Abstract
Activation of eukaryotic class II gene expression involves the formation of a transcription initiation complex that includes RNA polymerase II, general transcription factors, and SRB components of the holoenzyme. Negative regulators of transcription have been described, but it is not clear whether any are general repressors of class II genes in vivo. We reasoned that defects in truly global negative regulators should compensate for deficiencies in SRB4 because SRB4 plays a positive role in holoenzyme function. Genetic experiments reveal that this is indeed the case: a defect in the yeast homologue of the human negative regulator NC2 (Dr1·DRAP1) suppresses a mutation in SRB4. Global defects in mRNA synthesis caused by the defective yeast holoenzyme are alleviated by the NC2 suppressing mutation in vivo, indicating that yeast NC2 is a global negative regulator of class II transcription. These results imply that relief from repression at class II promoters is a general feature of gene activation in vivo.
Activation of class II gene transcription in eukaryotes involves the recruitment of a transcription initiation complex that includes the RNA polymerase II holoenzyme (1–6). The yeast RNA polymerase II holoenzyme is a large multisubunit complex containing RNA polymerase II, a subset of the general transcription factors, and SRB regulatory proteins (7–11). Mammalian RNA polymerase II holoenzymes have also been purified, and an SRB7 homologue has been identified as a component of those complexes (12–14).
For some class II genes, regulation appears to involve both positive and negative transcriptional regulators. The negative regulators that have been described include proteins purified for their ability to inhibit transcription in vitro (15–21) and genes identified because their products repress transcription from a subset of class II genes in vivo (21–29). For example, the human proteins NC1 (15, 16), NC2 or Dr1·DRAP1 (16, 17, 20), and DNA topoisomerase I (18, 19) repress basal transcription in vitro. The products of the yeast genes MOT1 (21–24), NOT1-4 (25–27), and SIN4 (28–29) negatively regulate at least a subset of yeast genes in vivo. Whether any of these negative regulators are generally employed for class II gene regulation in vivo is not yet clear.
The RNA polymerase II C-terminal domain and the associated SRB complex have been implicated in the response to transcriptional activators (7–9, 30, 31). Two holoenzyme components, SRB4 and SRB6, have been shown to play essential and positive roles in transcription at the majority of class II genes in Saccharomyces cerevisiae (32). We reasoned that a defect in SRB4 might be alleviated by defects in general negative regulators and that knowledge of such regulators could contribute to our understanding of the mechanisms involved in gene regulation in vivo. Here we show that a deficiency in yeast NC2 can compensate for the global transcriptional defects caused by mutations in the SRB4 and SRB6 subunits of the RNA polymerase II holoenzyme and that NC2 is a global negative regulator of class II transcription in vivo.
MATERIALS AND METHODS
Genetic Manipulations.
Yeast strains and plasmids are listed in Tables 1 and 2, respectively. Details of strain and plasmid constructions are available upon request. Yeast media and manipulation were as described (9). Extragenic suppressors of the temperature-sensitive phenotype of Z628 capable of growth at the restrictive temperature of 36°C were isolated. Dominant and recessive suppressors were identified by mating to Z811 and assaying growth at 36°C on yeast extract/peptone/dextrose (YPD). Complementation groups were established as described (9).
Table 1.
Yeast strains
| Strain | Genotype |
|---|---|
| Z579 | MATa ura3-52 his3Δ200 leu2-3,112 srb4Δ2::HIS3 [pCT127 (SRB4 LEU2 CEN)] |
| Z628 | MATa ura3-52 his3Δ200 leu2-3,112 srb4Δ2::HIS3 [pCT181 (srb4-138 LEU2 CEN)] |
| Z804 | MATa ura3-52 his3Δ200 leu2-3,112 srb4Δ2::HIS3 ncb1-1 [pCT181 (srb4-138 LEU2 CEN)] |
| Z805 | MATa ura3-52 his3Δ200 leu2-3,112 srb4Δ2::HIS3 ncb1-1 [pCT15 (SRB4 URA3 CEN)] |
| Z806 | MATα ura3-52 his3Δ200 leu2-3,112 ncb1Δ1::HIS3 [RY7135 (NCB1 URA3 CEN)] |
| Z807 | MATα ura3-52 his3Δ200 leu2-3,112 ncb1Δ1::HIS3 [RY7137 (NCB1 5′ FLAG tag LEU2 CEN)] |
| Z811 | MATα ura3-52 his3Δ200 leu2-3,112 srb4Δ2::HIS3 [pCT181 (srb4-138 URA3 CEN)] |
Table 2.
Plasmids
| Plasmid | Description |
|---|---|
| RY7133 | NCB1 in pGEX-4T-3 (Pharmacia) |
| RY7134 | NCB2 (amino acids 13–146) in pGEX-4T-3 |
| RY7135 | NCB1 (1.3 kb) URA3 CEN |
| RY7136 | ncb1Δ1::HIS3 in pBluescript II SK(+) |
| RY7137 | NCB1 5′ FLAG tag (IBI) in pUN105 |
| RY7138 | ncb1-1 (1.3 kb) URA3 CEN |
To determine whether the NCB1 gene is essential for cell viability, the entire coding region was deleted on one of the two chromosomes of a diploid cell, using a single step disruption method (33) and the plasmid RY7136, which carries the deletion allele ncb1Δ1. Southern analysis was used to confirm that a single copy of the NCB1 gene had been deleted. These heterozygous diploid cells were sporulated, and tetrad analysis was performed on YPD plates and scored for growth at a variety of temperatures. Spores with the ncb1Δ1 allele did not produce colonies, indicating that NCB1 is essential for cell viability.
DNA Methods.
DNA manipulations were performed as described (34). PCR amplifications to produce RY7133, RY7134, RY7136, RY7137, and RY7138 were performed with Vent DNA polymerase (New England Biolabs) as described by the manufacturer. The glutathione S-transferase (GST) fusions were constructed as described (13), and the ncb1Δ1 allele was constructed as described (35).
Cloning and Sequence Analysis.
The genomic clone of NCB1 was isolated by complementation of Z804 with a wild-type genomic library (35). The wild-type gene was further localized by subcloning fragments of the genomic insert and repeating the screen. The clone with the smallest insert, RY7135, was sequenced. The genomic clone of NCB1 was used to confirm the identity of each member of the complementation group and to identify additional members. RY7138 was created from RY7135 in vivo by transforming Z804 with linearized RY7135 lacking NCB1 coding DNA and then isolating the plasmid from a transformant that had repaired the plasmid with the mutant ncb1-1 sequence from the chromosome (33). NCB1 and ncb1-1 were completely sequenced on each strand using DNA from RY7135 and RY7138, respectively. Double-stranded sequencing with dideoxynucleotides and Sequenase (United States Biochemical) was carried out as described by the manufacturer using T3 and T7 promoter primers and internal oligonucleotide primers. Sequence comparison analysis was performed at the National Center for Biotechnology Information using the blast network service (36). The ncb1-1 mutant allele contained a single base pair deletion at nucleotide 340, causing a frameshift and a translational stop at nucleotides 347–349 (see Fig. 1b). Unlike the RY7135 plasmid, RY7138 did not prevent growth at 36°C when transformed into Z804, indicating that the correct gene was cloned.
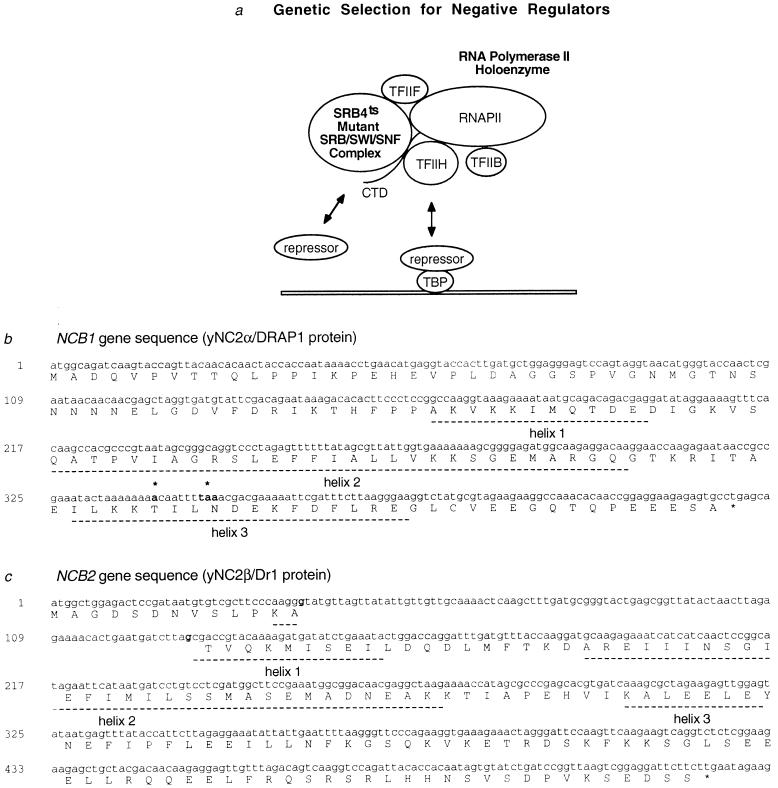
Figure 1.
Isolation of putative global negative regulators. (a) Genetic selection for suppressors of the temperature-sensitive (ts) SRB4 mutant RNA polymerase II holoenzyme. (b) Sequence of NCB1 (ORF YER159c on chromosome V, GenBank accession no. U18917U18917). The suppressing allele, ncb1-1, was isolated by gap-repair techniques and sequenced. The suppressing mutation, a single base pair deletion at nucleotide 340, is noted in boldface type. The deletion results in a frameshift causing a translational stop at nucleotides 347–349, also noted in boldface type. Underlined regions indicate homology to α-helices in the histone H2A histone fold (42–44). (c) Sequence of NCB2 (ORF D9509.16 on chromosome IV, GenBank accession no. U32274U32274) with the first and last nucleotide of the intron sequence noted in boldface type. Underlined regions indicate homology to α-helices in the histone H2B histone fold (42–46).
Antibodies.
Recombinant yNC2α and yNC2β proteins were purified for generating polyclonal antibodies in rabbits. Recombinant proteins were derived from Escherichia coli containing pGEX-4T-3 (Pharmacia) constructs RY7133 and RY7134 as described (37). The antibodies were used to detect yNC2α and yNC2β in Western blots at a dilution of 1:250 or 1:500.
Purification of Yeast NC2.
TATA box-binding protein (TBP) affinity chromatography was performed as described (38) starting with 1.2 kg of cell pellet. Approximately 60% of the total cellular amount of each NC2 subunit was eluted in 1 M KOAc. One-half of the 1 M KOAc eluate (80 ml; 3.3 mg) was dialyzed against buffer T plus 0.003% Nonidet P-40. The dialyzed sample was applied at 1 ml/min to a 1-ml HiTrap SP cartridge (Pharmacia), which was washed with 10 ml of buffer A (20 mM K-Hepes, pH 7.6/1 mM EDTA/10% glycerol and protease inhibitors) plus 100 mM KOAc. Bound proteins were eluted with a 10-ml gradient of buffer A from 100 to 1,000 mM KOAc at 0.25 ml/min. Peak NC2 fractions were pooled, frozen in liquid nitrogen, and stored at −70°C until use. One-half of the peak NC2 fractions (1 ml, 80 μg) was diluted with 2.7 ml of buffer B (20 mM Tris·OAc, pH 7.8/1 mM EDTA/10% glycerol) and applied to a DEAE 5PW 5/5 column (TosoHaas, Montgomeryville, PA) at 0.5 ml/min. The column was washed with 5 ml of buffer B plus 100 mM KOAc, and bound proteins were eluted with a 12-ml gradient of buffer B from 100 to 1,000 mM KOAc. The peak of NC2 contained 50 μg of total protein. SDS/PAGE and silver staining were performed as described (8).
Construction of FLAG-Tagged NC2α Yeast Strain.
Plasmid RY7137 was constructed by amplifying the NCB1 gene (including regulatory sequences) with two sets of overlapping primers to add a FLAG epitope (IBI) to the N terminus of yNC2α. The two PCR products were gel-purified and combined, and the entire FLAG-tagged NCB1 gene was amplified with primers adding 5′ HindIII and 3′ BamHI cloning sites. The final PCR product was cloned into plasmid pUN105 (39). RY7137 was transformed into a Z806, a yeast strain containing the ncb1Δ1 deletion, by plasmid-shuffle techniques (40) to produce Z807. The FLAG-tagged NCB1 was fully functional and able to complement the ncb1Δ1 deletion.
Purification of FLAG-Tagged Yeast NC2 and in Vitro Transcription Assays.
Yeast strain Z807 was grown in YPD to late log phase and harvested by centrifugation. The cell pellet (500 g) was resuspended in 500 ml of 150 mM KOAc/60 mM K-Hepes, pH 7.6/3 mM EDTA and protease inhibitors. The mixture was poured slowly into a bath of liquid nitrogen, the excess liquid nitrogen was decanted, and the frozen cells were blended for 4 min in a Waring blender. The blended cells were stored at −70°C until use. The frozen mixture was thawed at 55°C and centrifuged at 12,000 rpm for 30 min in a GSA (Sorvall) rotor. One volume (600 ml) of buffer A plus 100 mM KOAc and 300 g of damp-dry BioRex 70 (Bio-Rad) resin were added to the supernatant. After stirring for 2 h, the BioRex 70 resin was washed with 1 liter of buffer A plus 0.1 M KOAc on a Buchner funnel. The washed resin was packed into a 5 cm i.d. column and washed with 0.5 liter of buffer A plus 0.1 M KOAc at a flow rate of 10 ml/min. Bound proteins were eluted with buffer A plus 1 M KOAc. Fractions containing protein (115 ml at 4.1 mg/ml) were pooled, frozen in liquid nitrogen, and stored at −70°C until use. BioRex 70 (32 ml) eluate was thawed and mixed with 160 ml of buffer B plus protease inhibitors. The diluted eluate was centrifuged at 12,000 rpm for 30 min in a GSA rotor. The supernatant was applied to a 2-ml FLAG antibody M2 affinity column (IBI), the column was washed with 100 ml of buffer B plus 150 mM KOAc and 10 ml of buffer B plus 50 mM KOAc, and bound proteins were eluted with buffer B plus 50 mM KOAc plus 50 μM FLAG peptide. The eluate (8 ml) was filtered through a 0.2-μm filter and applied to a Mono Q PC 1.6/5 column (Pharmacia) at a flow rate of 0.1 ml/min, the column was washed with 1 ml buffer B plus 50 mM KOAc plus 1 mM DTT, and bound proteins were eluted with a 2-ml gradient of buffer B plus 1 mM DTT from 50 to 2,000 mM KOAc. SDS/PAGE, silver staining, and Western blot analysis were as described in the Fig. 2 legend. In vitro transcription reactions were performed with a yeast CYC1 promoter template as described (41) except that 3′ O-MeGTP was added to 40 μM, T1 RNase was omitted, and ethanol precipitations were performed with 400 instead of 600 μl.
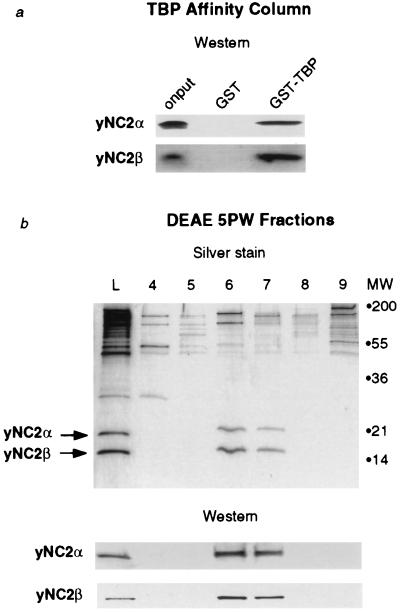
Figure 2.
Yeast NC2 binds to TBP and can be purified as a two-subunit complex. (a) Western blot analyses of TBP column onput and eluates with antibodies against yNC2α and yNC2β. Bound proteins were eluted with 2 M KCl. (b) Silver-stained SDS/polyacrylamide gel and Western blot analyses of fractions from the final step of the purification (DEAE 5PW).
Poly(A)+ Blots and S1 Analyses.
Aliquots of cells were removed from culture at the times indicated, total RNA was prepared, and poly(A)+ blots, quantitation, and S1 protection analysis were carried out as described (32).
RESULTS
Yeast ncb1-1 Is an Extragenic Suppressor of the srb4-138 Mutation.
Since SRB4 plays an essential and positive role in class II transcription, we reasoned that a defect in SRB4 might be alleviated by defects in general negative regulators (Fig. 1a). To identify mutations that compensate for a defect in SRB4, 78 spontaneous extragenic suppressors of the temperature-sensitive phenotype of the srb4-138 allele were isolated. Sixteen of the suppressors were dominant, and 62 were recessive. Five complementation groups were established among the recessive suppressors. One of the recessive suppressing genes was cloned by complementation using a wild-type genomic DNA library and was sequenced. The sequence is identical to the ORF YER159c, which predicts a 142-aa protein with a molecular weight of 15,500 (Fig. 1b). A search of the GenBank database (June 7, 1996) revealed that the predicted protein has 39% identity over 99 aa to the NC2α (DRAP1) subunit of human NC2 (Dr1·DRAP1), which binds to TBP and represses transcription in vitro (16, 17, 20, 42–44, 47–50). The gene encoding the putative yeast NC2α protein was named NCB1. Deletion analysis revealed that NCB1 is essential for cell viability (data not shown). The mutation present in the suppressing allele, ncb1-1, produces a 27-residue C-terminal truncation in the yeast NC2α protein (Fig. 1b). Since NCB1 is an essential gene, the truncation mutation must cause a partial functional defect in the NC2α protein.
Human NC2 consists of two subunits, NC2α and NC2β, both of which are necessary for maximal TBP binding and repression of transcription in vitro (42, 43). To determine if there is a yeast homologue of the NC2β subunit, the GenBank database was searched on June 7, 1996 with the human NC2β amino acid sequence. An ORF, D9509.16, that predicts a 146-aa protein with a molecular weight of 16,700 (Fig. 1c) was identified. The predicted protein has 37% identity to human NC2β. The gene, named NCB2, contains consensus sequence predicting an intron. Both DNA and cDNA clones containing the coding sequence for NCB2 were isolated and sequenced, and the intron structure produces a somewhat different amino acid sequence than that predicted by GenBank (Fig. 1c). The human NC2 subunits each contain sequences predicting a histone fold structure (42, 43, 51, 52); the yeast NC2 subunits also exhibit this sequence relationship (Fig. 1 b and c; ref. 44). Interestingly, the C-terminal truncation in the ncb1-1 suppressing allele removes part of the histone fold in the yeast NC2α subunit (Fig. 1b). Deletions in the human NC2 histone folds have been shown to decrease subunit association, TBP binding, and transcriptional repression (42, 43).
Yeast NC2 Binds to TBP and Can Be Purified as a Two-Subunit Complex.
If the two yeast gene products are genuine homologues of human NC2, they would be expected to copurify as a complex and to bind to TBP. To determine whether this is the case, a yeast whole-cell extract was subjected to GST and GST–TBP affinity chromatography (Fig. 2a). Western blot analyses of the column eluates confirmed that both yeast NC2α and NC2β proteins were specifically retained on the GST–TBP column. The eluate from the GST–TBP column was further purified over two ion-exchange columns. Silver staining and Western blot analyses showed that yeast NC2α and NC2β coeluted over both columns and that the proteins appear to be in equal stoichiometry (Fig. 2b and data not shown). Yeast NC2α is not present in a purified RNA polymerase II holoenzyme preparation, so NC2 is unlikely to be a component of the holoenzyme (data not shown). These data confirm that the yeast NC2α and NC2β proteins are stoichiometric subunits of a complex that can bind specifically to TBP.
Highly Purified Yeast NC2 Inhibits Transcription by RNA Polymerase II Holoenzyme in Vitro.
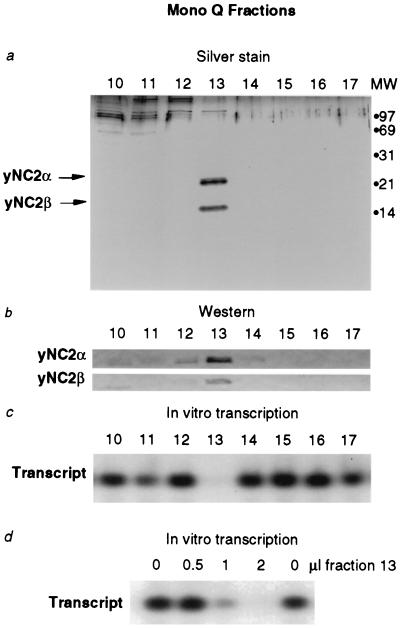
The observation that a defective form of yeast NC2 can compensate for a weakened RNA polymerase II holoenzyme suggests that yeast NC2 normally functions to repress holoenzyme activity. We tested the ability of purified yeast NC2 to repress transcription by yeast RNA polymerase II holoenzyme in vitro. A preparation of yeast NC2 from a strain containing an epitope-tagged NC2α subunit (Fig. 3 a and b) gave us material of higher yield and purity than from the TBP affinity column. In vitro transcription reactions were performed with a yeast CYC1 promoter template, holoenzyme, and fractions from the final column of this yeast NC2 purification (Fig. 3 c and d). Repression of transcription correlated with the peak of yeast NC2 protein. When an equimolar amount of NC2 was added to RNA polymerase II holoenzyme and TBP, 50% of the maximal inhibition was observed (Fig. 3d). The repression of RNA polymerase II holoenzyme transcription by yeast NC2 is consistent with the ability of a partial loss-of-function NC2 mutation to suppress a holoenzyme mutation.
Figure 3.
Highly purified yeast NC2 inhibits transcription by RNA polymerase II holoenzyme in vitro. (a) Analysis of NC2 Mono Q fractions by SDS/PAGE and silver staining. (b) Western blot analyses of Mono Q fractions. (c) Influence of Mono Q fractions on in vitro transcription by RNA polymerase II holoenzyme. (d) Inhibition of in vitro transcription by RNA polymerase II holoenzyme with increasing amounts of purified yeast NC2. Assuming a molecular weight of 64,000 for NC2, 0.5 pmol was required for 50% inhibition of an equimolar amount of RNA polymerase II holoenzyme (estimated molecular weight of 2 million) and TBP.
NC2 Functions at the Majority of Class II Promoters in Vivo.
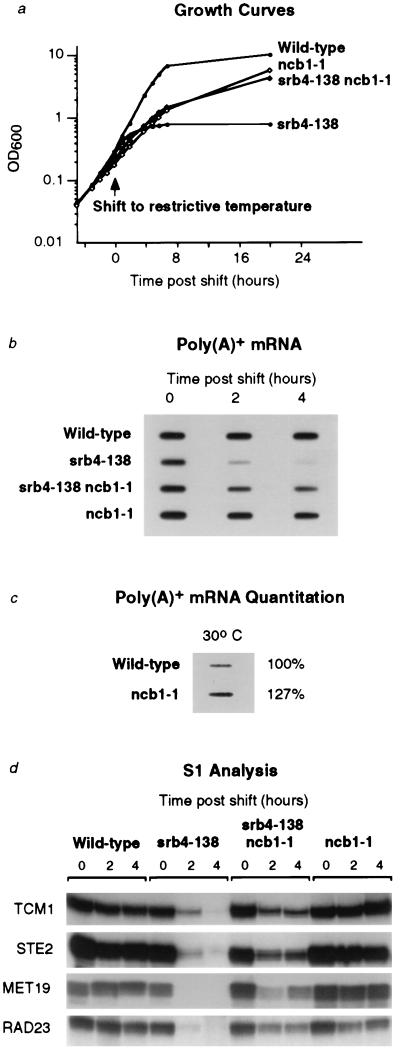
The observation that loss of NC2 function in yeast cells can compensate for a defect in the SRB4 component of the holoenzyme, together with previous evidence that SRB4 functions globally at class II promoters (32), suggests that NC2 may repress transcription at class II promoters in general. To determine whether yeast NC2 functions at the majority of class II promoters in vivo, we investigated whether the shutdown of mRNA synthesis observed in cells with the SRB4 temperature-sensitive mutant allele srb4-138 is reversed by the loss of NC2 function (Fig. 4). Upon shifting cells to the restrictive temperature, the growth rate of the srb4-138 strain was severely reduced, whereas the srb4-138 ncb1-1 suppressor strain was only modestly affected (Fig. 4a). The levels of poly(A)+ mRNA in these cells were measured immediately before and at several times after the shift to the restrictive temperature (Fig. 4b). There was a significant decrease in the mRNA population in the srb4-138 strain, as observed previously (32). In contrast, there was only a modest decrease in mRNA levels in the srb4-138 ncb1-1 strain after the temperature shift. Thus, the ncb1-1 mutation suppresses the general defect in transcription of class II messages caused by the srb4-138 mutation. Furthermore, the ncb1-1 mutant in an otherwise wild-type background showed 27% higher levels of poly(A)+ mRNA compared with the wild-type strain under normal conditions (Fig. 4c). This result is consistent with the partial loss-of-function of a class II global negative regulator. S1 analysis of individual class II transcripts confirmed that the decline in specific mRNAs in the srb4-138 strain is reversed in the srb4-138 ncb1-1 strain (Fig. 4d). These results, together with previous evidence that NC2 functions as a repressor of multiple promoters tested in vitro, argue that NC2 is a general negative regulator of class II gene transcription.
Figure 4.
Loss of yeast NC2 function compensates for the global defect in class II gene expression caused by the SRB4 mutant holoenzyme. (a) ncb1-1 mutation suppresses the growth defect of the srb4-138 mutant strain at the restrictive temperature. Growth of wild-type (Z579), srb4-138 (Z628), srb4-138 ncb1-1 (Z804), and ncb1-1 (Z805) strains in YPD medium at 30°C and after shifting to the restrictive temperature of 35.5°C. (b) The global decline in mRNA levels at the restrictive temperature in srb4-138 mutant strain is alleviated by the ncb1-1 mutation. (c) Global levels of mRNA are increased in the ncb1-1 strain relative to the wild-type strain. (d) The decrease in synthesis of individual class II messages at the restrictive temperature in the srb4-138 mutant strain is reversed by the ncb1-1 mutation.
DISCUSSION
Our results indicate that NC2 is an essential and conserved negative regulator of class II gene transcription. These results extend the known negative regulatory effects of NC2 on core RNA polymerase II in vitro (16, 17, 20, 42–44, 47–50) and confirm that this regulator can inhibit transcription by RNA polymerase II in vivo.
Since SRB4 has an essential and positive role in transcription at the majority of class II genes in yeast (32), we reasoned that suppressors of a temperature-sensitive SRB4 mutant should include negative regulators. In principle, such regulators could repress most class II genes or they could repress SRB4 specifically. Several lines of evidence argue that NC2 acts globally as a repressor of most, if not all, class II genes. NC2 can repress transcription in vitro from a wide variety of mammalian, viral, and yeast promoters (16, 20, 42–44, 50). The NC2 suppressor mutation that compensates for reduced class II gene transcription in vivo due to loss of SRB4 also compensates for the global defect due to loss of SRB6 (data not shown). Loss of function of NC2 in otherwise wild-type cells results in increased levels of poly(A)+ mRNA. These data do not prove that NC2 regulates all class II genes, but they are most consistent with a global role for this repressor.
Mechanism of Yeast NC2 Repression.
Much is already known about the biochemistry of NC2 repression; NC2 binds to TBP on promoter DNA and subsequently inhibits the binding of TFIIA and TFIIB in vitro (16, 17, 42, 43, 50). NC2 binds to the same basic region of TBP as TFIIA (50), suggesting that NC2 physically blocks TFIIA from binding to TBP.
The other proteins known to have global negative regulatory properties are the histones (53, 54), which share notable structural features with NC2/Dr1·DRAP1. The presence of the histone fold motif in NC2 raises the intriguing possibility of interactions with other histone fold-containing proteins. Histones H2A, H2B, H3, and H4 all contain histone folds (51), as do the yeast HAP3 and HAP5 activator proteins (CBF proteins in mammals; ref. 52) and several TAFIIs (45, 46). These TAFIIs and histones are able to interact with each other in vitro through their histone fold regions (45). Thus, NC2 might introduce a nucleosome-like structure at the promoter, either by itself or with other histone fold-containing proteins.
Transcription Activation and Relief from Repression.
The SRB components of the RNA polymerase II holoenzyme contribute to the response to transcriptional activators (7–9). We have shown that a partial loss in NC2 function compensates for deficiencies in SRB4 and SRB6 functions. These results indicate that relief from NC2 inhibition is a required step during transcription initiation at most class II promoters in vivo. Evidence consistent with this view has recently emerged from a study of SUC2 gene regulation. Prelich and Winston (55) isolated yeast mutations that suppress a deletion of the upstream activating sequence in the SUC2 promoter. The mutant genes that compensated for the absence of SUC2 activator function included several histones, certain SPT genes, and other unidentified genes called BUR genes (Bypass UAS Requirement). The bur6 mutant allele was recently found to be a partial loss-of-function mutation in NCB1 (56). The observation that BUR6 is identical to NCB1 indicates that a loss in NC2 function can compensate for the loss of an activator. These data support the model that activators function to recruit the transcription apparatus, which must overcome negative regulation by NC2 to initiate transcription.
Acknowledgments
We thank R. Roeder, P. Sharp, C. Thompson, A. Hoffmann, T. Lee, H. Madhani, and S. Liao for advice and discussions; J. Madison and E. Shuster for strains and plasmids; L. Ziaugra, E. Jennings, and V. Tung for technical assistance; F. Lewitter for assistance with database searches; and G. Prelich, A. Goppelt, M. Meisterernst, J. Kim, and D. Reinberg for sharing data prior to publication. E.L.G. and D.M.C. are predoctoral fellows of the Howard Hughes Medical Institute. J.C.R. is a postdoctoral fellow of the Damon Runyon-Walter Winchell Cancer Research Foundation. This work was supported by National Institutes of Health grants to R.A.Y.
ABBREVIATIONS
- TBP
TATA box-binding protein
- GST
glutathione S-transferase
Footnotes
References
- 1.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 2.Koleske A J, Young R A. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- 3.Halle J P, Meisterernst M. Trends Genet. 1996;12:161–163. doi: 10.1016/0168-9525(96)30035-8. [DOI] [PubMed] [Google Scholar]
- 4.Orphanides G, Lagrange T, Reinberg D. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 5.Roeder R G. Trends Biochem Sci. 1996;21:327–334. [PubMed] [Google Scholar]
- 6.Struhl K. Cell. 1996;84:179–182. doi: 10.1016/s0092-8674(00)80970-8. [DOI] [PubMed] [Google Scholar]
- 7.Kim Y J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 8.Koleske A J, Young R A. Nature (London) 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 9.Hengartner C J, Thompson C M, Zhang J, Chao D M, Liao S M, Koleske A M, Okamura S, Young R A. Genes Dev. 1995;9:897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- 10.Liao S M, Zhang J, Jeffery D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, van Vuuren H J, Young R A. Nature (London) 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 11.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 12.Ossipow V, Tassan J P, Nigg E A, Schibler U. Cell. 1995;83:137–146. doi: 10.1016/0092-8674(95)90242-2. [DOI] [PubMed] [Google Scholar]
- 13.Chao D M, Gadbois E G, Murray P J, Anderson S F, Sonu M S, Parvin J D, Young R A. Nature (London) 1996;380:82–85. doi: 10.1038/380082a0. [DOI] [PubMed] [Google Scholar]
- 14.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. Nature (London) 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 15.Meisterernst M, Roy A L, Lieu H M, Roeder R G. Cell. 1991;66:981–993. doi: 10.1016/0092-8674(91)90443-3. [DOI] [PubMed] [Google Scholar]
- 16.Meisterernst M, Roeder R G. Cell. 1991;67:557–567. doi: 10.1016/0092-8674(91)90530-c. [DOI] [PubMed] [Google Scholar]
- 17.Inostroza J A, Mermelstein F H, Ha I, Lane W S, Reinberg D. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 18.Kretzschmar M, Meisterernst M, Roeder R G. Proc Natl Acad Sci USA. 1993;90:11508–11512. doi: 10.1073/pnas.90.24.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merino A, Madden K, Lane W, Champoux J, Reinberg D. Nature (London) 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 20.Kim J, Parvin J D, Shykind B M, Sharp P A. J Biol Chem. 1996;271:18405–18412. doi: 10.1074/jbc.271.31.18405. [DOI] [PubMed] [Google Scholar]
- 21.Auble D T, Hahn S. Genes Dev. 1993;7:844–856. doi: 10.1101/gad.7.5.844. [DOI] [PubMed] [Google Scholar]
- 22.Davis J L, Kunisawa R, Thorner J. Mol Cell Biol. 1992;12:1879–1892. doi: 10.1128/mcb.12.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piatti S, Tazzi R, Pizzagalli A, Plevani P, Lucchini G. Chromosoma. 1992;102:S107–S113. doi: 10.1007/BF02451793. [DOI] [PubMed] [Google Scholar]
- 24.Auble D T, Hansen K E, Mueller C G, Lane W S, Thorner J, Hahn S. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 25.Collart M A, Struhl K. EMBO J. 1993;12:177–186. doi: 10.1002/j.1460-2075.1993.tb05643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collart M A, Struhl K. Genes Dev. 1994;8:525–537. doi: 10.1101/gad.8.5.525. [DOI] [PubMed] [Google Scholar]
- 27.Collart M A. Mol Cell Biol. 1996;16:6668–6676. doi: 10.1128/mcb.16.12.6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Y W, Dohrmann P R, Stillman D J. Genetics. 1995;140:47–54. doi: 10.1093/genetics/140.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song W, Treich I, Qian N, Kuchin S, Carlson M. Mol Cell Biol. 1996;16:115–120. doi: 10.1128/mcb.16.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scafe C, Chao D, Lopes J, Hirsch J P, Henry S, Young R A. Nature (London) 1990;347:491–494. doi: 10.1038/347491a0. [DOI] [PubMed] [Google Scholar]
- 31.Gerber H P, Hagmann M, Seipel K, Georgiev O, West M A, Litingtung Y, Schaffner W, Corden J L. Nature (London) 1995;374:660–662. doi: 10.1038/374660a0. [DOI] [PubMed] [Google Scholar]
- 32.Thompson C M, Young R A. Proc Natl Acad Sci USA. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothstein R. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 35.Thompson C M, Koleske A J, Chao D M, Young R A. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 36.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 37.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 38.Reese J C, Apone L, Walker S S, Griffin L A, Green M R. Nature (London) 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- 39.Elledge S J, Davis R W. Gene. 1988;70:303–312. doi: 10.1016/0378-1119(88)90202-8. [DOI] [PubMed] [Google Scholar]
- 40.Boeke J D, Trueheart J, Natsoulis G, Fink G R. Methods Enzymol. 1987;154:164–75. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 41.Koleske A J, Chao D M, Young R A. Methods Enzymol. 1996;273:176–184. doi: 10.1016/s0076-6879(96)73018-5. [DOI] [PubMed] [Google Scholar]
- 42.Goppelt A, Stelzer G, Lottspeich F, Meisterernst M. EMBO J. 1996;15:3105–3116. [PMC free article] [PubMed] [Google Scholar]
- 43.Mermelstein F, Yeung K, Cao J, Inostroza J A, Erdjument-Bromage H, Eagelson K, Landsman D, Levitt P, Tempst P, Reinberg D. Genes Dev. 1996;10:1033–1048. doi: 10.1101/gad.10.8.1033. [DOI] [PubMed] [Google Scholar]
- 44.Goppelt A, Meisterernst M. Nucleic Acids Res. 1996;24:4450–4456. doi: 10.1093/nar/24.22.4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoffmann A, Chiang C M, Oelgeschlager T, Xie X, Burley S K, Nakatani Y, Roeder R G. Nature (London) 1996;380:356–359. doi: 10.1038/380356a0. [DOI] [PubMed] [Google Scholar]
- 46.Xie X, Kokubo T, Cohen S L, Mirza U A, Hoffmann A, Chait B T, Roeder R G, Nakatani Y, Burley S K. Nature (London) 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- 47.Kraus V B, Inostroza J A, Yeung K, Reinberg D, Nevins J R. Proc Natl Acad Sci USA. 1994;91:6279–6282. doi: 10.1073/pnas.91.14.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White R J, Khoo B C, Inostroza J A, Reinberg D, Jackson S P. Science. 1994;266:448–450. doi: 10.1126/science.7939686. [DOI] [PubMed] [Google Scholar]
- 49.Yeung K C, Inostroza J A, Mermelstein F H, Kannabiran C, Reinberg D. Genes Dev. 1994;8:2097–2109. doi: 10.1101/gad.8.17.2097. [DOI] [PubMed] [Google Scholar]
- 50.Kim T K, Zhao Y, Ge H, Bernstein R, Roeder R G. J Biol Chem. 1995;270:10976–10981. doi: 10.1074/jbc.270.18.10976. [DOI] [PubMed] [Google Scholar]
- 51.Arents G, Burlingame R W, Wang B C, Love W E, Moudrianakis E N. Proc Natl Acad Sci USA. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baxevanis A D, Arents G, Moudrianakis E N, Landsman D. Nucleic Acids Res. 1995;23:2685–2691. doi: 10.1093/nar/23.14.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kingston R E, Bunker C A, Imbalzano A N. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 54.Wolffe A P. Cell. 1994;77:13–16. doi: 10.1016/0092-8674(94)90229-1. [DOI] [PubMed] [Google Scholar]
- 55.Prelich G, Winston F. Genetics. 1993;135:665–676. doi: 10.1093/genetics/135.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prelich, G. (1997) Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]