Abstract
Mutations in the genes encoding the interacting proteins AML1 and CBFβ are the most common genetic abnormalities in acute leukaemia, and congenital mutations in the related AML3 gene are associated with disorders of osteogenesis. Furthermore, the interaction of AML1 with CBFβ is essential for haematopoiesis. We report the 2.6 Å resolution crystal structure of the complex between the AML1 Runt domain and CBFβ, which represents a paradigm for the mode of interaction of this highly conserved family of transcription factors. The structure demonstrates that point mutations associated with cleidocranial dysplasia map to the conserved heterodimer interface, suggesting a role for CBFβ in osteogenesis, and reveals a potential protein interaction platform composed of conserved negatively charged residues on the surface of CBFβ.
Keywords: chromosomal translocation/cleidocranial dysplasia/core binding factor/mutation/osteogenesis
Introduction
Leukaemias are characterized by the presence of recurrent chromosomal translocations (Rabbitts, 1994). The genes associated with these chromosomal breakpoints in acute leukaemias frequently encode transcription factors that play pivotal roles in normal development and in leukaemogenesis (Cleary, 1991; Rabbitts, 1991). The core binding factors (CBFs) are representative of this phenomenon. These heterodimeric transcription factors consist of a DNA-binding α-subunit, and a non-DNA-binding β-subunit (Ogawa et al., 1993b). Three mammalian genes encode the α-subunit: AML1/CBFA2/PEPBP2αB (herein called AML1), AML2/CBFA3/PEBP2αC and AML3/CBFA1/PEBP2αA/Osf2 (herein called AML3). All α-subunits share an evolutionarily conserved region of 128 amino acids known as the Runt domain, which mediates both DNA binding and heterodimerization to the β-subunit. The Drosophila gene runt, which is the founding member of the α-subunit family, is required for segmentation, sex determination and neurogenesis. Only one gene (CBFB) is known to encode the mammalian β-subunit, CBFβ, which associates with all three α-subunits (Ogawa et al., 1993a). However, two CBFB homologues, brother and big brother, have been identified in Drosophila (Golling et al., 1996).
The AML1 gene encodes a 453 amino acid protein with an N-terminal transcriptional inhibitory domain (residues 1–49), the Runt domain (residues 50–177), and C-terminal transcriptional inhibition (residues 178–290) and activation (residues 291–453) domains (Ito, 1999). It was cloned from one of the most frequently acquired cytogenetic abnormalities in acute myeloid leukaemia (AML), the translocation t(8;21)(q22;q22), and was subsequently shown to be involved in the recurrent chromosomal translocation t(12;21)(p13;q22) associated with childhood acute lymphoblastic leukaemias, and the translocation t(3;21)(q26;q22) associated with therapy-related leukaemias and myelodysplasia (reviewed in Look, 1997). In all of these translocations, the AML1 Runt domain becomes fused with new protein domains encoded by exons from the partner chromosome, thereby retaining the ability to heterodimerize with the CBFβ protein. Additionally, nonsense, missense and frameshift mutations in the AML1 gene are associated with sporadic AML (Osato et al., 1999), and congenital mutations in AML1 have been described in individuals with the rare autosomal dominant disease, familial platelet disorder (FDP), in which there is a congenital predisposition to the development of AML (Song et al., 1999). Interestingly, these disease-associated mutations in the sporadic and congenital disorders are clustered within the Runt domain of AML1. In man, mutations in AML3 are associated with cleidocranial dysplasia (CCD), an autosomal dominant disorder of skeletal morphogenesis (Lee et al., 1997; Mundlos et al., 1997), and again, the majority of the point mutations associated with this disorder cluster within the Runt domain (Lee et al., 1997; Quack et al., 1999; Zhou et al., 1999). Aml3 is essential for osteoblast differentiation and bone development in the mouse (Komori et al., 1997; Otto et al., 1997). Thus, the conserved Runt domain of this family of transcription factors is a key target for disease-associated mutations in man.
It is significant that the gene encoding CBFβ, the β-subunit of the core binding factors family, is also involved in chromosomal translocations in AML as a result of inv(16)(p13q22), t(16;16) and del(16)(q22), which fuse the N-terminal 165 amino acids of CBFβ in-frame with a C-terminal portion of the smooth muscle myosin heavy chain in 15% of AML (Liu et al., 1993). Thus, together, the heterodimeric CBF transcription factor genes AML1 and CBFB are the most frequently mutated genes in human acute leukaemia, accounting for 25% of AML and 20% of paediatric common B-cell acute lymphoblastic leukaemia (Look, 1997).
AML1 binds as a monomer to the core DNA sequence TGT/cGGT, which is present in a number of different viral and cellular promoters and enhancers, as well as haematopoietic cell-specific genes (Rodan and Harada, 1997). The Runt domain binds to the major groove of DNA (Thornell et al., 1988), and dimerization with CBFβ enhances the DNA-binding activity of AML1 without itself contacting DNA (Kamachi et al., 1990). Binding of CBFβ to the Runt domain protects Cys81 from oxidation by diamide, but does not hinder access of much larger reducing molecules to this site (Akamatsu et al., 1997a). These data suggest that the Runt domain undergoes a conformational change on binding to CBFβ, which results in enhanced DNA binding and alters the susceptibility of Cys81 to oxidation. However, direct evidence for this has not been obtained.
We have determined the structure of the Runt domain of AML1 bound to CBFβ to understand the mode of interaction between the two subunits, to investigate the mechanism whereby CBFβ enhances DNA binding by the Runt domain, and to understand the molecular consequences of physiologically relevant mutations. We describe the 2.6 Å resolution crystal structure of the human AML1–CBFβ complex, providing insights into the significance of human disease mutations associated with acute leukaemia and cleidocranial dysplasia.
Results
Formation of the Runt domain–CBFβ complex, crystallization and structure determination
Fragments of the human AML1 and CBFβ proteins were co-expressed in Escherichia coli. The strategy of co-expression was crucial to obtaining a soluble functional heterodimeric complex without a requirement for mutagenesis or refolding. The expressed fragments of human AML1 (residues 50–183, out of 451), corresponding to the Runt domain, and CBFβ (residues 2–135, out of 182) form a stable complex in solution that is fully active in sequence-specific DNA binding as determined by electrophoretic mobility shift assays (data not shown).
Two crystal forms of the complex were obtained, one with P61 symmetry and the second with P21 symmetry. The P21 symmetry crystals diffracted to higher resolution and were used in the structure determination. The structure was solved by the method of multiple anomalous dispersion (MAD) (Hendrickson et al., 1990), using isomorphous crystals produced from seleno-methionine (SeMet)-substituted protein. Crystallographic phases were determined using data sets collected at two wavelengths, measured at the ESRF, Grenoble, from crystals maintained at 100 K (Table I). Phases were extended to 2.6 Å resolution, and the resulting electron density map was of sufficient quality to build the initial model. The structure has been refined to a free R-factor of 29.4%, with no residues in disallowed regions of the Ramachandran plot (Table I).
Table I. Data collection, structure determination and refinement statistics.
| Data collection and MIR phasing statistics | |||||||
| Data set | Resolution (Å) | Observations/unique reflections | Completeness (last shell) % | Rmergea (last shell) | |||
| SeMet λ1 | 3.2 | 78580/60509 | 88.9 (84.1) | 0.047 (0.15) | |||
| SeMet λ2 | 3.2 | 86114/63424 | 92.8 (85.4) | 0.066 (0.25) | |||
| Native |
2.6 |
259515/71198 |
99.3 (99.2) |
0.084 (0.28) |
|
|
|
| Structure refinement statistics | |||||||
| Resolution (Å) | Protein atoms | Waters | Rcrystb | Rfreeb (% data used) | R.m.s.d. from idealityc | ||
| Bonds | Angles | Dihedrals | |||||
| 25.0–2.6 | 9551 | 121 | 26.46 | 29.43 (3) | 0.0072 | 1.3921 | 24.76 |
aRmerge = ΣhklΣi|Ii(hkl) – <I(hkl)>|/ΣhklΣi Ii(hkl).
bRcryst and Rfree = Σ|Fobs – Fcalc|/ΣFobs; Rfree calculated with the percentage of the data shown in parentheses.
cR.m.s.ds for bond angles and lengths in regard to Engh and Huber parameters.
Overall structure of the Runt domain–CBFβ heterodimeric complex
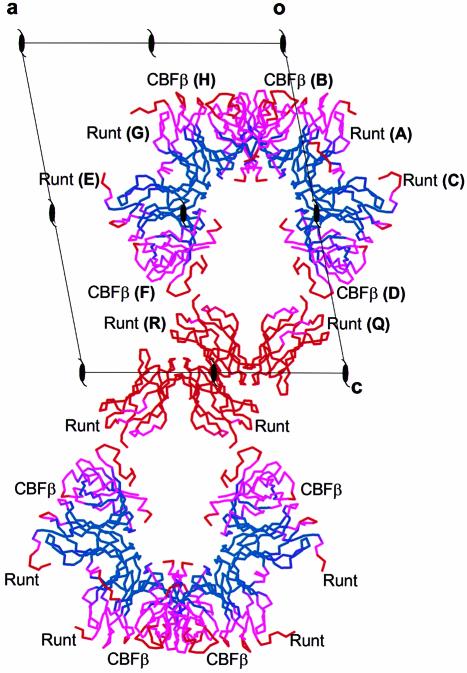
The asymmetric unit of the crystal contains six Runt domain subunits and four CBFβ subunits arranged as two dimers of Runt domain–CBFβ heterodimers [(Runt domain–CBFβ)2], and a Runt domain homodimer (Figure 1). The molecules pack in alternate layers of heterodimers and homodimers. The final atomic model is most complete for the heterodimer CD, which includes residues 54–177 of AML1, and residues 2–135 from the CBFβ subunit. Residues 52–53 and 178–183 of the Runt domain are disordered, as are residues 75–80 from CBFβ, and are not included in the model. One-hundred and twenty-one water molecules are also included. The Runt domain homodimer QR (Figure 1) has electron density at the interfaces where CBFβ binds in the heterodimers, but not sufficient to indicate an ordered CBFβ subunit. Attempts to refine with a model fitted into this density resulted in a higher free R-factor. This density may represent partial occupancy by CBFβ subunits. We have noted the formation of (Runt domain–CBFβ)2 dimers mediated by interactions between the Runt domain N-terminal residues in two different crystal forms (P21 and P61 symmetry) and in three independent examples in the asymmetric unit of the P21 crystal form. Gel filtration studies (data not shown) suggest that the formation of (Runt domain–CBFβ)2 dimers is not a consequence of crystal packing, but reflects homodimerization of the Runt domain–CBFβ complex in solution.
Fig. 1. B-factor distribution and crystal packing of Runt domain–CBFβ heterodimers. Six Runt domain subunits and four CBFβ subunits are packed in alternating layers in the crystal. One layer is composed of two dimers of heterodimers (F+E, G+H; B+A, C+D), and the second comprises a single Runt domain homodimer (Q+R). The subunits are represented as Cα traces, and are coloured according to temperature (B) factors. Colours are graded blue (≤30 Å2) through to red (≥70 Å2). Runt domain subunits are labelled A, C, E, G, Q and R; CBFβ subunits are labelled B, D, F and H. The view is down the b-axis with a, c and crystallographic 21 axes indicated.
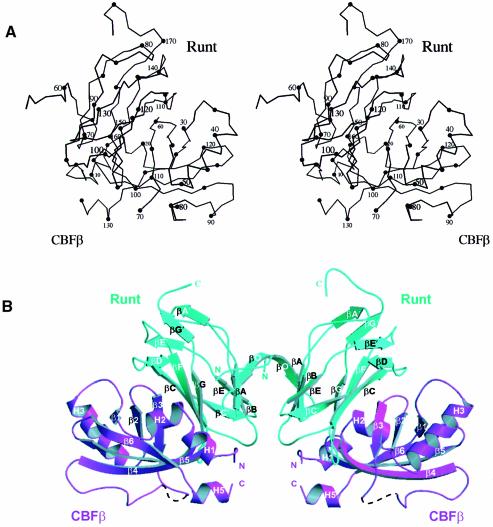
The overall structure of the (Runt domain–CBFβ)2 dimer is shown in Figure 2. Although the fold of the individual subunits of the complex is consistent with recent NMR studies (Berardi et al., 1999; Goger et al., 1999; Huang et al., 1999; Nagata et al., 1999), specific regions of the Runt domain differ in conformation (discussed later). The Runt domain of AML1 forms a 12-stranded (10 antiparallel and two parallel strands) β-barrel that adopts an s-type immunoglobulin (Ig) fold (Bork et al., 1994). CBFβ is a mixed α/β structure, consisting of a partly open six-stranded β-barrel with α-helices packed against the top and bottom. Although structural comparisons with the DALI database show that the β-barrel component of CBFβ has overall structural similarity to a number of functionally unrelated proteins, it appears that the fold is distinct, as opposed to a possible relationship to the OB fold (Goger et al., 1999). As classified in the SCOP protein structure database (Murzin et al., 1995), the OB fold consists of a five-stranded β-barrel, with Greek key topology and a shear number of 8 or 10. CBFβ forms a partly opened six-stranded β-barrel with a unique combination of a meandering up and down topology of the β-strands and a shear number of 10. Consequently, this particular β-barrel structure can be classified as a novel fold from the distinct combination of topology and shear number.
Fig. 2. Structure of the Runt domain–CBFβ heterodimeric complex. (A) A stereoscopic diagram of the Cα trace of the AML1 Runt domain, residues 54–178, bound to CBFβ residues 2–135, prepared with MOLSCRIPT (Kraulis, 1991). The numbering corresponds to the amino acid sequences of human AML1 (Miyoshi et al., 1991) and human CBFβ (Liu et al., 1993). (B) Ribbon diagram of a dimer of Runt domain–CBFβ heterodimers (two per asymmetric unit). Runt domain, cyan; CBFβ, magenta. The Runt domain β-strands are labelled βO to βG, consistent with the established immunoglobulin fold nomenclature and with Nagata et al. (1999), except for the extensions to strands βA and βG, which have been labelled βA′ and βG′, respectively. The β-strands of CBFβ are labelled β1–6, and the helices are numbered H1–5, consistent with previous nomenclature (Goger et al., 1999). Only one hydrogen bond corresponding to a short potential 310-helix was seen in the region corresponding to helix H4, which was therefore not represented. CBFβ residues 73–78 are disordered and are shown as a dashed line.
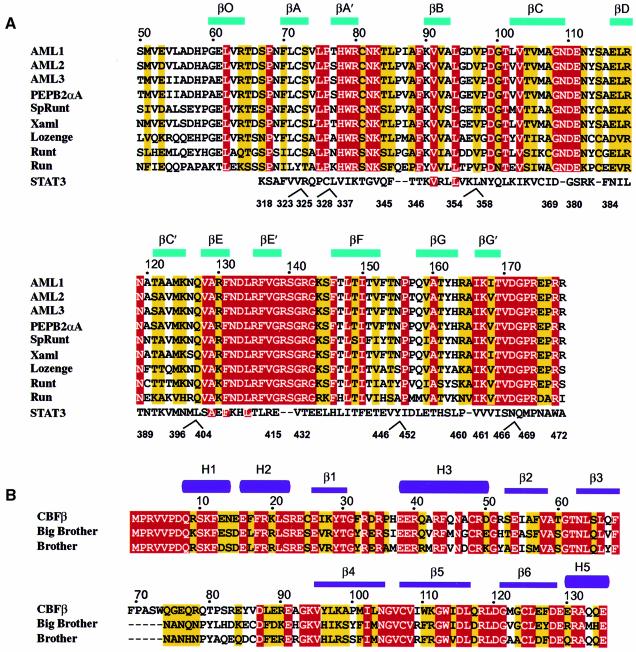
There are extensive heterodimeric contacts at the interface between the Runt domain and CBFβ subunits, and homodimeric contacts between the N-termini of the Runt domains, but no contacts between the CBFβ subunits. The temperature factors for the (Runt domain–CBFβ)2 structure (Figure 1) suggest that the Runt domain forms a relatively stable core, whereas the CBFβ subunit is more mobile. Figure 3 shows the secondary structure elements for the conserved Runt domain and CBFβ determined from our structure, aligned to the protein sequences of various family members.
Fig. 3. Amino acid sequence alignment of the Runt domain and CBFβ. (A) The amino acid sequence accession numbers for the Swiss-Prot and DDBJ/EMBL/GenBank databases are given in parentheses. The sequence information is derived from human AML1 (Q01196), residues 50–178; human AML2 (Q13761), residues 54–182; human AML3 (Q08775), residues 102–220; murine PEBP2αA (Q08775, D14636), residues 50–178; sea-urchin SpRunt-1 (Q26628), residues 57–185; frog Xaml1 (O73725), residues 50–178; fruit-fly Lozenge (Q24183), residues 278–406 and Runt (Q24709), residues 106–234; nematode Run (O01834), residues 10–138. The last line of the alignment shows the structural similarity of the Runt domain to murine STAT3β (P42227), residues 318–472, with manually introduced sequence gaps indicated. Identical residues are highlighted in red, conservatively substituted residues are highlighted in yellow. Sequences were aligned using CLUSTAL_W (Thompson et al., 1994) and manually adjusted. (B) Sequence alignment of human CBFβ (Q13951), residues 1–135, and the fruit-fly homologues Brother (Q24039) and Big Brother (Q24040). Residues 70–74 in CBFβ have no equivalent in Drosophila, and are indicated by dashed lines.
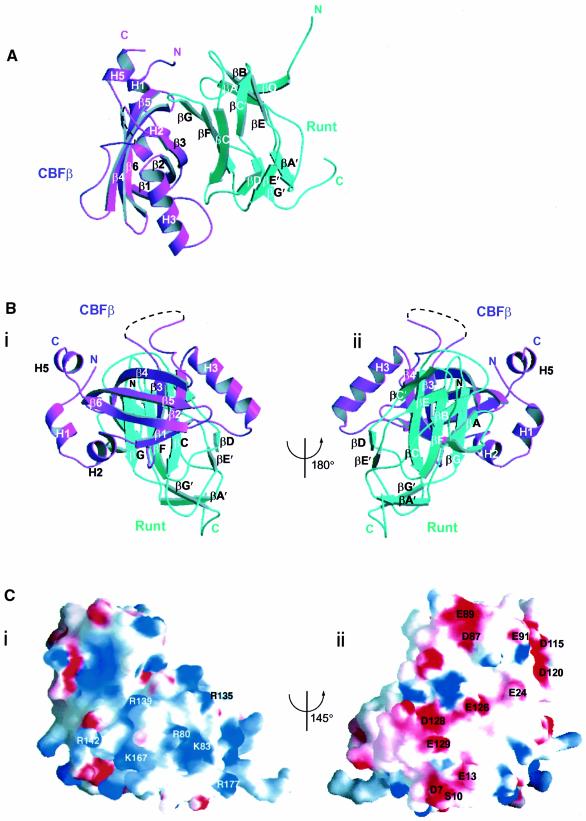
The root mean square deviations (r.m.s.ds) of the Cα backbone traces between different Runt domain subunits in the asymmetric unit range between 0.15 and 0.35 Å, and for the four CBFβ subunits, the values range between 0.16 and 0.36 Å. The overall dimensions of a single Runt domain–CBFβ heterodimer are ∼41 × 50 × 27 Å. The Runt domain and CBFβ interact along a large continuous curved interface (Figure 4A), and are oriented such that the long axes of the two β-barrel domains are orthogonal to one another (Figure 4B). When viewed from the perspective of Figure 4B, with the C-terminus of the Runt domain oriented downward, it is evident that CBFβ makes contact only with the upper part of the Runt β-barrel. CBFβ makes no direct contact with Runt domain loops βA′–B, βE′–F or the C-terminus. Residues within these loops have been shown biochemically to be essential for DNA binding (Kagoshima et al., 1996; Osato et al., 1999). The α-helices H5, H1 and H2 of CBFβ lie on the right lateral aspect of the upper half of the Runt β-barrel, and helix H3 lies on the upper left lateral aspect (Figure 4Bii). Our data differ from the proposed mode of interaction between the Runt domain and CBFβ, based on chemical shift analysis (Nagata et al., 1999). It was suggested that CBFβ is oriented in the heterodimer with helices H1, H2 and H5 up, and helix H3 down, placing CBFβ residues Gln74, Gln79 and Arg83 on the same face of the heterodimer as the proposed DNA-binding surface of the Runt domain (loops βA′–B and βE′–F and the C-terminus). In fact, the crystal structure demonstrates that the CBFβ is rotated by 90° relative to the previous proposal (Figure 4B), so that the evolutionarily non-conserved CBFβ loop β3–β4 (residues 68–93) makes no contribution either to the heterodimer interface or to the DNA binding surface of the molecule.
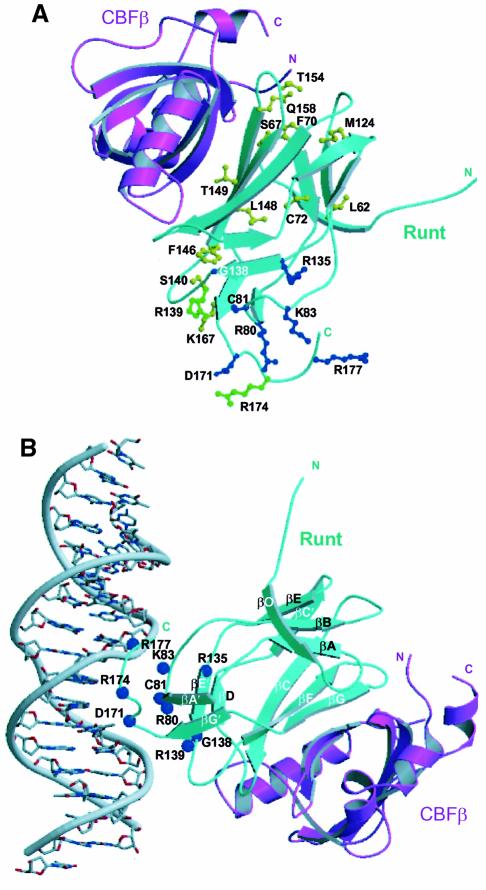
Fig. 4. The mode of interaction between the Runt domain and CBFβ. (A) Runt domain (cyan) and CBFβ (magenta) viewed perpendicular to the long axis of CBFβ. The concave surface of the Runt domain β-sheet, formed from strands βG, βF and βC, packs against the complementary convex strand β3 of CBFβ. (B) Two views of the Runt domain–CBFβ structure (i and ii), related by a 180° rotation about the vertical axis. The long axes of the CBFβ and Runt domain β-barrels are orthogonal to one another. (C) Electrostatic surface potential of the Runt domain--CBFβ heterodimer. The two views are related by a 145° rotation about the vertical axis. Positive areas are shaded blue; negative areas are shaded red. This figure was prepared using GRASP (Nicholls et al., 1991). (i) Positive surface. Labelled residues are mutated in cleidocranial dysplasia, familial platelet disorder and sporadic acute myeloid leukaemia. (ii) Negative surface. The evolutionarily conserved, negatively charged residues that are labelled are all located on the surface of CBFβ.
The conformation of the C-terminus of the Runt domain (residues 169–177), which is essential for DNA binding, is clearly defined in the crystal structure. The C-terminus forms a loop that extends towards the N-terminus of the Runt domain, passing below strand βA′ (Figure 4B). All the loops on the lower face of the Runt domain β-barrel, as orientated in Figure 4B (βC–D, βE′–F, the C-terminus and βA′–B), are linked to one another and are well buttressed on one side as a result of the interaction of loop βC–D with CBFβ. Runt domain loop βC–D, which has not been implicated in DNA binding, makes a number of contacts with CBFβ through the side chain of Tyr113. Finally, we find that the three Cys residues in CBFβ (Cys25, Cys107 and Cys124) are not related to the heterodimer interface as proposed (Huang et al., 1999), suggesting that these residues are not directly related to the modulation of oxidation state-dependent behaviour of AML1.
Electrostatic surface potential of the Runt domain–CBFβ heterodimer
There are two contrasting surfaces on the heterodimer in terms of the electrostatic surface potential (Figure 4Ci and ii). The strongly positive surface corresponds to the position of loops βA′–B, βE′–F and the C-terminus of the Runt domain. This supports the biochemical and human mutation data, which directly implicate these regions of the Runt domain in DNA binding (Lenny et al., 1995; Kagoshima et al., 1996; Osato et al., 1999). In contrast, rotation by 145° from this region reveals a strikingly negative surface (Figure 4Cii), corresponding to the upper outer surface of CBFβ. Five areas on the surface of CBFβ make up this charged surface: the end of strand β6 and the residues prior to helix H5; residues in loop β3–β4; the β5–β6 loop; strand β1; and helix H1. The majority of these residues are conserved in evolution (Figure 3B), suggesting a conserved biological function.
The Runt domain–CBFβ interface
The Runt domain and CBFβ subunits interact over a large continuous curved interface (Figure 4A), such that a total of 1900 Å2 in solvent-accessible surface area is buried [assuming default radii of GRASP and a 1.4 Å solvent probe (Nicholls et al., 1991)]. The interaction surface of the Runt domain is concave, and packs against a complementary convex surface on CBFβ. The curved heterodimerization surface of the Runt domain shown in Figure 4A involves loops βF–G, βO–A and βB–C at the top of the β-barrel; strands βC, βF and βG; and loop βC–D at the bottom of the β-barrel. The regions of CBFβ involved in heterodimerization are the N-terminal loop and helix H1; strand β1 and loop β1–H3; strands β2, β3 and the connecting loop β2–β3, which together form the central convex interaction surface; strand β4 and the proximal part of loop β4–β5.
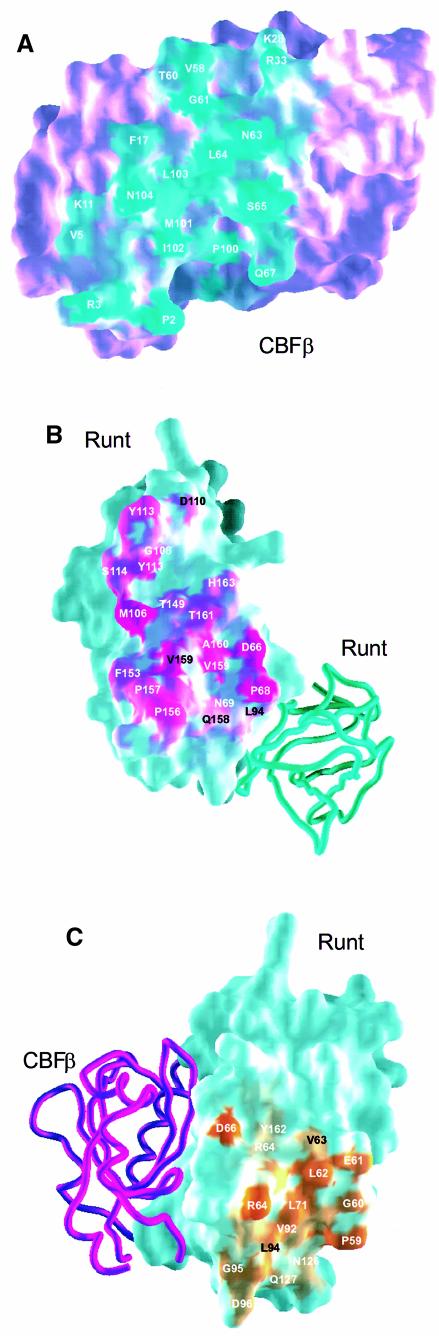
Figure 5A shows the residues on the surface of CBFβ that are buried in the Runt domain at the interface. Comparing interface residues between human CBFβ and the Drosophila homologues, 13 residues are identical and six involve similar substitutions (Figure 3B). Only two residues have non-conservative substitutions between Drosophila and human. Mutagenesis studies demonstrated a requirement for the N-terminal 5–6 residues of CBFβ for heterodimerization to the Runt domain (Golling et al., 1996; Kagoshima et al., 1996). We find that the Runt domain–CBFβ interaction involves the N-terminus of CBFβ (residues 2–5). The Runt domain loop βO–A, loop βB–C and strand βG are all involved in contacts with the N-terminus of CBFβ, with four potential hydrogen bonding interactions mediated by CBFβ Arg3. Significant chemical shifts at Gln74, Gln79 and Arg83 were noted on binding of a Runt domain–DNA complex to CBFβ (Goger et al., 1999), but the crystal structure demonstrates that this is not a result of burying these residues at the heterodimer interface, and that they do not come to lie proximal to the putative DNA-binding loops βE′–F or βA′–B of the Runt domain (see below).
Fig. 5. Interaction surfaces within the (Runt domain–CBFβ)2 complex. (A) Heterodimerization surface of CBFβ. Solvent-accessible surface is shown in purple; residues buried in the Runt domain surface are shown in cyan. All interface residues apart from Q67 and P100 are conserved (see also Figure 3). (B) Heterodimerization surface of the Runt domain. Solvent-accessible surface is shown in cyan; residues buried in the interface with CBFβ are shown in magenta. The Runt domain homodimer binding partner is shown in worm representation (cyan). (C) Homodimerization surface of the Runt domain. Solvent-accessible surface is shown in cyan; buried residues are shown in orange. The related heterodimeric CBFβ subunit is shown in worm form (magenta).
Two-thirds of the residues on the surface of the Runt domain that are buried in the CBFβ subunit (Figure 5B) are either conserved or conservatively substituted between members of the α-subunit family (Figure 3A). Structurally, the conserved cis-Pro156 induces a kink in Runt domain loop βF–G, which makes important contacts with residues in CBFβ. The Runt domain strand βG (residues 159–161) pairs with CBFβ strand β4 (residues 102–104) to form a short antiparallel β-sheet extension. The β-sheet extension between the two subunits is stabilized by a cluster of conserved hydrophobic residues (Runt domain Val159; CBFβ Met101, Ile102 and Leu103). At the opposite end of the heterodimer interface, Runt domain Ser114 and Tyr113 provide a large surface area of interaction with CBFβ. These residues lie in a solvent-accessible polar environment, consistent with previous spectroscopic analysis (Crute et al., 1996). CBFβ residues Glu111 and Asp110 also contribute to this polar environment. Runt domain Met106 makes a significant contribution to the buried surface area in the central part of the heterodimer interface. The functional importance of this residue to the interaction is supported by biochemical studies, which demonstrate that a M106V mutation abolishes the interaction between AML1 and CBFβ in vitro (Akamatsu et al., 1997b). There are a total of 42 contacts between the two subunits, 10 of which represent potential hydrogen-bonding interactions. Bridging contacts are also mediated indirectly via water molecules near the interface. We have identified eight water molecules buried at the heterodimer interface, the role of which is presumably to optimize the complementarity of the interaction interfaces.
The structure of the heterodimer interface is consistent with the in vitro Runt domain mutants M106V, G108R and N109D, which show loss of heterodimerization, but preservation of DNA-binding activity (Akamatsu et al., 1997b). Mutations in the Runt domain residues 66–69 located on the βO–A loop also abolish heterodimerization (Lenny et al., 1995). Recent in vivo experiments in Drosophila demonstrated that the G108R Runt protein mutant was dysfunctional in several in vivo assays, showing that the interaction of Runt with the Drosophila CBFβ homologues is essential in vivo for the function of the transcription complex (Li and Gergen, 1999).
Homodimer interface of the Runt domain
The crystal structure identifies two homodimeric interactions between the Runt domains, one interface between the Runt domains in the dimer of heterodimers, and a smaller interface involving a subset of these interactions within the homodimer QR (890 Å2 solvent-accessible surface area buried as opposed to 1300 Å2). The residues buried at the interface are shown in Figure 5C. The N-terminal residues (59–66) make the most prominent contribution to this interface, which is stabilized further by contacts between strands βA and βB, and loops βB–C and βC′–E. Runt domain Asp66 contributes to both the heterodimer and homodimer interfaces. The homodimeric interaction in the dimer of heterodimers is mediated by a short edge to edge anti-parallel β-sheet formed from the pairing of residues 60–62 at the N-terminal end of the Runt domain, and is stabilized by a hydrophobic cluster involving Pro59, Leu62, Val63, Leu71, Val92 and Leu94 from each subunit. The hydrophobic core of the interface shows strong sequence conservation (Figure 3A).
Comparison with NMR data shows conformational differences in the Runt domain
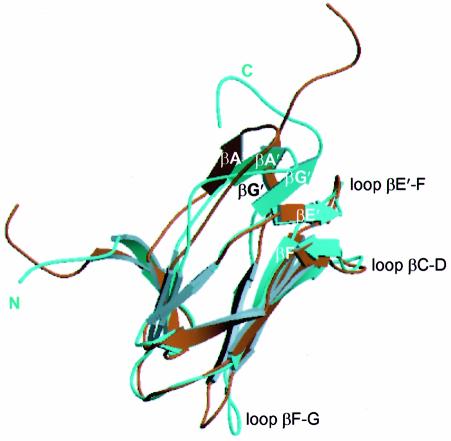
Interaction with CBFβ is essential for the in vivo function of AML1 (Wang et al., 1996). In vitro, CBFβ decreases the dissociation constant (Kd) of the isolated Runt domain for its cognate DNA-binding site 5- to 10-fold (Kagoshima et al., 1996). The molecular basis for this enhanced DNA-binding affinity of the Runt domain in the presence of CBFβ is not yet established. The availability of structures of the Runt domain–CBFβ binary complex (this work), together with the co-ordinates for the NMR structures of the Runt domain (performed in the presence of DNA) (Nagata et al., 1999) and the bundle of NMR structures of the uncomplexed CBFβ subunit (Goger et al., 1999; Huang et al., 1999), allows us to address this issue. Although the experimental restraints for the NMR structures are not available, examination of the bundle of NMR structures gives some idea of the flexibility of the structure and the accuracy of the model. We found no significant differences in the crystal structure of CBFβ with respect to the available NMR co-ordinates (Goger et al., 1999; Huang et al., 1999) (PDB codes 2jhb and 1cl3), apart from the disorder in the flexible non-conserved loop β3–β4 (residues 75–80) in the heterodimer. The Cα traces for the structures of the CBFβ- and DNA-bound forms of the Runt domain superimpose well, except in the region of the putative DNA-binding loops, where the structures differ significantly (Figure 6). In the Runt domain–CBFβ complex, the C-terminal residues of the Runt domain are ordered, defining the conformation of this essential DNA-binding region. The parallel β-sheet, βA′–βG′, is shifted significantly towards the βE′–F loop, and is stabilized in this position by the potential main-chain hydrogen bonding interactions between strand βG′ (Ile168) and loop βE′--F (Arg139 and Gly141). Cys81 forms potential main- and side-chain hydrogen bonding interactions linking loop βA′–B (Cys81) both to strand βG′ (Thr169) and to strand βE′ (Val137). Relative to the DNA-bound Runt domain conformation, Cys81 shows the largest change in relative position of any Cα (7.2 Å). This change in chemical environment may alter the susceptibility of Cys81 to oxidation (Akamatsu et al., 1997a).
Fig. 6. The Runt domain complexed to CBFβ has a novel conformation. Least squares superposition of the Cα trace of the Runt domain crystal structure onto the most closely related NMR conformer [PDB code 1cmo, number 38, (Nagata et al., 1999)]. The analysis was performed for all the NMR conformers, and similar results were obtained. The NMR structure is shown in brown and the crystal structure is shown in cyan. The crystal and NMR structures differ in the relative positions of the βA′–βG′ sheet and the βF--G loop. The C-terminus is well defined in the crystal structure.
To accommodate the movement of the βA′–βG′ sheet, there is a displacement in the main-chain conformation of residues 85–89 (Pro86 is shifted by 5.9 Å), and an upward kink at Ala165 (shifted by 6.5 Å), relative to the NMR structure. The kink at Ala165 is stabilized by main-chain potential hydrogen bonding with Tyr162. Tyr162 projects into the hydrophobic core of the molecule, but the residues immediately adjacent (Thr161 and His163) are buried in the CBFβ subunit at the heterodimer interface, suggesting a mechanism for induced conformational change transmitted from the heterodimer interface by stabilization of the orientation of the Tyr162 side chain. There is also a change in the conformation of the Runt domain βF–G loop (residues 153–159), which is twisted through a 90° angle in the crystal structure, resulting in displacement of Pro156 by 6.15 Å relative to its position in the NMR structures. This region of the Runt domain forms extensive contacts with CBFβ (Figures 4A and 5B).
These comparisons demonstrate ordering of the C-terminus of the Runt domain in the presence of CBFβ, and reveal conformational flexibility within the Runt domain that can alter the relative orientation of a specific DNA-binding subdomain. This suggests that CBFβ may enhance DNA binding by the AML1 Runt domain through the stabilization of this conformation.
Discussion
The structure demonstrates that the interaction between the AML1 Runt domain and CBFβ is mediated by highly conserved residues from each subunit, and therefore provides a structural paradigm for heterodimerization by other Runt domain family members. In addition, the molecular consequences of several point mutations associated with human disease can be explained from the structure of the complex. Furthermore, interaction of the Runt domain and CBFβ appears to have conformational consequences for a region of the Runt domain directly implicated in DNA binding.
Mutations in cleidocranial dysplasia map to heterodimer interface
Congenital point mutations within the Runt domain of AML1 and AML3 have been attributed to the pathogenesis of familial platelet disorder, which progresses to acute myeloid leukaemia (AML1) (Song et al., 1999), and to a disorder of osteogenesis, cleidocranial dysplasia (AML3) (Lee et al., 1997; Mundlos et al., 1997). Somatic mutations in AML1 have also been demonstrated in acute myeloid leukaemia (Osato et al., 1999).
The majority of point mutations associated with AML involve residues directly implicated in DNA binding. In contrast, recently described missense mutations associated with skeletal dysgenesis map to the heterodimer interface (Quack et al., 1999; Zhou et al., 1999) (Figure 7A). Three mutations affect residues identified in our structure as buried at the heterodimer interface: T149A, Q158R and S67R (Figure 7A), which map to the Runt domain strands βF, strand βG and loop βO–A, respectively. The T149A mutation is associated with variable expressivity in a single family, ranging from classical CCD, to dental anomalies alone, while the Q158R and S67R mutations are associated with classical CCD (Zhou et al., 1999). The T149A mutation would be expected to disrupt a potential hydrogen bond with CBFβ Asn63. Q158 interacts with CBFβ, but also stabilizes the conformation of Runt domain loops βO–A and βF–G. The Q158L mutation in vitro results in impairment of DNA binding and loss of heterodimerization (Lenny et al., 1995). These mutations demonstrate that the integrity of Runt domain loops βF–G and βO–A is crucial both for interaction with CBFβ and for DNA binding. This may be due to indirect stabilization of the DNA-binding surface, or to an effect on the overall stability of the Runt domain. The Runt domain M124R mutation associated with CCD (Lee et al., 1997) disrupts DNA binding, but maps to a region away from the potential DNA-binding surface. This mutation introduces a bulky, charged residue into the hydrophobic core of the Runt domain, and would be expected to destabilize the structure.
Fig. 7. Location of human disease mutations on the AML1–CBFβ structure. (A) A ribbon diagram of the Runt domain–CBFβ heterodimer in which residues mutated in cleidocranial dysplasia (CCD) are shown in yellow, familial platelet disorder (FPD) and acute myeloid leukaemia (AML) are shown in blue. Mutations common to CCD and FPD/AML are shown in green. The ribbon representation of the Runt domain is in cyan, and that of CBFβ is in magenta. (The AML-associated biallelic point mutations G138D, R135G, D171G are a personal communication from P.Fenaux.) (B) Model orienting the Runt domain–CBFβ heterodimer with respect to B-form DNA. The model was generated by least squares superposition of the co-ordinates of the STAT3β–DNA complex (PDB code, 1bg1) (Becker et al., 1998) onto the AML1 Runt domain. The amino acid residues mutated in human disease mutations are indicated as blue spheres. The Runt domain is shown in cyan, and CBFβ in magenta. The model is consistent with: the electrostatic surface potential of the heterodimer (Figure 4C); in vitro mutagenesis data (Lenny et al., 1995; Kagoshima et al., 1996; Osato et al., 1999); chemical footprinting analysis (Thornell et al., 1988; Melnikova et al., 1993); NOE data from NMR studies (Nagata et al., 1999); and the observations that CBFβ does not contact DNA directly, or extend the Runt domain footprint on DNA (Kamachi et al., 1990).
Our structure defines the conformation of the Runt domain C-terminus, which is essential for nuclear localization and DNA binding (Kagoshima et al., 1996; Osato et al., 1999; Quack et al., 1999). Mapping mutations associated with FPD, AML and CCD onto the structure clearly delineates the potential DNA-binding surface (Figure 7A). One of the mutations described in FPD, R139Q, which is also associated with CCD, has indeed been shown to confer loss of DNA-binding function in vitro (Zhou et al., 1999). Arg174 was the residue most frequently affected by mutation in the AML3 Runt domain in a recent CCD study (Quack et al., 1999), which also demonstrated that the mutation R174Q abolishes DNA binding in vitro, and that R174Q and R174W disrupt the function of the nuclear localization signal, preventing AML3 accumulation in the nucleus.
Insight into the DNA-binding function of the AML1–CBFβ heterodimer
The structural similarity of the AML1 Runt domain to transcription factors p53, NF-κB p52, NF-κB p65, NFAT1, the T-domain and the STAT proteins has been shown (Berardi et al., 1999; Nagata et al., 1999). The co-ordinates from our crystal structure were used to search the protein database (Dali server), which demonstrated that the Runt domain is most closely related structurally to the transcription factor STAT3β. Structural alignment of the Runt domain with STAT3β gives an r.m.s.d. of 2.1 Å over 82 residues (see Figure 3A). The structural similarity is prominent even within the loop regions of the Runt domain, most strikingly within the C-terminus. The smaller size of the Runt domain DNA-binding module is related to the shorter length of the putative DNA-binding loops. In contrast to STAT3β, which binds DNA as a dimer, the Runt domain appears to bind as a monomer (Ogawa et al., 1993a).
We have generated a model for major groove DNA binding by the Runt domain–CBFβ heterodimer, by superposition of the STAT3β–DNA co-crystal complex (Becker et al., 1998) onto the AML1 Runt domain (Figure 7B). The orientation of the Runt domain–CBFβ complex with respect to the DNA is consistent with the location of human disease mutations (Figure 7A) and other data (see Figure 7 legend). Three loops from the Runt domain β-barrel are predicted to make close contacts with the DNA. The βA′–B loop and the C-terminus are predicted to participate in core sequence recognition in the major groove. The conformation of the C-terminus, which interacts with loop βA′–B through the small parallel βA′–βG′ sheet, is a conserved structural feature between the related Ig fold transcription factors. Our model predicts that the βE′–F loop would make DNA backbone and minor groove contacts. The analogous segment in STAT3β is involved in DNA backbone interactions (Becker et al., 1998).
Berardi et al. noted nuclear Overhauser effects (NOEs) to DNA from Runt domain loop βF–G in their NMR studies (Berardi et al., 1999). However, in the crystal structure of the heterodimeric complex, residues on the surface of Runt domain loop βF–G (cis-Pro156, Pro157, Gln158) are buried within the CBFβ subunit (Figure 5B). Our model is compatible with existing footprinting data that demonstrate that the AML1–CBFβ complex protects ∼10 bp of DNA (Thornell et al., 1988). We therefore propose that the core binding factor heterodimer binds DNA through Runt domain loops βE′–F and βA′–B, and the C-terminus.
The role of the CBFβ interaction
The interaction of CBFβ with the Runt domain enhances its DNA-binding affinity (Kagoshima et al., 1996). We observe significant differences in the conformation of loop βA′–B and the C-terminus of the Runt domain in our structure compared with the NMR data (Nagata et al., 1999). The AML1–CBFβ heterodimer adopts the same conformation in multiple copies within the asymmetric unit of the crystal, demonstrating that the structure is not an artifact of specific crystal contacts. The shift of 7.2 Å in the position of Cys81 offers a structural explanation for biochemical data that demonstrate protection from oxidation for this residue following binding of the Runt domain to CBFβ (Akamatsu et al., 1997a). This residue is predicted to contact the DNA backbone (consistent with the C81D mutation that abolishes DNA binding (Akamatsu et al., 1997a), analogous to the role of conserved Cys residues in other Ig fold transcription factors (Ghosh et al., 1995). This indicates that the conformational changes observed in the heterodimer are biologically relevant.
Conformational differences at the DNA-binding surface are consistent with reported changes in the NMR spectra of the Runt domain upon binding CBFβ (Berardi et al., 1999). STAT3β superimposes well onto the essential DNA-binding C-terminus of AML1, and NMR studies have demonstrated NOEs between Arg80 and a cytidine residue in the major groove of the DNA, directly implicating Runt domain loop βA′–B in DNA recognition (Nagata et al., 1999). The analogous segment in five other transcription factors binds to DNA in the major groove. Therefore, constraints on the flexibility of the βA′–B loop and the C-terminus would be expected to have an important influence on DNA-binding affinity. Our data therefore support the hypothesis that one role of CBFβ is to stabilize the relative orientation and conformation of a modular DNA-binding subdomain consisting of loop βA′–B and the C-terminus, in the context of the ternary complex with DNA.
The solvent-exposed surface of CBFβ in the heterodimer may have additional functions in transcription. CBFβ may act as a platform to recruit other proteins to modulate the transcriptional activity of AML1 in vivo. Our structural data reveal a negatively charged surface composed of conserved residues that may provide a surface for such protein interactions (Figure 4C). The significance of the Runt domain homodimerization in our structure is at present unclear, but may indicate a potential binding site for other proteins in vivo. Physical interaction has been demonstrated, for example, between the AML1 Runt domain and the transcription factors C/EBPα and PU.1 (Petrovick et al., 1998). Thus, the structure gives insight into the surfaces of the AML1–CBFβ heterodimer, which may recruit other tissue-specific factors to stimulate lineage-restricted transcription.
Materials and methods
Protein expression and purification
The DNA sequence encoding human AML1, residues 50–183, was amplified from plasmid pET/RM7 (gift of Dr F.Calabi) by the polymerase chain reaction (PCR). The PCR fragment was cloned into the mini-pRSET vector (gift from O.Perisic), adding 16 residues at the N-terminus (MRGSHHHHHHGLVPRG). The DNA sequence encoding human CBFβ, residues 1–135, was amplified by PCR from cDNA synthesized from RNA purified from HEL cells. The fragment was cloned downstream of a Shine–Dalgarno sequence in pBluescript, and subsequently subcloned into the mini-pRSET vector downstream of the AML1 coding sequence. The pmini-α/β expression plasmid was transfected into E.coli C41 (DE3) cells (Miroux and Walker, 1996). Transformed cells were grown at 37°C in 2× TY medium containing ampicillin at 100 µg/ml to an OD600 of 0.6 and induced for 4 h with 1 mM isopropyl-β-d-thiogalactopyranoside. The protein complex was purified on a combination of Ni2+-NTA affinity resin (Qiagen), Resource S (Pharmacia) and HiPrep Sephacryl S100 (Pharmacia) columns, with no subsequent cleavage of the His-tag from the AML1 fragment. The protein was concentrated to 12 mg/ml in a solution containing 20 mM HEPES pH 6.6, 300 mM NaCl, 10 mM dithiothreitol (DTT) and 1 mM EDTA, and stored in liquid nitrogen.
Preparation of SeMet-substituted AML1–CBFβ
SeMet-substituted protein was expressed from the pmini-α/β plasmid in the E.coli strain 834 (DE3). Log phase cells (OD600 = 0.3) precultured in 2× TY were diluted 1:100 into 2× M9 minimal medium supplemented with 0.4% glucose, 19 amino acids at 40 µg/ml, seleno-l-methionine (Sigma) at 40 µg/ml, and vitamins at 1 µg/ml (Ramakrishnan and Biou, 1997). Bacteria grew at 37°C and were induced after 8 h (OD600 = 0.6). Induction was at 25°C overnight (OD600 = 1.1). The method of protein purification was identical to that of the unsubstituted protein. SeMet substitution was assayed by MALDI mass spectrophotometry, and was consistent with complete substitution of methionine residues with SeMet, but loss of the N-terminal methionine of CBFβ.
Crystallization
Crystals of the complex were grown as hanging drops against a reservoir containing 100 mM imidazole, 6% PEG 8000, 5% sucrose and 5 mM DTT at 21°C, using 1 µl of 12 mg/ml protein solution and 1 µl of crystallization solution. Macroseeding was used to obtain diffraction quality crystals, which grew to their final size over 2 weeks. Crystals were harvested by transfer to a cryobuffer containing 30% sucrose, 12% PEG 8000, 5% MPD, 200 mM NaCl, 100 mM imidazole pH 6.5 for 30 s, and cryocooled in liquid nitrogen. Crystals were prepared from SeMet-substituted protein under similar conditions, except that 5% MPD was incorporated as an additive in the initial drops. Crystals belonging to space group P61 were also obtained using a crystallization solution with 400 mM NaCl, 50 mM HEPES pH 7.5, 5 mM DTT, 5% PEG 8000. These were cryocooled after transferring to a buffer containing 30% glycerol, 100 mM HEPES pH 7.5, 200 mM NaCl, 5% PEG 8000.
Data collection and structure determination
Crystals used for native protein data collection were typically 400 × 200 × 40 µm or larger, had P21 symmetry, with unit cell dimensions of a = 103.2 Å, b = 79.4 Å, c = 130.1 Å, β = 101.6°, a solvent content of 60%, and diffracted to 2.6 Å. SeMet-substituted crystals had unit cell dimensions a = 104.0 Å, b = 79.3 Å, c = 130.9 Å, β = 101.5°, were somewhat smaller and diffracted to only 2.8 Å resolution. Diffraction data for the native crystal were collected at ESRF beamline ID14-3 (Grenoble) using a MAR CCD detector and the data for the SeMet-substituted crystal were collected at ID14-4 using an ADSC CCD detector, using crystals maintained at 100 K (Table I). Native data were processed using MOSFLM (Leslie, 1992). Each data set was collected as 180 1° oscillations. For the SeMet-substituted crystal, data sets were collected at two wavelengths, λ1 = 0.9793 Å and λ2 = 0.9795 Å, corresponding to the peak and inflection points in the fluorescence spectrum of a SeMet-substituted crystal. Data sets for the SeMet-substituted crystal were integrated with HKL2000 and scaled with SCALEPACK (Otwinowski and Minor, 1997). The native data were scaled and merged with SCALA (CCP4, 1994). Final merging of the SeMet data was carried out with SOLVE (Terwilliger and Berendzen, 1999). The R-factor for dispersive differences between data collected at λ1 and data collected at λ2 was 0.046. The R-factor for anomalous differences was 0.077 for λ1 and 0.066 for λ2. SOLVE (Terwilliger and Berendzen, 1999) was used to locate 21 Se sites. The SOLVE Z-scores for the Patterson correlation, cross-validation Fourier, native Fourier and mean figure of merit for the 21-site solution were 7.4, 52, 21 and 9.4, respectively. The overall Z-score was 66. The mean figure of merit was 0.47. SeMet data to 3.5 Å resolution were used to provide an initial estimate for the phases of the native data set. Density modification was carried out to improve the phases and extend them to 2.6 Å resolution (Brünger et al., 1998). A model was built into the 2.6 Å resolution electron density maps using the program O (Jones et al., 1991), and refined using CNS (Brünger et al., 1998). The average B-factor for all atoms is 55.9 Å2. The structure shows good stereochemistry with no residues in disallowed regions of the Ramachandran plot.
Acknowledgments
Acknowledgements
We thank G.Leonard and S.McSweeney for help with MAD data collection at synchrotron beamline ID14-4 at ESRF. We also thank staff at the ESRF synchrotron beamlines ID2b and ID14-3, station X11 at EMBL Hamburg, Station 9.6 at Daresbury SRS and Elettra, Italy for help with data collection. We thank F.Calabi for clone pETRM7, J.Walker for C41 cells, O.Perisic for clone mini-pRSET and for advice, and A.Murzin for discussion on protein structure. We also thank M.Montgomery, I.Arechaea and D.Stock for advice, L.Chapman, L.Fairall, D.Rhodes, D.Neuhaus and G.Varani for advice and discussion, and P.Fenaux for sharing unpublished data. A.J.W. was supported by an MRC Clinician Scientist Fellowship.
References
- Akamatsu Y., Ohno,T., Hirota,K., Kagoshima,H., Yodoi,J. and Shigesada,K. (1997a) Redox regulation of the DNA binding activity in transcription factor PEBP2. The roles of two conserved cysteine residues. J. Biol. Chem., 272, 14497–14500. [DOI] [PubMed] [Google Scholar]
- Akamatsu Y., Tsukumo,S., Kagoshima,H., Tsurushita,N. and Shigesada,K. (1997b) A simple screening for mutant DNA binding proteins: application to murine transcription factor PEBP2α subunit, a founding member of the Runt domain protein family. Gene, 185, 111–117. [DOI] [PubMed] [Google Scholar]
- Becker S., Groner,B. and Müller,C.W. (1998) Three-dimensional structure of the Stat3β homodimer bound to DNA. Nature, 394, 145–151. [DOI] [PubMed] [Google Scholar]
- Berardi M.J., Sun,C., Zehr,M., Abildgaard,F., Peng,J., Speck,N.A. and Bushweller,J.H. (1999) The Ig fold of the core binding factor α Runt domain is a member of a family of structurally and functionally related Ig-fold DNA-binding domains. Struct. Fold Des., 7, 1247–1256. [DOI] [PubMed] [Google Scholar]
- Bork P., Holm,L. and Sander,C. (1994) The immunoglobulin fold. Structural classification, sequence patterns and common core. J. Mol. Biol., 242, 309–320. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) Collaborative Computational Project Number 4: A suite of programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cleary M.L. (1991) Oncogenic conversion of transcription factors by chromosomal translocations. Cell, 66, 619–622. [DOI] [PubMed] [Google Scholar]
- Crute B.E., Lewis,A.F., Wu,Z., Bushweller,J.H. and Speck,N.A. (1996) Biochemical and biophysical properties of the core-binding factor α2 (AML1) DNA-binding domain. J. Biol. Chem., 271, 26251–26260. [DOI] [PubMed] [Google Scholar]
- Ghosh G., van Duyne,G., Ghosh,S. and Sigler,P.B. (1995) Structure of NF-κB p50 homodimer bound to a κB site. Nature, 373, 303–310. [DOI] [PubMed] [Google Scholar]
- Goger M., Gupta,V., Kim,W.Y., Shigesada,K., Ito,Y. and Werner,M.H. (1999) Molecular insights into PEBP2/CBFβ-SMMHC associated acute leukemia revealed from the structure of PEBP2/CBFβ. Nature Struct. Biol., 6, 620–623. [DOI] [PubMed] [Google Scholar]
- Golling G., Li,L., Pepling,M., Stebbins,M. and Gergen,J.P. (1996) Drosophila homologs of the proto-oncogene product PEBP2/CBFβ regulate the DNA-binding properties of Runt. Mol. Cell. Biol., 16, 932–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson W.A., Horton,J.R. and LeMaster,D.M. (1990) Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J., 9, 1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Peng,J.W., Speck,N.A. and Bushweller,J.H. (1999) Solution structure of core binding factor β and map of the CBFα binding site. Nature Struct. Biol., 6, 624–627. [DOI] [PubMed] [Google Scholar]
- Ito Y. (1999) Molecular basis of tissue-specific gene expression mediated by the Runt domain transcription factor PEBP2/CBF. Genes Cells, 4, 685–696. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kagoshima H., Akamatsu,Y., Ito,Y. and Shigesada,K. (1996) Functional dissection of the α and β subunits of transcription factor PEBP2 and the redox susceptibility of its DNA binding activity. J. Biol. Chem., 271, 33074–33082. [DOI] [PubMed] [Google Scholar]
- Kamachi Y., Ogawa,E., Asano,M., Ishida,S., Murakami,Y., Satake,M., Ito,Y. and Shigesada,K. (1990) Purification of a mouse nuclear factor that binds to both the A and B cores of the polyomavirus enhancer. J. Virol., 64, 4808–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T. et al. (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell, 89, 755–764. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Lee B., Thirunavukkarasu,K., Zhou,L., Pastore,L., Baldini,A., Hecht,J., Geoffroy,V., Ducy,P. and Karsenty,G. (1997) Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nature Genet., 16, 307–310. [DOI] [PubMed] [Google Scholar]
- Lenny N., Meyers,S. and Hiebert,S.W. (1995) Functional domains of the t(8;21) fusion protein, AML-1/ETO. Oncogene, 11, 1761–1769. [PubMed] [Google Scholar]
- Leslie A.G.W. (1992) Recent changes to the MOSFLM package for film and image plate data. Joint CCP4 and ESF-EACMB Newsletter on Protein Crystallography, Daresbury Laboratory, Warrington, UK. [Google Scholar]
- Li L.H. and Gergen,J.P. (1999) Differential interactions between Brother proteins and Runt domain proteins in the Drosophila embryo and eye. Development, 126, 3313–3322. [DOI] [PubMed] [Google Scholar]
- Liu P., Tarle,S.A., Claxton,D.F., Marlton,P., Freedman,M., Siciliano,M.J. and Collins,F.S. (1993) Fusion between transcription factor CBFβ/PEBP2β and a myosin heavy chain in acute myeloid leukemia. Science, 261, 1041–1044. [DOI] [PubMed] [Google Scholar]
- Look A.T. (1997) Oncogenic transcription factors in the human acute leukemias. Science, 278, 1059–1065. [DOI] [PubMed] [Google Scholar]
- Melnikova I.N., Crute,B.E., Wang,S. and Speck,N.A. (1993) Sequence specificity of the core-binding factor. J. Virol., 67, 2408–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroux B. and Walker,J.E. (1996) Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol., 260, 289–298. [DOI] [PubMed] [Google Scholar]
- Miyoshi H., Shimizu,K., Kozu,T., Maseki,N., Kaneko,Y. and Ohki,M. (1991) t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc. Natl Acad. Sci. USA, 88, 10431–10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundlos S. et al. (1997) Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell, 89, 773–779. [DOI] [PubMed] [Google Scholar]
- Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- Nagata T., Gupta,V., Sorce,D., Kim,W.Y., Sali,A., Chait,B.T., Shigesada,K., Ito,Y. and Werner,M.H. (1999) Immunoglobulin motif DNA recognition and heterodimerization of the PEBP2/CBF Runt domain. Nature Struct. Biol., 6, 615–619. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Ogawa E., Inuzuka,M., Maruyama,M., Satake,M., Naito-Fujimoto,M., Ito,Y. and Shigesada,K. (1993a) Molecular cloning and characterization of PEBP2β, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2α. Virology, 194, 314–331. [DOI] [PubMed] [Google Scholar]
- Ogawa E., Maruyama,M., Kagoshima,H., Inuzuka,M., Lu,J., Satake,M., Shigesada,K. and Ito,Y. (1993b) PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc. Natl Acad. Sci. USA, 90, 6859–6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osato M. et al. (1999) Biallelic and heterozygous point mutations in the Runt domain of the AML1/PEBP2αB gene associated with myeloblastic leukemias. Blood, 93, 1817–1824. [PubMed] [Google Scholar]
- Otto F. et al. (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell, 89, 765–771. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Petrovick M.S., Hiebert,S.W., Friedman,A.D., Hetherington,C.J., Tenen,D.G. and Zhang,D.E. (1998) Multiple functional domains of AML1: PU.1 and C/EBPα synergize with different regions of AML1. Mol. Cell. Biol., 18, 3915–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quack I. et al. (1999) Mutation analysis of core binding factor A1 in patients with cleidocranial dysplasia. Am. J. Hum. Genet., 65, 1268–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbitts T.H. (1991) Translocations, master genes and differences between the origins of acute and chronic leukemias. Cell, 67, 641–644. [DOI] [PubMed] [Google Scholar]
- Rabbitts T.H. (1994) Chromosomal translocations in human cancer. Nature, 372, 143–149. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V. and Biou,V. (1997) Treatment of multiwavelength anomalous diffraction data as a special case of multiple isomorphous replacement. Methods Enzymol., 276, 538–557. [PubMed] [Google Scholar]
- Rodan G.A. and Harada,S. (1997) The missing bone. Cell, 89, 677–680. [DOI] [PubMed] [Google Scholar]
- Song W.J. et al. (1999) Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nature Genet., 23, 166–175. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL_W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornell A., Hallberg,B. and Grundström,T. (1988) Differential protein binding in lymphocytes to a sequence in the enhancer of the mouse retrovirus SL3-3. Mol. Cell. Biol., 8, 1625–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. et al. (1996) The CBFβ subunit is essential for CBFα2 (AML1) function in vivo. Cell, 87, 697–708. [DOI] [PubMed] [Google Scholar]
- Zhou G. et al. (1999) CBFA1 mutation analysis and functional correlation with phenotypic variability in cleidocranial dysplasia. Hum. Mol. Genet., 8, 2311–2316. [DOI] [PubMed] [Google Scholar]