flies. (B) Schematic representation of the tetracycline repressor-VP16 (TET-VP16)-mediated ectopic induction of SCR transgenes. TET-VP16 is produced upon heat shock. (C) Western blot showing the expression of HA-SCR(wt), HA-SCR(AA) and HA-SCR(DD) proteins in fly embryos. Lane 1, hs-SCR(wt); lane 2, TETo alone; lanes 3–5, HA-SCR(wt); lanes 6 and 7, and 8 and 9, HA-SCR(AA) and HA-SCR(DD), respectively, expressed from different transgenic lines for each construct. The arrow indicates the position of the distinct HA-SCR transgene products. The positions of protein size markers (in kDa) are indicated on the left.
SCR(AA), which mimics the dephosphorylated state, is functionally active in developing embryos, but not SCR(DD) mimicking the phosphorylated state
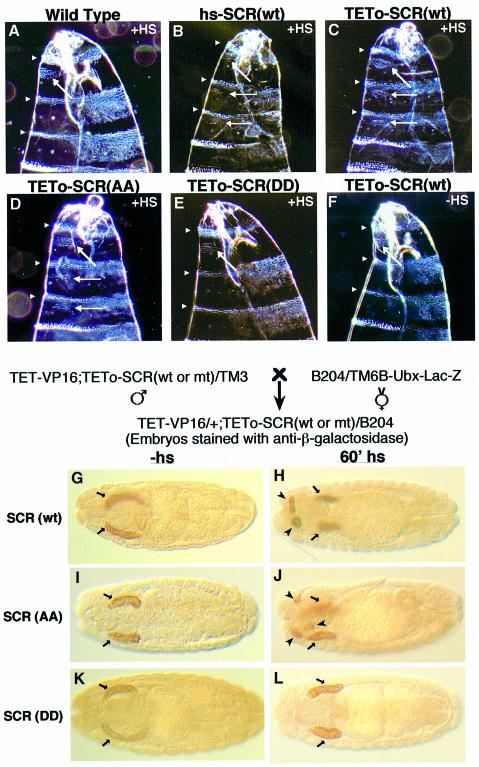
Homeotic cuticle transformations. Ectopic expression of SCR in developing embryos results in the anterior transformation of mesothoracic and metathoracic segments into the prothorax. As a result, ectopic T1 ‘beards’ of denticles appear on the ventral portion of posterior thoracic segments (T2 and T3) accompanied by head abnormalities (Gibson et al., 1990; Zeng et al., 1993). Three transgenic lines of TETo-SCR(wt) and TETo-SCR(AA) and two lines of TETo-SCR(DD) were analysed for their larval cuticular pattern. We have assessed the degree of transformations caused by TETo-SCR(wt) versus hs-SCR (Gibson et al., 1990). We found equally drastic transformations and head defects (Figure 5A–C). Heat shock treatment of developing embryos revealed that the anterior transformations and the head defects caused by the ectopic expression of SCR(AA) are indistinguishable from those caused by hs-SCR(wt) or SCR(wt) (Figure 5, compare D with B and C). In contrast, ectopic expression of SCR(DD) did not cause any transformation or head abnormalities. In fact, the pattern of cuticles of SCR(DD) embryos looked identical to wild-type embryos or transgenic embryos without heat shock (Figure 5, compare E with A and F).
Fig. 5. SCR(wt) and SCR(AA) but not SCR(DD) transgenes transform T2 and T3 towards T1 identity and cause head involution defects (A–F). Darkfield photomicrographs of the anterior ends of cuticles of (A) a wild-type embryo, (B) an embryo expressing SCR(wt) from a direct hs-SCR transgene (see Gibson et al., 1990), heat-shocked embryos from a cross between TET-VP16 and (C) TETo-SCR(wt), (D) TETo-SCR(AA) and (E) TETo-SCR(DD), and (F) an embryo of a cross between TET-VP16 and TETo-SCR(wt) without a heat shock. Arrowheads indicate segmental borders of the anterior region of larval cuticles. Arrows show the normal (A–F) and ectopic T1 beards (B–D). Note the head involution defects observed in embryos in (B), (C) and (D) compared with the normal head involution of embryos in (A), (E) and (F). Both SCR(wt) and SCR(AA) induce additional salivary glands in transgenic embryos (G–L). A scheme of the cross with the indicated genotypes is given above (G–L). B204 is a dCREB-A-lacZ enhancer trap line. Embryos were stained with anti-β-galactosidase to locate salivary glands. Embryos expressing dCREB-A-lacZ and SCR(wt) (G and H), SCR(AA) (I and J) or SCR(DD) transgenes (K and L) are shown. Normal and extra salivary glands are indicated by arrows and arrowheads, respectively.
Ectopic salivary gland formation. Ectopic expression of SCR in developing embryos induces the formation of additional salivary glands (Panzer et al., 1992). To visualize the salivary glands, we carried out the analysis in an enhancer trap line B204 background (Andrew et al., 1997). We observed ectopic salivary glands in addition to normal ones following heat shock-induced expression of both SCR(wt) and SCR(AA) (Figure 5, compare H and J with G and I, respectively). In contrast, heat shock-induced expression of SCR(DD) did not provoke this response (Figure 5, compare K and L). These in vivo findings demonstrate that the SCR protein is in the functionally active form when it mimics the constitutively dephosphorylated state, while it is inactive when it mimics the constitutively phosphorylated state.
The fact that SCR(DD) was inactive in our in vivo assays, despite being expressed at levels equivalent to SCR(AA) or SCR(wt) (Figure 4C), raised concerns that this particular SCR mutant might be impaired in its ability to localize to the nucleus. To check this possibility, developing embryos (0–12 h) transformed with different TETo-SCR transgenes were heat shocked and stained with an SCR-specific polyclonal antiserum. This analysis confirmed that all three SCR transgenes were expressed and localized to the nucleus. Also, the stability of the ectopically induced proteins over time was found to be similar (data not shown).
The in vitro DNA-binding activity of the SCR homeodomain is controlled by the phosphorylation status of its N-terminal arm
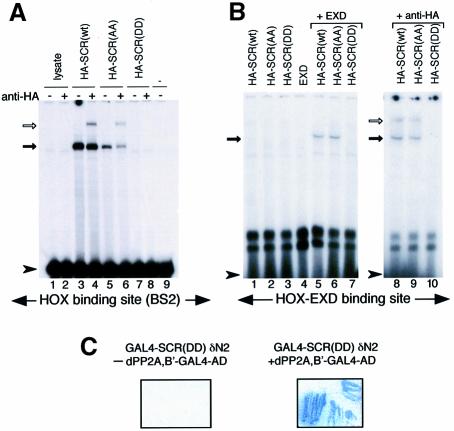
To test whether the (DD) mutation might affect the DNA-binding activity of SCR, we synthesized full-length wild-type and mutant SCR proteins with an HA tag by in vitro translation, and analysed in band-shift assays their ability to bind to a radiolabelled oligonucleotide pair representing BS2, a natural Hox-binding site (Muller et al., 1988). Both SCR(wt) and SCR(AA) were able to bind the BS2 site, although the SCR(AA)–BS2 complex was formed at reduced levels (Figure 6A). Complex formation was specific, since it was supershifted following incubation with an anti-HA antibody. In contrast, SCR(DD) did not bind BS2 (Figure 6A). We reasoned that SCR(DD) might require the function of EXD (a Hox cofactor) to form a stable complex with DNA. We therefore used a consensus HOX–EXD-binding site (Chang et al., 1996; Jaffe et al., 1997) as a probe for band-shift assays with SCR(wt) or mutants in the presence or absence of EXD. We could not detect any binding when SCR or EXD proteins were present alone. However, co-operative DNA binding was observed between EXD and SCR(wt) or EXD and SCR(AA). The specificity of the bands was confirmed by supershifting the complexes with anti-HA antibody (Figure 6B). Note that the retarded complexes formed by SCR(wt) + EXD versus SCR(AA) + EXD are indistinguishable in both the position and the intensity of the bands. On the other hand, no retarded complex was seen when SCR(DD) and EXD were present in a binding reaction (Figure 6B), demonstrating that the presence of EXD cannot overcome the DNA-binding defect of SCR(DD). Taken together, these results demonstrate that SCR(DD) does not bind DNA, providing an explanation at the molecular level for the observed functional inactivity of this SCR mutant in fly embryos. Importantly, GAL4–SCRδN2(DD), similarly to its wild-type version (GAL4–SCRδN2), fully interacts with dPP2A,B′ in a yeast two-hybrid system (Figure 6C, see also Figure 1). Combined with our previous data showing that an intact SCR homeodomain (N-terminal arm and three helices) is essential for the interaction with dPP2A,B′ (see Figure 1A and Results), these findings demonstrate that the lack of DNA-binding activity is clearly not due to a general disruption of homeodomain structure in mutant (TS→DD).
Fig. 6. SCR(DD) does not bind DNA either on its own or in the presence of EXD. (A) Band-shifts performed with BS2 as a probe using lysate alone (lanes 1 and 2), HA-SCR(wt) (lanes 3 and 4), HA-SCR(AA) (lanes 5 and 6) and HA-SCR(DD) (lanes 7 and 8), or in the absence of any lysate (lane 9). (B) Band-shifts with a consensus HOX–EXD-binding site. HA-SCR(wt) (lane 1), HA-SCR(AA) (lane 2), HA-SCR(DD) (lane 3) or EXD alone (lane 4). In the presence of EXD, HA-SCR(wt) (lane 5) and HA-SCR(AA) (lane 6) specifically bind the HOX–EXD motif. Solid arrows indicate the position of specific complexes. Supershifts of the specific complexes are indicated with unfilled arrows (lanes 4, 6, 8 and 9). The position of the unbound probes is marked with an arrowhead. Note that HA-SCR(DD) does not bind to DNA at all (lanes 7 and 8 in A and lanes 3, 7 and 10 in B). (C) The SCR(DD) homeodomain interacts with dPP2A,B′ in a yeast two-hybrid system: co-transformation of GAL4–SCR(DD)δN2 (323–378;TS→DD) and dPP2A,B′–GAL4 AD induces the expression of β-galactosidase detected as blue colour of the colonies (right panel). No β-galactosidase is induced when GAL4–SCR(DD)δN2 is present by itself (left panel).
dPP2A,B′ is essential for the functional activity of SCR
The above results show that SCR is functionally active in the dephosphorylated state and, upon phosphorylation, becomes inactive. We therefore wanted to know whether dPP2A,B′ was involved in converting a functionally inactive into an active form of SCR. To this end, we checked whether the two genes interact with each other genetically. Derepression of Scr in Polycomb group (Pc-G) mutant backgrounds, e.g. polyhomeotic (ph), causes transformations similar to the gain-of-function mutants that are characterized by occurrence of additional sex comb teeth on the second and third pair of legs of mutant male flies, e.g. in ph410/Y males (Fauvarque and Dura, 1993). If dPP2A,B′ interacts genetically with SCR, then the transformation might be attenuated in the legs of males that are hemizygous for ph410 and heterozygous for the deficiencies Df(3R)90D1-2;90F3-6 and Df(3R)90C2-D1;91A1-2, which remove one allele of the dPP2A,B′ gene (ph410/Y; Df/+). The data presented in Table I show that heterozygosity for these deficiencies clearly leads to suppression of ectopic sex comb teeth formation on the second and third pair of legs of ph410/Y flies, whereas the deficiency Df(3R)90F;91B1-2, which partially overlaps the above two deficiencies but leaves the dPP2A,B′ gene intact, has essentially no effect on the ectopic sex comb teeth phenotype (Table I, compare iv and v with ii and iii). Note that sex comb teeth are absent on mesothoracic and metathoracic legs in wild-type flies (Table I, i).
Table I. Occurrence of ectopic sex comb teeth on mesothoracic and metathoracic legs of males.
| Mean no. of sex comb teeth |
|||
|---|---|---|---|
| Mesothoracic leg | Metathoracic leg | ||
| i | Wild type | 0 | 0 |
| ii | ph410/y | 6.9 | 3.8 |
| iii | ph410/y; Df(3R)90F;91B1-2/+ | 6.4 | 4.0 |
| iv | ph410/y; Df(3R)90D1-2;90F3-6/+ | 3.6 | 1.6 |
| v | ph410/y; Df(3R)90C2-D1;91A1-2/+ | 0.8 | 0.5 |
(i) Wild type (OregonR), (ii) hemizygous for ph410 (polyhomeotic mutation) and (iii–v) heterozygous for the deficiency and hemizygous for ph410. A minimum of 50 legs was scored in each analysis. The data are represented as the mean number of sex comb teeth per leg.
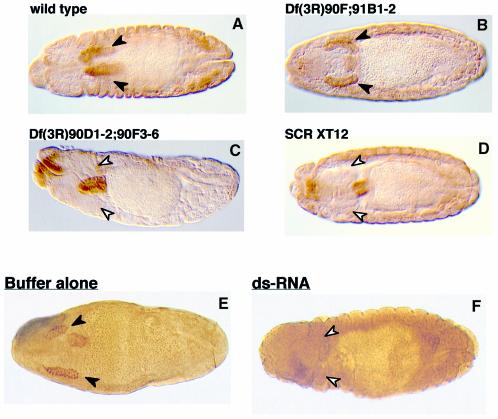
Salivary glands are completely missing in SCR loss-of-function mutant embryos (Panzer et al., 1992). If dPP2A,B′ were essential for the functional activity of SCR, no salivary glands should develop in embryos lacking the dPP2A,B′ gene. Salivary glands are indeed absent in fly embryos with a homozygous deficiency [Df(3R)90D1-2;90F3-6] encompassing the dPP2A,B′ gene locus (Figure 7C), thus resembling SCR null embryos (SCR XT12) (Figure 7D). In contrast, embryos with a deficiency Df(3R)90F;91B1-2 that leaves the dPP2A,B′ gene intact have normal salivary glands, like the wild-type embryos (Figure 7A and B). These in vivo findings strongly suggest that the functional activity of SCR is dependent on the availability of dPP2A,B′. However, since deficiencies remove several genes in addition to the gene of interest, these data do not rigorously exclude the possibility that the absence of some other gene(s) located in the vicinity of dPP2A,B′ was responsible for or at least contributed to the observed effects. Unfortunately, null mutants of dPP2A,B′ are not yet available. To overcome these limitations, we employed a recently developed technique, termed dsRNA-mediated genetic interference (RNAi), which has been shown to allow specific ablation of single gene functions in both Caenorhabditis elegans and Drosophila (Fire et al., 1998; Kennerdell and Carthew, 1998). The method is based on selective inhibitory effects of dsRNA molecules homologous to transcripts of the gene of interest. Injection of embryos with dPP2A,B′ dsRNA (nucleotides 875–1620) resulted in a complete loss of salivary glands. Note that only those embryos that had undergone germ band retraction were screened for the presence of salivary glands, because only from this stage onwards are the embryonic salivary glands sufficiently developed. We obtained the loss-of-function phenotype in 40% of the RNAi-injected embryos analysed. In contrast, salivary glands developed normally in all the embryos injected with buffer alone (Figure 7E and F). Our genetic interference data therefore prove that expression of dPP2A,B′ is essential for the functional activity of SCR.
Fig. 7. dPP2A,B′ is essential for SCR activity in developing fly embryos. Normal salivary glands appear in wild-type embryos (A) and in the embryos with a deficiency that does not remove the dPP2A,B′ gene (B). In contrast, salivary glands are missing in the deficient embryos that lack the dPP2A,B′ gene (C) and in the SCR null embryos (D). Wild-type embryos (0–60 min) were injected either with buffer (E) or with dPP2A,B′ dsRNA (F). Normal salivary glands are shown by filled arrowheads. The positions where salivary glands are expected to form, but fail to appear, are indicated by unfilled arrowheads.
Discussion
Interaction of SCR with dPP2A,B′
We have isolated in a yeast two-hybrid screen a cDNA encoding a novel Drosophila homologue of the regulatory subunit B′ (PR61) of the serine-threonine-specific PP2A. dPP2A,B′ interacts specifically with the SCR homeodomain. Our studies have shown that this interaction requires the presence of the N-terminal flexible arm of the SCR homeodomain, but neither the YPWM motif nor the linker region located between this motif and the homeodomain proper. Interestingly, the corresponding region of the ANTP homeodomain was unable to interact with this novel protein, although the respective region of ANTP differs from SCR in just four amino acids that provide functional specificity to these two homeodomain proteins—a finding that highlights the extraordinary degree of specificity of the reported interaction.
PP2A exists as a multisubunit enzyme complex in a variety of organisms and cell types. The enzyme complex is composed of a catalytic and a scaffold subunit, which together form a core dimer that then associates with one of a number of regulatory subunits to constitute a trimeric enzyme complex. Regulatory subunits of PP2A are encoded by at least three unrelated gene families: B (PR55), B′ (PR61) and B′′ (PR72). Each family consists of several members, which in addition can give rise to a number of splice variants, thereby greatly increasing the variety of distinct trimeric enzyme complexes (Groves et al., 1999). Several lines of evidence suggest that the regulatory subunits of PP2A may serve as specific adaptors that confer substrate specificity to the core domain of PP2A (reviewed in Schönthal, 1998). Specific interaction of dPP2A,B′ with the SCR homeodomain, as documented here, therefore reflects its potential of reversibly recruiting SCR into the PP2A complex.
Control of SCR activity by phosphorylation/dephosphorylation of specific homeodomain residues
The two phosphorylatable residues (T and S) within the N-terminal arm of the SCR homeodomain appear to be conserved, since at least one such site has been found in all SCR homologues from other species, except for PS12-B of Atlantic salmon [compare sequences constituting N-terminal of homeodomains in Gehring et al. (1994b)]. We note that the homeodomain of PS12-B in fact seems more closely related to ANTP than to SCR. Our in vivo results suggest that in developing embryos, SCR is functionally inactive when the N-terminal arm of its homeodomain is phosphorylated and is active upon dephosphorylation. These results may have important implications for the functional specificity of homeotic proteins in general: since ANTP has a glutamine instead of threonine at position 6, which is well conserved in all the SCR homologues, we propose that the differential modification of this residue plays an important role in determining the functional specificity of these two homeotic proteins.
Reciprocal findings with regard to the homeobox-containing protein Fushi-tarazu (FTZ) were reported recently (Dong et al., 1998). Substitution of threonine at position 7 of the homeodomain by aspartate gave rise to an active form of FTZ when expressed as a transgene. In contrast, a FTZ (T7→A) mutant transgene failed to do so. These results were interpreted to indicate that phosphorylation of threonine at position 7 was required for normal protein activity. Unfortunately, the results were somewhat blurred since the inactive form of FTZ (T7→A) exhibited residual activity in some assays and a high variability in the penetrance of the phenotypes. Also, the activated form of FTZ (T7→D) rescued to a lesser extent than the wild-type form. In contrast, in our experimental system, SCR(AA) was as active as SCR wild type, while SCR(DD) was inactive. Taken together, these findings highlight the functional importance of the phosphorylation site(s) within the N-terminal arm of the homeodomain. However, in our case, it is dephosphorylation of specific residues that leads to activation, while phosphorylation gives rise to an inactive form.
Dependence of the in vitro DNA-binding activity of SCR on phosphorylation/dephosphorylation events
Our gel retardation analyses demonstrate that the conversion of just two residues, threonine and serine at positions 6 and 7 of the SCR homeodomain, into aspartate is sufficient to abrogate in vitro DNA-binding activity, whereas a conversion into alanine is tolerated. These in vitro findings perfectly mirror the in vivo activity of the respective wild-type and mutant SCR transgenes.
Since we have not tested a whole range of potential SCR-binding sites, we cannot exclude the possibility that mutant DD can bind to a unique set of target sequences. We also cannot rule out that SCR(DD) may require the presence of an as yet unknown cofactor(s) to interact with specific DNA motifs. Depending upon the distribution and activity of this hypothetical cofactor(s), Thr6-/Ser7-phosphorylated SCR could then be functional in certain cells or certain promoter contexts. However, our in vivo data do not support this view, since we have not observed any effect of the expressed SCR(DD) transgene in developing fly embryos. We therefore propose that SCR(DD) is functionally inactive in vivo due to a general inability to bind specific DNA recognition motifs. It is very likely that electrostatic interference of a large, negatively charged phosphate group and/or steric hindrance abrogates DNA binding when positioned at or near the protein–DNA interface. We exclude the possibility that substitution of TS by DD leads to a non-specific disruption of SCR protein structure, since the TS→DD-substituted homeodomain still maintains the competence to interact with dPP2A,B′—exactly as one would predict for a transcription factor that is turned off in its phosphorylated form and requires association with dPP2A,B′ in order to be turned on.
Several reports have described situations where phosphorylation/dephosphorylation events modulate the ability of a homeobox-containing transcription factor to bind its DNA recognition motif (see Hunter and Karin, 1992; Bourbon et al., 1995; Hunter, 1995; Jaffe et al., 1997; Coqueret et al., 1998). These reports focus on in vitro studies and lack data regarding the in vivo relevance of the respective mutations or, as in the case of ANTP (Jaffe et al., 1997), the in vivo data refer to residues located outside the homeodomain. Our study firmly establishes the functional relevance of a phosphorylation site(s) within the highly conserved homeodomain of a Hox protein and, at the same time, correlates the phosphorylation status of this site with the functional status of the respective protein and its ability to recognize and bind specific target sequences.
Control of SCR activity by PP2A: towards a phosphorylation/dephosphorylation-based model of SCR function
The data from our functional knockout of dPP2A,B′ by dsRNA interference prove unequivocally that expression of dPP2A,B′ is essential for the functional activity of SCR.
Genetic studies in Drosophila have shown that Ras-1 activity positively modulates the function of Hox proteins such as proboscipedia (PB) and SCR (Boube et al., 1997)—a finding which suggested that covalent modifications triggered by Ras-1-mediated signals might influence the activity of PB and SCR. In an independent genetic screen, the catalytic subunit of dPP2A was identified as a component operating downstream of Ras-1 (Wassarman et al., 1996). Our observation that the functional activity of SCR is dependent upon the presence of dPP2A,B′ seems to provide a missing link between the studies mentioned above, suggesting that Ras-1 might influence the activity of SCR via dPP2A.
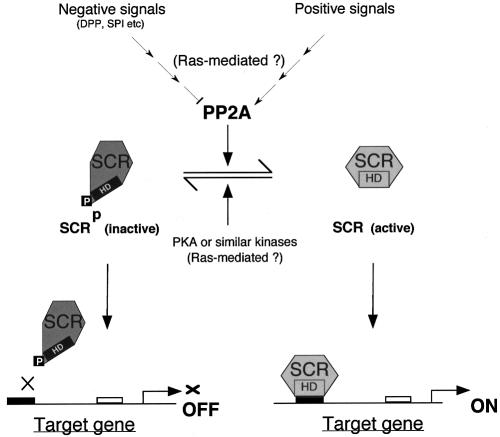
Based on our data, we propose the following model to describe the regulation of SCR activity (see also Figure 8): in a cell, where SCR function is not required continuously, the protein is locked in an inactive state by phosphorylation of residues 6 and/or 7 within the N-terminal arm of the homeodomain. The fact that, in older embryos, SCR is present but is no longer able to induce the expression of its target gene forkhead, may be a case in point (Ryoo and Mann, 1999). In response to positive signals, SCR-specific protein phosphatase (dPP2A) becomes activated, possibly through a signalling cascade involving Ras-1. In the absence of positive signals, or when negative signals, e.g. DPP and SP1 (Panzer et al., 1992; Andrew et al., 1997) prevail, specific dPP2A activity is inhibited and, as a result, SCR can no longer be maintained in its dephosphorylated state. PKA or PKA-like enzymes will phosphorylate residues 6/7 of the SCR homeodomain, thus abrogating the ability of SCR to bind to its target genes. A delicate balance between the activities of SCR-specific PP2A and specific protein kinases would thus allow a cell to fine-tune SCR activity. The proposed model centres on the phosphorylation/dephosphorylation events in the N-terminal arm of the SCR homeodomain, which was the focus of our study. Of course, this does not rule out the possibility that phosphorylation/dephosphorylation at other sites and/or other post-translational modifications also regulate SCR function.
Fig. 8. Proposed model for the regulation of SCR function.
Materials and methods
Yeast two-hybrid screen
A Drosophila third instar larvae cDNA library in the pACT-yeast vector (Durfee et al., 1993) was used to screen for protein candidates that interact specifically with the SCR bait (construct GAL4–SCR; Figure 1A). The PCY3 strain of yeast that is identical to PCY7 (Chevray and Nathans, 1992), except that it lacks the integrated GAL4 (DB)-bz-JUN fragment, was used as the host. The two-hybrid screen was performed as described (Bartel and Fields, 1995). Specific positive clones were subjected to restriction analysis and were sequenced. Homology searches were performed using the server at NCBI/NIH using Blastn, tblastn and blastp programs.
Isolation of full-length dPP2A,B′
A Drosophila embryo (2–14 h old) cDNA library in the Uni-Zap XR Vector (Stratagene) was kindly provided by B.Hemmings. Approximately 107 plaque-forming units were screened. Inserts were excised following the instructions of Stratagene. The inserts were sequenced using the ‘Big Dye Terminator Cycle Sequencing Kit’ (Perkin Elmer) and the ABI 373A stretch automated sequencer (Perkin Elmer/Applied Biosystems).
Recombinant constructs
Details of the individual plasmid constructs are available upon request. All constructs made by PCR amplification or using synthetic oligonucleotides were verified by sequencing. The GAL4 (DBD) fusion proteins, GAL4–SCR (297–378), GAL4–SCRδN1 (308–378), GAL4–SCRδN2 (323–378), GAL4–SCR(DD)δN2 (323–378; TS→DD), GAL4–SCR(A)4 (323–378; RQRTS→AQAAA), GAL4–SCRδN1δC (308–368), GAL4–ANTPδN (297–364) and GAL4–ANTPδN2 (297–351), were expressed from the yeast expression vector pAS (Durfee et al., 1993). For in vitro expression of HA-SCR(wt), HA-SCR(AA) (TS at 329/330→AA) and HA-SCR(DD) (TS at 329/330→DD), and for in vivo expression of HA-SCR, the full-length cDNAs encoding wild-type and the mutated versions of SCR with an HA tag were cloned in the Bluescript (Stratagene) and pSI (Promega) vectors, respectively. Full-length extradenticle cDNA was cloned in pSG5 (Stratagene). GST fusion proteins, GST alone, GST–HA-SCRHD(wt) (314–417) and GST–HA-SCRHD(AA) (314–417; TS at 329/330→AA), were produced using the pGEX-4T-1 vector system (Pharmacia). For the generation of transgenic flies, cDNA fragments encoding full-length HA-SCR(wt), HA-SCR(AA) and HA-SCR(DD) were subcloned downstream of tetracycline operator sequences of WTP2 to yield the constructs TETo-SCR(wt), TET-o-SCR(AA) and TETo-SCR(DD). WTP2 is identical to the WTP vector (Bello et al., 1998), except for the polylinker that is derived from the pCasPeR-hs vector (Thummel and Pirrotta, 1992).
In vivo labellings with 32Pi
COS-1 cells were electroporated with 5 µg of expression plasmid (pSI and pSI-HA-SCR), and 1 mCi of [32P]orthophosphate (Amersham) was added. The cells were processed and immunoprecipitated using 3F10 antibody and GammaBindPlus Sepharose (Pharmacia) essentially as described (Chen et al., 1999). The retained complexes were incubated at 30°C for 45 min in the presence of either 2 U of PP2A catalytic subunit (Promega) or 1 U of calf intestine alkaline phosphatase (Boehringer Mannheim) in a total volume of 25 µl. The complexes were resolved by SDS–PAGE and detected by western blot analysis using alkaline phosphatase staining to determine the levels of SCR in each sample. To detect potential phosphorylation signals, the membrane was autoradiographed.
Protein kinase and phosphatase assays
GST fusion proteins were purified according to the manufacturer’s instructions (Pharmacia). Purified and dialysed recombinant GST, GST–HA-SCRHD(wt) and GST–HA-SCRHD(AA) fusion proteins (3 µg) were used as substrates for in vitro PKA assays. The reactions were performed at 30°C for 30 min in the presence of 2 µg of catalytic subunit of cAMP-dependent PKA (Promega) and 1 µCi of [γ-32P]ATP and were re-purified on glutathione–Sepharose. The purified products were resolved by SDS–PAGE followed by western blotting using 3F10 antibody. GST–HA-SCRs were detected by alkaline phosphatase staining prior to autoradiography to normalize the protein content in each lane. Radioactive PKA and PKC (Promega) assays were carried out on biotinylated peptides (Neosystems) containing either the wild-type (TS) or mutant form (TS→AA) of the N-terminal arm of the SCR homeodomain as substrates. We used the phosphorylated peptides TKRQR(pT)SYT and TKRQRT(pS)YT (Neosystems) as substrates for phosphatase reactions driven by the catalytic subunit of PP2A (Promega). All the above enzyme reactions were performed and measured following the manufacturer’s protocols (Promega).
Fly stocks and transformants
Transformant lines were created by standard procedures (Spradling and Rubin, 1982; Spradling, 1986). The details of the driver hsp70-Tet-VP16 line (Tet-VP16) have been described earlier (Bello et al., 1998). The different responder lines carry TETo-SCR(wt), TETo-SCR(AA) or the TETo-SCR(DD) as transgenes. The in vivo studies were performed either on the F1 offspring of the cross between the driver and the responder line or with stable lines of the following genotypes: Tet-VP16;TETo-SCR(wt), Tet-VP16;TETo-SCR(AA)/TM3 and Tet-VP16;TETo-SCR(DD). Salivary glands were visualized in Tet-VP16;TETo-SCR (wild-type or mutant version)/B204 (Andrew et al., 1997) transheterozygous embryos with an anti-β-galactosidase antibody (Promega). To study genetic interactions, the following deficiencies were brought in a ph410 background (Dura et al., 1987): Df(3R)90F;91B1-2, Df(3R)90D1-2;90F3-6 and Df(3R)90C2-D1;91A1-2.
Heat shock induction of transgenes
To compare cuticle patterns, embryos were collected for 2 h and allowed to develop for a further 6.5 h (counting from the midpoint of collection). The embryos were then given a heat shock at 37°C for 1 h for TETo-SCR and 20 min for hs-SCR, after which they were allowed to recover and to develop at 25°C for ∼30 h. Cuticles were fixed and prepared as described (Cadigan et al., 1994). To examine the induction of ectopic salivary glands, embryos were collected for 3.5 h and kept to mature for 1 h. Heat shock was given at 37°C for 1 h. Thereafter, embryos were returned to 25°C and harvested 12 h later for staining with antibodies.
Antibody staining
The rabbit polyclonal serum against SCR (LeMotte et al., 1989), rat polyclonal serum against dCREB-A and mouse monoclonal antibody against β-galactosidase (Promega) were used at dilutions of 1:200, 1:1000 and 1:500, respectively. The stainings were performed as described (Grossniklaus et al., 1992).
Western blotting
Proteins were separated by SDS–PAGE (Sambrook et al., 1989). Mouse or rat monoclonal antibodies (12CA5 and 3F10, respectively) specific for the HA tag were diluted 100- to 1000-fold and the horseradish peroxidase- or alkaline phosphatase-conjugated secondary antibodies were diluted 1000- to 2000-fold in blocking solution [1% bovine serum albumin (BSA), 0.05% Tween-20 in Tris-buffered saline]. Specific bands were visualized using the ECL detection kit (Amersham) or by immunostaining using alkaline phosphatase. In addition, 32P-radiolabelled blots were autoradiographed.
Gel retardation assays
Proteins were produced in rabbit reticulocyte lysates using TNT Quick Coupled Transcription/Translation Systems (Promega). Quantification of proteins produced in vitro was deduced by substituting cold methionine with [35S]methionine in an aliquot of the reaction mix. The labelled products were analysed by SDS–PAGE followed by autoradiography. Gel retardation assays (20 µl) contained equal amounts of in vitro translated proteins, 2 µg of poly(dI–dC), 40 000 c.p.m. of 5′-32P-end-labelled double-stranded BS2 (5′-GAGCTGAGAAAAAGCCATTAGAGAAGC-3′) or the HOX–EXD site (5′-GGGGTGATTTATGGGCGCTG-3′) in 20 mM HEPES pH 7.2, 50 mM NaCl, 50 mM KCl, 1 mM dithiothreitol, 50 µg/ml BSA, 1 mM EDTA and 4% Ficoll-400. The protein–DNA complexes were resolved on pre-electrophoresed polyacrylamide gels (5%).
Synthesis and injection of dsRNA
A 745 bp DNA template from position 875 to 1620 of dPP2A,B′ cDNA was generated by PCR amplification using a set of specific primers each with a T7 promoter sequence at its 5′ end. dsRNA was synthesized in a single tube reaction using a transcription kit (Boehringer Mannheim). The dried dsRNA pellet was suspended in the injection buffer and was analysed by agarose gel electrophoresis.
dsRNA (test) and the buffer (control) injections of 0- to 60-min-old embryos were performed as described (Kennerdell and Carthew, 1998) except that an Eppendorf microinjector 5242 was used. The injected embryos were aged either for 15–18 h at 18°C or for 12–14 h at 24°C. Embryos were fixed for 30 min at room temperature. The vitelline membrane of each embryo was removed manually with a pair of needles. De-vitellinized embryos were then processed and stained for dCREB-A.
Accession number
The DDBJ/EMBL/GenBank accession No. for the dPP2A,B′ sequence is AJ277140.
Acknowledgments
Acknowledgements
This paper is dedicated to Edmond H.Fischer. We thank F.Girard for generating some of the transformant flies used in this study, S.Vincent, S.Flister and M.Seimiya for suggestions on antibody stainings, cuticle preparations and injections, respectively, and U.Kloter for the chromosomal mapping of the dPP2A,B′ gene. We are grateful to S.Ali, M.O.Fauvarque, B.Hemmings and the members of his laboratory for very helpful discussions. We thank D.Andrew, M.O.Fauvarque, J.M.Dura, K.Moses, T.Volk, G.Rubin and the Bloomington fly stock centre for providing the fly strains, A.Vigano for EXD-PSG5, D.Andrew for dCREB-A antibody, and S.Elledge for the yeast two-hybrid library and vectors. We are grateful to H.J.Fehling for many helpful suggestions and for careful editing of the original manuscript. We also thank A.Percival-Smith for his critical comments. The present work was supported by grants from the Kantons Basel-Stadt and Basel-Land and from the Swiss National Science Foundation.
References
- Andrew D.J., Baig,A., Bhanot,P., Smolik,S.M. and Henderson,K.D. (1997) The Drosophila dCREB-A gene is required for dorsal/ventral patterning of the larval cuticle. Development, 124, 181–193. [DOI] [PubMed] [Google Scholar]
- Bartel P.L. and Fields,S. (1995) Analyzing protein–protein interactions using two-hybrid system. Methods Enzymol., 254, 241–263. [DOI] [PubMed] [Google Scholar]
- Bello B., Resendez-Perez,D. and Gehring,W.J. (1998) Spatial and temporal targeting of gene expression in Drosophila by means of a tetracycline-dependent transactivator system. Development, 125, 2193–2202. [DOI] [PubMed] [Google Scholar]
- Boube M., Benassayag,C., Seroude,L. and Cribbs,D.L. (1997) Ras1-mediated modulation of Drosophila homeotic function in cell and segment identity. Genetics, 146, 619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon H.M., Martin-Blanco,E., Rosen,D. and Kornberg,T.B. (1995) Phosphorylation of the Drosophila engrailed protein at a site outside its homeodomain enhances DNA binding. J. Biol. Chem., 270, 11130–11139. [DOI] [PubMed] [Google Scholar]
- Cadigan K.M., Grossniklaus,U. and Gehring,W.J. (1994) Localized expression of sloppy paired protein maintains the polarity of Drosophila parasegments. Genes Dev., 8, 899–913. [DOI] [PubMed] [Google Scholar]
- Chang C.P., Brocchieri,L., Shen,W.F., Largman,C. and Cleary,M.L. (1996) Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox locus. Mol. Cell. Biol., 16, 1734–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Pace,P.E., Coombes,R.C. and Ali,S. (1999) Phosphorylation of human estrogen receptor α by protein kinase A regulates dimerization. Mol. Cell. Biol., 19, 1002–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevray P.M. and Nathans,D. (1992) Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc. Natl Acad. Sci. USA, 89, 5789–5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coqueret O., Martin,N., Berube,G., Rabbat,M., Litchfield,D.W. and Nepveu,A. (1998) DNA binding by cut homeodomain proteins is down-modulated by casein kinase II. J. Biol. Chem., 273, 2561–2566. [DOI] [PubMed] [Google Scholar]
- Dong J., Hung,L.H., Strome,R. and Krause,H.M. (1998) A phosphorylation site in the ftz homeodomain is required for activity. EMBO J., 17, 2308–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dura J.M., Randsholt,N.B., Deatrick,J., Erk,I., Santamaria,P., Freeman,J.D., Freeman,S.J., Weddell,D. and Brock,H.W. (1987) A complex genetic locus, polyhomeotic, is required for segmental specification and epidermal development in D.melanogaster. Cell, 51, 829–839. [DOI] [PubMed] [Google Scholar]
- Durfee T., Becherer,K., Chen,P.L., Yeh,S.H., Yang,Y., Kilburn,A.E., Lee,W.H. and Elledge,S.J. (1993) The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev., 7, 555–569. [DOI] [PubMed] [Google Scholar]
- Fauvarque M.O. and Dura,J.M. (1993) Polyhomeotic regulatory sequences induce developmental regulator-dependent variegation and targeted P-element insertions in Drosophila. Genes Dev., 7, 1508–1520. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Furukubo-Tokunaga K., Flister,S. and Gehring,W.J. (1993) Functional specificity of the Antennapedia homeodomain. Proc. Natl Acad. Sci. USA, 90, 6360–6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring W.J., Qian,Y.Q., Billeter,M., Furukubo-Tokunaga,K., Schier,A.F., Resendez-Perez,D., Affolter,M., Otting,G. and Wuthrich,K. (1994a) Homeodomain–DNA recognition. Cell, 78, 211–223. [DOI] [PubMed] [Google Scholar]
- Gehring W.J., Affolter,M. and Burglin,T. (1994b) Homeodomain proteins. Annu. Rev. Biochem., 63, 487–526. [DOI] [PubMed] [Google Scholar]
- Gibson G., Schier,A., LeMotte,P. and Gehring,W.J. (1990) The specificities of Sex combs reduced and Antennapedia are defined by a distinct portion of each protein that includes the homeodomain. Cell, 62, 1087–1103. [DOI] [PubMed] [Google Scholar]
- Graba Y., Aragnol,D. and Pradel,J. (1997) Drosophila Hox complex downstream targets and the function of homeotic genes. BioEssays, 19, 379–388. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U., Pearson,R.K. and Gehring,W.J. (1992) The Drosophila sloppy paired locus encodes two proteins involved in segmentation that show homology to mammalian transcription factors. Genes Dev., 6, 1030–1051. [DOI] [PubMed] [Google Scholar]
- Groves M.R., Hanlon,N., Turowski,P., Hemmings,B.A. and Barford,D. (1999) The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandomly repeated HEAT motifs. Cell, 96, 99–110. [DOI] [PubMed] [Google Scholar]
- Hunter T. (1995) Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell, 80, 225–236. [DOI] [PubMed] [Google Scholar]
- Hunter T. and Karin,M. (1992) The regulation of transcription by phosphorylation. Cell, 70, 375–387. [DOI] [PubMed] [Google Scholar]
- Jaffe L., Ryoo,H.D. and Mann,R.S. (1997) A role for phosphorylation by casein kinase II in modulating Antennapedia activity in Drosophila. Genes Dev., 11, 1327–1340. [DOI] [PubMed] [Google Scholar]
- Kennelly P.J. and Krebs,E.G. (1991) Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J. Biol. Chem., 266, 15555–15558. [PubMed] [Google Scholar]
- Kennerdell J.R. and Carthew,R.W. (1998) Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell, 95, 1017–1026. [DOI] [PubMed] [Google Scholar]
- LeMotte P.K., Kuroiwa,A., Fessler,L.I. and Gehring,W.J. (1989) The homeotic gene Sex Combs Reduced of Drosophila: gene structure and embryonic expression. EMBO J., 8, 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R.S. (1995) The specificity of homeotic gene function. BioEssays, 17, 855–863. [DOI] [PubMed] [Google Scholar]
- Muller M., Affolter,M., Leupin,W., Otting,G., Wuthrich,K. and Gehring,W.J. (1988) Isolation and sequence-specific DNA binding of the Antennapedia homeodomain. EMBO J., 7, 4299–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzer S., Weigel,D. and Beckendorf,S.K. (1992) Organogenesis in Drosophila melanogaster: embryonic salivary gland determination is controlled by homeotic and dorsoventral patterning genes. Development, 114, 49–57. [DOI] [PubMed] [Google Scholar]
- Ryoo H.D. and Mann,R.S. (1999) The control of trunk Hox specificity and activity by Extradenticle. Genes Dev., 13, 1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schönthal A.H. (1998) Role of PP2A in intracellular signal transduction pathways. Front. Biosci., 3, D1262–D1273. [DOI] [PubMed] [Google Scholar]
- Spradling A.C. (1986) P Element-mediated Transformation. IRL Press, Oxford, UK. [Google Scholar]
- Spradling A.C. and Rubin,G.M. (1982) Transposition of cloned P elements into Drosophila germ line chromosomes. Science, 218, 341–347. [DOI] [PubMed] [Google Scholar]
- Thummel C.S. and Pirrotta,V. (1992) New pCaSpeR P element vectors. Drosoph. Inf. Serv., 71, 50. [Google Scholar]
- Wassarman D.A., Solomon,N.M., Chang,H.C., Karim,F.D., Therrien,M. and Rubin,G.M. (1996) Protein phosphatase 2A positively and negatively regulates Ras1-mediated photoreceptor development in Drosophila. Genes Dev., 10, 272–278. [DOI] [PubMed] [Google Scholar]
- Zeng W., Andrew,D.J., Mathies,L.D., Horner,M.A. and Scott,M.P. (1993) Ectopic expression and function of the Antp and Scr homeotic genes: the N terminus of the homeodomain is critical to functional specificity. Development, 118, 339–352. [DOI] [PubMed] [Google Scholar]
- Zhao J.J., Lazzarini,R.A. and Pick,L. (1993) The mouse Hox-1.3 gene is functionally equivalent to the Drosophila Sex combs reduced gene. Genes Dev., 7, 343–354. [DOI] [PubMed] [Google Scholar]