Abstract
The eukaryotic translation initiation factor 4E (eIF4E) plays an important role in the control of cell growth. eIF4E binds to the mRNA 5′ cap structure m7GpppN (where N is any nucleotide), and promotes ribosome binding to the mRNA in the cytoplasm. However, a fraction of eIF4E localizes to the nucleus. Here we describe the cloning and functional characterization of a new eIF4E-binding protein, referred to as 4E-T (eIF4E-Transporter). We demonstrate that 4E-T is a nucleocytoplasmic shuttling protein that contains an eIF4E-binding site, one bipartite nuclear localization signal and two leucine-rich nuclear export signals. eIF4E forms a complex with the importin αβ heterodimer only in the presence of 4E-T. Overexpression of wild-type 4E-T, but not of a mutant defective for eIF4E binding, causes the nuclear accumulation of HA-eIF4E in cells treated with leptomycin B. Taken together, these results demonstrate that the novel nucleocytoplasmic shuttling protein 4E-T mediates the nuclear import of eIF4E via the importin αβ pathway by a piggy-back mechanism.
Keywords: cap-binding protein/eIF4E localization/initiation factor/shuttling protein/transport
Introduction
Regulation of translation plays an important role in the control of gene expression. In eukaryotes, translation regulation occurs primarily at the initiation step, which is rate limiting under most circumstances (reviewed in Mathews et al., 1996). Translation initiation is facilitated by the mRNA 5′ cap structure, m7GpppN, which is present on all nuclear encoded mRNAs (Shatkin, 1976). The stimulatory effect of the cap on translation is mediated by eukaryotic translation initiation factor 4E (eIF4E), which directly contacts the cap structure (Sonenberg et al., 1979; Marcotrigiano et al., 1997). eIF4E exists either as a monomer or as a subunit in a complex called eIF4F. eIF4F is a heterotrimeric complex composed of eIF4E, eIF4A and eIF4G, where eIF4A is an ATP-dependent RNA helicase (Gingras et al., 1999). eIF4G is a modular scaffolding protein, which binds to eIF4E, eIF4A and the ribosome-associated initiation factor, eIF3, thus bridging the mRNA and the ribosome (Lamphear et al., 1995; Imataka and Sonenberg, 1997; reviewed in Gingras et al., 1999). There are two functional homologs of eIF4G, eIF4GI and eIF4GII, which function interchangeably as part of the eIF4F complex (Gradi et al., 1998).
eIF4E is a major target for translational control. It is the least abundant of the initiation factors (Duncan et al., 1987) and is considered to be the limiting factor for cap-dependent translation initiation (Mader and Sonenberg, 1995). eIF4E overexpression in NIH 3T3 cells causes malignant transformation (Lazaris-Karatzas et al., 1990), whereas depletion of eIF4E by antisense RNA partially reverses the transformed phenotype of ras-overexpressing cells (Rinker-Schaeffer et al., 1993). eIF4E is phosphorylated on Ser209 by the serine/threonine kinase Mnk1 (Waskiewicz et al., 1997; Pyronnet et al., 1999). Phosphorylation of eIF4E enhances its affinity for the cap structure (Minich et al., 1994), and increased eIF4E phosphorylation correlates with elevated translation rates in response to extracellular stimuli (Gingras et al., 1999). Cell stimulation also induces the phosphorylation and subsequent dissociation of eIF4E-binding proteins (4E-BPs) from eIF4E (Pause et al., 1994). The 4E-BP family of translation repressors comprises three members (Pause et al., 1994; Poulin et al., 1998) that compete with eIF4G for the interaction with eIF4E, and thus inhibit translation by preventing the assembly of the eIF4F complex (Haghighat et al., 1995).
Cell fractionation and immunofluorescence analysis demonstrated that between 12 and 33% of total eIF4E is localized to the nucleus of mammalian cells (Lejbkowicz et al., 1992). Electron microscope studies showed that eIF4E is also partially nuclear in Saccharomyces cerevisiae (Lang et al., 1994). Nuclear eIF4E co-localizes with splicing factors in speckles, suggesting that eIF4E may also play a role in pre-mRNA maturation or another function in the nucleus (Dostie et al., 2000). However, nothing is known about the nuclear import or export mechanism of eIF4E. Nucleocytoplasmic transport of macromolecules occurs through the nuclear pore complex (NPC) and is usually an energy-dependent, signal- mediated process (reviewed in Mattaj and Englmeier, 1998; Nakielny and Dreyfuss, 1999). Various types of nuclear localization signals (NLSs), nuclear export signals (NESs) and their receptors have been described. The best characterized NLSs are those of SV40 large T and nucleoplasmin, which consist of either a single or a bipartite cluster of basic amino acids, respectively (Kalderon et al., 1984; Robbins et al., 1991). Basic NLSs are recognized in the cytoplasm by importin α, an adaptor protein that is associated with the receptor protein, importin β. Interaction of the import cargo with the importin αβ complex triggers docking of the heterotrimeric complex to the NPC via importin β (Adam and Adam, 1994; Gorlich et al., 1995). The best described nuclear export signal is the leucine-rich NES, which was first identified in the human immunodeficiency type 1 (HIV-1) Rev protein (Fischer et al., 1995), and in the cellular protein kinase A inhibitor, PKI (Wen et al., 1995). Leucine-rich NESs are recognized directly by the nuclear export receptor CRM1/exportin 1 (Fornerod et al., 1997; Stade et al., 1997).
Translocation of transport cargoes through the NPC requires, in addition to receptors, several other proteins including the GTPase Ran/TC4 and p10/NTF2 (Mattaj and Englmeier, 1998). Ran/TC4 cycles between a GDP-bound and GTP-bound state, with Ran⋅GDP being predominantly cytoplasmic and Ran⋅GTP nuclear. Ran⋅GTP interacts directly with importins and exportins, and promotes directionality of transport by triggering the disassembly and assembly of import and export complexes, respectively (Gorlich et al., 1996; reviewed in Moore, 1998). When complexed with Ran⋅GTP, importin β releases the import cargo, whereas its association with the export receptor CRM1/exportin 1 is required for the formation of a stable export cargo–receptor complex (reviewed in Mattaj and Englmeier, 1998; Nakielny and Dreyfuss, 1999).
Here we report the cloning and functional characterization of a new eIF4E-binding protein termed 4E-T (eIF4E-Transporter). We show that 4E-T is a novel nucleocytoplasmic shuttling protein that targets eIF4E for nuclear import.
Results
Cloning of 4E-T cDNA
Two partial human 4E-T cDNA clones were isolated from a placental cDNA expression library by far-western screening using a [32P]HMK-eIF4E probe. To obtain a full-length cDNA, the longest (1.5 kb) cDNA was used to screen a human fetal brain cDNA library. Two overlapping partial clones were ligated to form a contiguous cDNA of 3520 bp (Figure 1A). Northern blotting analysis, using probes from three different regions, revealed one prominent mRNA of ∼3.8 kb (Figure 1B and data not shown). A similar sized RNA was also detected in all tissues tested including heart, brain, placenta, lung, liver, skeletal muscle, kidney and pancreas (data not shown). 4E-T cDNA encodes a 5′-untranslated region (UTR) of 52 nucleo tides, an open reading frame (ORF) of 2958 nucleotides and a 3′-UTR of 513 nucleotides. The ORF encodes a protein of 985 amino acids with a predicted molecular mass of 108.2 kDa. Because expression of the cDNA in HeLa cells generates a protein that co-migrates with the endogenous protein (see below; Figure 3), it is most probable that the first AUG is the authentic initiation codon, inasmuch as the length of the cDNA closely resembles that of the mRNA. Based on the results described below, we have named the protein encoded by this cDNA 4E-T.
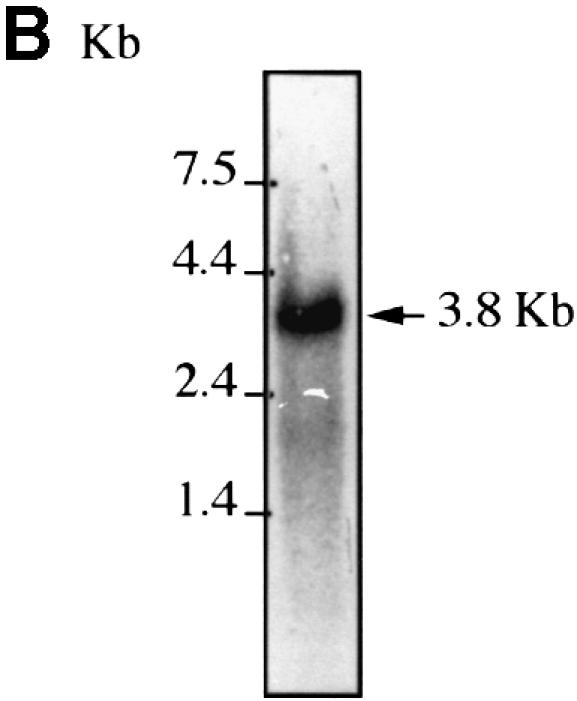
Fig. 1. 4E-T cDNA. (A) Nucleotide and amino acid sequence of human 4E-T. The first methionine is circled. The stop codon and polyadenylation signal are underlined and the eIF4E-binding site is boxed. The nuclear localization signal and nuclear export signals are highlighted with gray and black boxes, respectively. The 4E-T sequence has been deposited in the DDBJ/EMBL/GenBank database (accession No. AF240775). (B) Northern blotting analysis of 4E-T. HeLa poly(A)+ RNA (5 µg) was probed with a 4E-T 5′ end probe as described in Materials and methods.
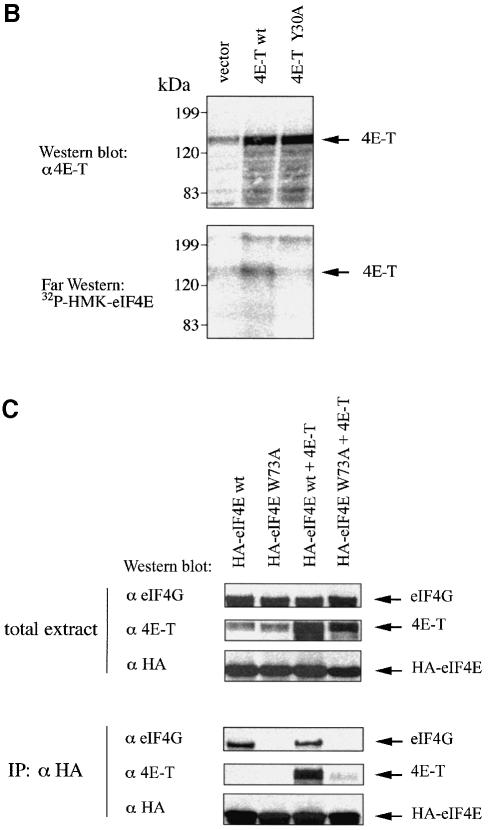
Fig. 3. 4E-T and eIF4E interact through conserved binding sites. (A) Amino acid sequence alignment of the eIF4E-binding site of 4E-T with those found in human eIF4GI (AF012088), human eIF4GII (AF012072), human 4E-BP1 (L36055), human 4E-BP2 (L36056), human 4E-BP3 (AF038869), Drosophila eIF4G (AF030155), S.cerevisiae eIF4GI (p39935), S.cerevisiae eIF4GII (p39936), S.cerevisiae p20 (X15731) and wheat iso-eIF4G (M95747). Residues which are critical for eIF4E binding are boxed. Solid and dashed lines represent identical and conserved residues, respectively. (B) 4E-T associates with eIF4E through a conserved motif. Untagged 4E-T wt and Y30A mutant were overexpressed in HeLa cells using the vaccinia virus system. Top: immunoblotting with anti-4E-T antibody. Bottom: far-western with [32P]HMK-eIF4E. (C) eIF4E binds to 4E-T and eIF4G through a shared sequence. HeLa cells were infected with vTF7-3 and transiently transfected with the plasmids indicated. Total extract [25% of input (50 µg)] was analyzed by western blotting with the indicated antibodies. Immunoprecipitation (from 200 µg of extract) of HA-eIF4E with anti-HA 12CA5 was performed as described in Materials and methods, and analyzed by western blotting with the antibodies indicated.
4E-T interacts with eIF4E in vitro and in vivo
To study the expression of 4E-T in cells, two different antisera against a glutathione S-transferase (GST)–4E-T N-terminal fusion protein were raised in rabbits, and antibodies from one antiserum were affinity purified. Both crude antisera and purified antibodies recognized a protein migrating at ∼140 kDa, which is higher than the predicted mass of 108 kDa in HeLa cells (Figure 2A, right) and in other cells, including NIH 3T3, COS-7, CV-1 and 293 (data not shown). An ∼140 kDa polypeptide was also recognized by a [32P]HMK-eIF4E probe in a far-western assay (Figure 2A, middle). As expected (Mader et al., 1995), eIF4E also bound to eIF4G in this assay. The 4E-T protein expressed from the 4E-T cDNA in HeLa cells also migrates aberrantly on SDS–PAGE and co-migrates with endogenous 4E-T (see below; Figure 3B and C). The slower mobility of 4E-T may be due to phosphorylation (see Discussion) or to its high proline content, as previously suggested for proline-rich proteins (See and Jakowski, 1989).
Fig. 2. Interaction of 4E-T with eIF4E. (A) eIF4E interacts with 4E-T as determined by far-western. HeLa cell extract (100 µg) was resolved by SDS–PAGE and stained with Coomassie Blue R-250 (left), or transferred to nitrocellulose and probed by far-western with a [32P]HMK-eIF4E probe (middle) or by western blotting with anti-4E-T antibody (right). (B) eIF4E specifically co-immunoprecipitates with 4E-T. HeLa cell extract (1 mg) was incubated with either pre-immune sera or anti-4E-T antibody as described in Materials and methods. Total extract [5% of input (50 µg)] and total immunoprecipitates from 1 mg of extract were resolved by SDS–PAGE, and analyzed by far-western with a [32P]HMK-eIF4E probe or by immunoblotting with anti-actin or anti-eIF4E monoclonal antibodies. (C) 4E-T interacts with eIF4E on a cap affinity resin. HeLa cell extract (1 mg) was incubated with resin or m7GDP resin, and processed as described in Materials and methods. Total extract [5% of input (50 µg)], resin, m7GDP resin-bound material from 1 mg of extract and flow through (5%) were resolved by SDS–PAGE, and analyzed by far-western with a [32P]HMK-eIF4E probe or by immunoblotting with anti-actin or anti-eIF4E monoclonal antibodies.
To determine whether 4E-T interacts with eIF4E in vivo, a co-immunoprecipitation experiment was performed with HeLa cell extracts. eIF4E specifically co-immunoprecipitated with 4E-T (Figure 2B). eIF4G and 4E-BP1 did not co-immunoprecipitate with 4E-T, suggesting that when complexed with 4E-T, eIF4E can no longer interact with eIF4G or 4E-BP1. Furthermore, 4E-T co-precipitated with eIF4E from HeLa extracts using an m7GDP resin (Figure 2C). Actin, tested as a negative control, did not co-immunoprecipitate with eIF4E, and was not retained on the m7GDP resin (Figure 2B and C). Taken together, these results demonstrate that 4E-T interacts with eIF4E both in vitro and in vivo.
4E-T and eIF4E interact through conserved binding sites
Interaction of the eIF4Gs and 4E-BPs with eIF4E is mutually exclusive (Haghighat et al., 1995) and occurs through the conserved eIF4E recognition motif Tyr-X-X-X-X-Leu-φ (where φ is a hydrophobic amino acid and X is any amino acid; Mader et al., 1995). Mutation of either Tyr or Leu-φ to alanine residues abolishes the binding of eIF4GI and 4E-BP1 to eIF4E (Mader et al., 1995). There is one putative eIF4E-binding site (Tyr-Thr-Lys-Glu-Glu-Leu-Leu) in the N-terminus of 4E-T (Figure 3A). To demonstrate that the putative eIF4E-binding site of 4E-T mediates its interaction with eIF4E, wild-type 4E-T (4E-T wt) or 4E-T Y30A mutant proteins were expressed in HeLa cells using the vaccinia virus system (Poulin et al., 1998). Expression of transfected 4E-T was assessed by immunoblotting with purified anti-4E-T antibody (Figure 3B, top), and the interaction with eIF4E was analyzed by far-western using a [32P]HMK-eIF4E probe. 4E-T wt, but not the Y30A mutant, associated with eIF4E, indicating that 4E-T interacts with eIF4E through the conserved eIF4E recognition motif (Figure 3B, bottom).
A phylogenetically conserved Trp73 is located on the convex dorsal surface of eIF4E (Marcotrigiano et al., 1997), and interacts with the invariant Leu and φ residues of the eIF4E-binding motif of eIF4G and 4E-BP1 (Marcotrigiano et al., 1999). The eIF4E W73A mutant protein does not interact with eIF4GI in vivo or in vitro (Pyronnet et al., 1999). To determine whether eIF4E interacts with 4E-T through the conserved Trp73, HeLa cells were infected with the recombinant vaccinia virus vTF7-3 (Fuerst et al., 1986), and then transfected with either hemagglutinin (HA)-eIF4E wt or the W73A mutant, alone or together with 4E-T wt. 4E-T co-immunoprecipitated with eIF4E wt, but not with the W73A mutant (Figure 3C). Interaction of HA-eIF4E wt with endogenous 4E-T was not detectable in this assay, presumably because of the small amount of extract used. Taken together, these results demonstrate that eIF4E and 4E-T interact through a conserved binding site.
4E-T contains one functional bipartite NLS
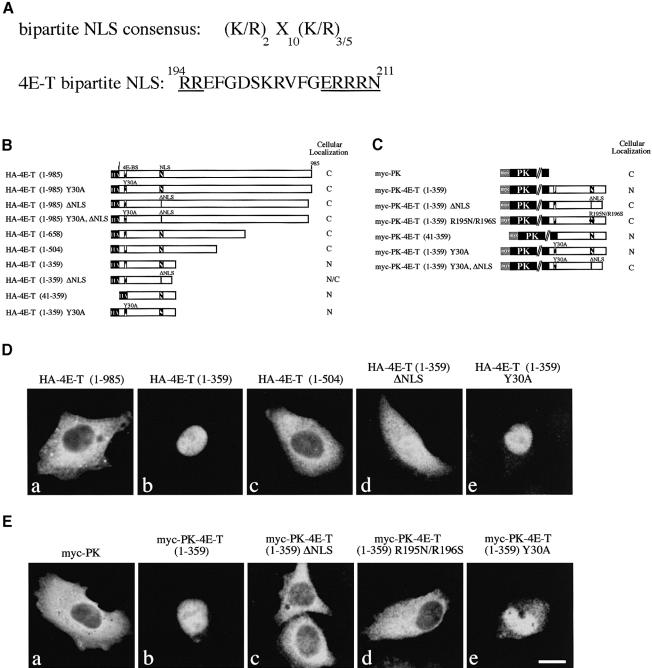
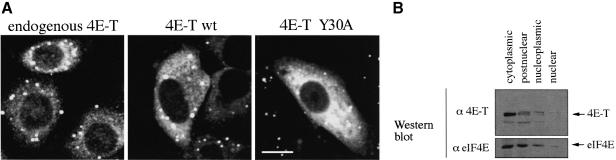
Sequence analysis of 4E-T revealed a putative bipartite NLS (Robbins et al., 1991; Figure 5A; see also Figure 1) in its N-terminus that could be important for its physiological function. Consequently, the cellular localization of 4E-T was examined. 4E-T localizes predominantly to the cytoplasm in HeLa cells as demonstrated by indirect immunofluorescence with anti-4E-T (Figure 4A, left). Overexpressed untagged 4E-T wt and Y30A mutant also localize to the cytoplasm in HeLa cells (Figure 4A, middle, right; cells overexpressing 4E-T were identified by their stronger fluorescence intensity as compared with the endogenous signal) and COS-7 cells (data not shown). Similarly, the localization of HA-tagged 4E-T wt and Y30A mutant is cytoplasmic (Figures 5, 6 and 7). However, in a small percentage of cells (∼5%), endogenous and transfected 4E-T is found in the nucleus (data not shown). HeLa cell fractionation shows that 4E-T is predominantly in the cytoplasmic and post-nuclear fraction. eIF4E is present in somewhat higher amounts in the nucleoplasmic fraction than was demonstrated earlier (Figure 4B; Lejbkowicz et al., 1992).
Fig. 5. 4E-T contains a functional bipartite NLS. (A) Putative bipartite NLS of 4E-T. Invariant residues are underlined. (B) and (C) Schematic maps of HA-4E-T and myc-PK-4E-T expression constructs used for NLS mapping. HeLa cells were transfected with either HA-4E-T or myc-PK-4E-T expression constructs. Cells were fixed at 36 h post-transfection and immunostained with either anti-HA 12CA5 (D) or anti-myc 9E10 antibody (E). The transfected construct is indicated above each panel. Bar, 10 µm.
Fig. 4. 4E-T localizes predominantly to the cytoplasm. (A) Indirect immunofluorescence analysis with anti-4E-T. Localization of endogenous 4E-T in HeLa cells (left). HeLa cells were transiently transfected with either 4E-T wt (middle) or Y30A (right). At 36 h post-transfection, cells were fixed and immunostained with anti-4E-T antibody as described in Materials and methods. Bar, 10 µm. (B) Cell fractionation. HeLa cells were fractionated as described in Materials and methods, and an equal cell volume from each fraction was resolved by SDS–PAGE. Fractions were analyzed by western blotting with anti-4E-T (top) or anti-eIF4E monoclonal antibody (bottom).
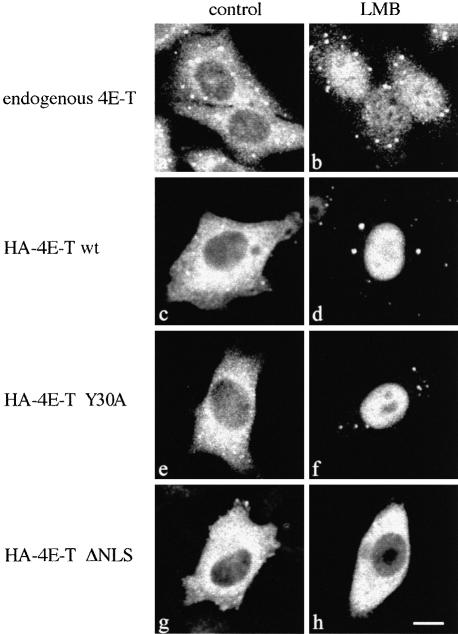
Fig. 6. 4E-T is a shuttling protein. HeLa cells were incubated with medium in the absence (A) or the presence of leptomycin B (LMB) (B) for 5 h prior to fixation and immunostaining with anti-4E-T as described in Materials and methods. (C–H) HeLa cells were transfected with either HA-4E-T wt, Y30A or ΔNLS expression constructs. At 36 h post-transfection, the medium was replaced with medium either lacking (control) or containing LMB. Cells were processed as described above, except that anti-HA 12CA5 antibody was used for immunoblotting. Bar, 10 µm.
Fig. 7. 4E-T contains two functional NESs. (A) Putative NESs of 4E-T. Invariant residues are boxed. HeLa cells were transfected with either HA-4E-T (B) or myc-NPc-4E-T expression constructs (C). Cells were fixed at 36 h post-transfection and immunostained with either anti-HA 12CA5 (B) or anti-myc 9E10 antibody (C). Transfected construct is indicated above each panel. (D) and (E) Schematic maps of HA-4E-T and myc-NPc-4E-T expression constructs used for NES mapping. Bar, 10 µm.
Our results show that although 4E-T contains a putative NLS, it is predominantly cytoplasmic. It is possible that the NLS is functional, but that other segments in 4E-T could cause its cytoplasmic localization. To determine whether the putative NLS motif is functional, several HA-4E-T deletion mutants were constructed and analyzed by indirect immunofluorescence (Figure 5B and D). While the full-length HA-4E-T localized to the cytoplasm (Figure 5D, panel a), the HA-4E-T 1–359 fragment, which contains the putative NLS, localized to the nucleus (panel b). However, in contrast, longer fragments of 4E-T (1–504) localized to the cytoplasm in a manner similar to the full-length protein (panel c). The exclusive nuclear localization of 4E-T (1–359) is not the result of free diffusion because of its low molecular weight, inasmuch as a deletion of the putative NLS (HA-4E-T 1–359 ΔNLS) results in an even distribution of this fragment between the nucleus and the cytoplasm (panel d). Mutation of Y30A did not affect the nuclear targeting of HA-4E-T 1–359 (panel e), indicating that the interaction with eIF4E is not required for nuclear import of 4E-T.
To establish further that the putative NLS of 4E-T is functional, we examined its ability to confer nuclear import on a heterologous cytoplasmic protein. The 4E-T 1–359 fragment was fused to myc-tagged chicken muscle pyruvate kinase (myc-PK; Siomi and Dreyfuss, 1995; Figure 5C and E). The 4E-T fragment 1–359, but not 4E-T 1–359 ΔNLS, caused myc-PK to accumulate in the nucleus (Figure 5E, panels a, b and c). Furthermore, mutation of the first two arginines in the NLS motif in 4E-T 1–359 (R195N/R196S) prevented the import of myc-PK into the nucleus (panel d). This demonstrates the presence of a bipartite NLS in the 4E-T 1–359 fragment. Y30A mutation did not affect the nuclear localization of myc-PK-4E-T 1–359 (panel e). These results indicate that 4E-T contains one functional bipartite NLS, and demonstrate that nuclear import of 4E-T does not require interaction with eIF4E.
4E-T contains two functional leucine-rich NESs
One possible explanation for the predominant cytoplasmic localization of 4E-T could be that its nuclear import is regulated, either by phosphorylation, as was shown for example for NF-AT4 (Zhu et al., 1998), or through its interaction with cytoplasmic proteins, as was demonstrated for NF-κB (Henkel et al., 1992). It is also possible that the NLS of 4E-T is always active, but that 4E-T shuttles between the nucleus and the cytoplasm. To determine whether 4E-T is a shuttling protein, HeLa cells were treated with the nuclear export inhibitor leptomycin B (LMB). LMB is a Streptomyces metabolite that inhibits export of leucine-rich NES-containing proteins by interacting with the export receptor CRM1/exportin 1, and preventing its association with the NES substrate (Kudo et al., 1998). Endogenous and transfected HA-4E-T wt, but not HA-4E-T ΔNLS, accumulated in the nucleus upon LMB treatment (Figure 6). LMB also caused the nuclear accumulation of HA-4E-T Y30A, confirming that association with eIF4E is not required for nuclear import of 4E-T. This result shows that 4E-T is a nucleocytoplasmic shuttling protein and suggests that 4E-T contains a leucine-rich NES.
There are two putative NESs in the middle region of 4E-T (Figure 7A). To determine whether these motifs are functional, HA-4E-T deletion mutants were constructed (Figure 7D), transfected into HeLa cells and analyzed by indirect immunofluorescence. 4E-T 321–658, which contains both putative NESs, is predominantly cytoplasmic (Figure 7B, panel c), whereas HA-4E-T 1–359 is nuclear (panel b). HA-4E-T 659–985 is distributed evenly between the cytoplasm and the nucleus (panel d). These data show that nuclear export activity is present only in the middle region of 4E-T. To determine which of the putative NES motifs is functional, deletion and point mutants of 4E-T 321–658 were fused to the nuclear myc-tagged nucleoplasmin core protein (myc-NPc; Michael et al., 1997), and examined for their ability to confer export activity on NPc (Figure 7E). Fusion of 4E-T fragments 321–658, 321–504 and 505–658 to the NPc conferred on it nuclear export (Figure 7C), indicating that 4E-T contains at least two independent functional NESs. Deletion of NES1 in 4E-T 321–504 or NES2 in 4E-T 505–658 abolished the nuclear export activity of these fragments (Figure 7E). This result confirms that both NESs are functional, and indicates that there are no additional NESs in the 4E-T 321–658 fragment. Point mutations of all the invariant residues in NES1 and NES2 (4E-T NES1;NES2 mutations) also abolished nuclear export of 4E-T 1–985 and 4E-T 321–658 (Figure 7B and C). Y30A mutation or deletion of the eIF4E-binding site did not affect nuclear export of 4E-T (Figures 6 and 7D). These results show that 4E-T contains two separate and independent functional NESs, and indicate that nuclear export of 4E-T does not require binding to eIF4E.
4E-T mediates the nuclear import of eIF4E
Although eIF4E is sufficiently small to diffuse passively through the NPC, the nucleocytoplasmic transport of most low molecular weight proteins and that of all known RNAs is believed to be an active, signal-mediated process (Mattaj and Englmeier, 1998). Also, since eIF4E is complexed with either eIF4Gs or 4E-BPs in the cytoplasm, it is very likely that eIF4E requires an active process for its import into the nucleus. To determine whether 4E-T mediates the nuclear import of eIF4E, we first examined the ability of eIF4E to form a quaternary complex with 4E-T, importin α and importin β. Purified recombinant proteins were pre-incubated before the addition of glutathione–Sepharose resin to precipitate the complex via GST–importin β. 4E-T associated with importin β only in the presence of importin α (Figure 8, compare lanes 5 and 6), and eIF4E interacted with importin α and importin β only in the presence 4E-T (compare lanes 7 and 8). A lower amount of importin α was precipitated with GST–importin β when 4E-T was present. We found that this was dependent on the order in which the recombinant proteins were added to the reaction mixture. When importin α was pre-incubated with importin β, this effect was not observed, but noticeably less eIF4E was pulled-down in the quaternary complex. This effect may be due to some extent to improper folding of the recombinant proteins or may reflect the physiological order in which the complex is formed in the cell. This assay shows that the interaction of eIF4E with the importin αβ complex requires 4E-T, and that interaction of eIF4E and importin α with 4E-T is therefore not mutually exclusive. Thus, although nuclear import of 4E-T does not require an interaction with eIF4E, it is plausible that 4E-T mediates the nuclear import of eIF4E by a piggy-back mechanism (Turpin et al., 1999).
Fig. 8. eIF4E forms a complex with importin α and β only in the presence of 4E-T. The purified proteins indicated were incubated with glutathione–Sepharose beads as described in Materials and methods. Precipitates were analyzed by western blotting with anti-4E-T, anti-GST, S-protein (Novagen) and anti-eIF4E (No. 5853) as indicated. Input represents 25% of total protein used for pull-down; flow through represents 10% of unbound material.
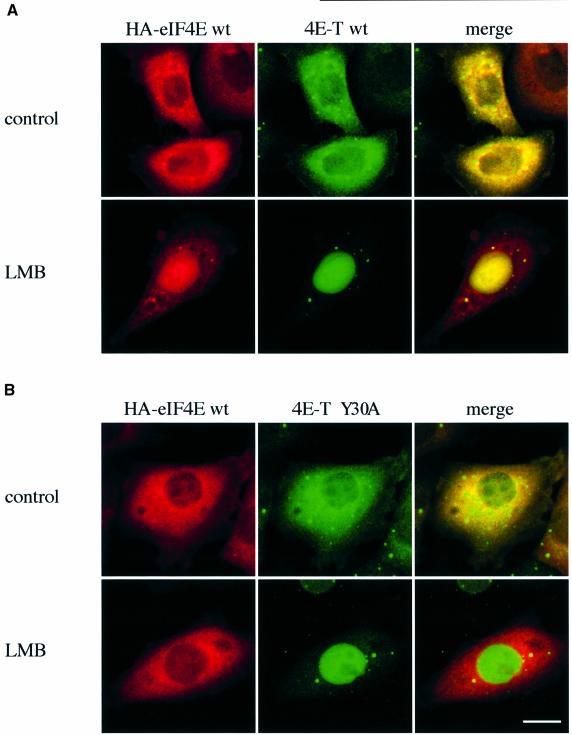
To demonstrate that 4E-T mediates the nuclear import of eIF4E in vivo, HeLa cells were co-transfected with HA-eIF4E together with 4E-T wt or 4E-T Y30A, the mutant defective in eIF4E binding (see Figure 3B). The localization of the proteins was analyzed by double-label immunofluorescence using anti-HA and anti-4E-T antibodies. HA-eIF4E was shown previously to localize partially to the nucleus (Lejbkowicz et al., 1992), while in another report, it localized to the cytoplasm (Waskiewicz et al., 1999). We found that this discrepancy is most probably due to different eIF4E expression levels in the two studies. Transfection with low amounts of DNA results in cytoplasmic accumulation, while transfection with high amounts results in a predominant nuclear accumulation of HA-eIF4E (data not shown). Similarly, the localization of a green fluorescent protein (GFP)–Cdc25 fusion protein was also shown to be dependent on the expression levels (Kumagai and Dunphy, 1999). The cytoplasmic retention of eIF4E, which is expressed at low levels, occurs presumably because of its interaction with cytoplasmic binding partners such as the eIF4Gs and the 4E-BPs. To demonstrate the effect of 4E-T on eIF4E nuclear transport, HA-eIF4E was expressed at low levels, where it localizes predominantly to the cytoplasm (Figure 9A and B, left). Expression of 4E-T caused the nuclear accumulation of HA-eIF4E in cells treated with LMB (Figure 9A). However, in sharp contrast, expression of 4E-T Y30A, which accumulated in the nucleus in the presence of LMB, failed to cause the import of eIF4E into the nucleus. That the localization of HA-eIF4E was not affected by 4E-T Y30A substantiates the idea that 4E-T is required for the nuclear import of eIF4E. Taken together, our results show that 4E-T mediates the nuclear import of eIF4E by a piggy-back mechanism.
Fig. 9. 4E-T imports HA-eIF4E into the nucleus. HeLa cells were co-transfected with either HA-eIF4E and 4E-T wt (A), or HA-eIF4E and 4E-T Y30A (B). At 36 h post-transfection, the medium was replaced with fresh medium (control) or medium containing leptomycin B (LMB), and cells were incubated for 5 h prior to fixation. The localization of HA-eIF4E was determined by indirect immunofluorescence with anti-HA 12CA5 and Texas red-conjugated anti-mouse IgG (left), and that of 4E-T with anti-4E-T and fluorescein-conjugated anti-rabbit IgG (middle). The co-localization of HA-eIF4E and 4E-T appears yellow (right). Bar, 10 µm.
Discussion
Here we provide evidence that the nuclear import of murine eIF4E occurs via the importin αβ pathway by a piggy-back mechanism, which requires a novel nucleocytoplasmic shuttling protein, 4E-T. Several other proteins lacking a nuclear import signal rely on the use of the NLS of binding partners for import into the nucleus. For example, the interleukin (IL)-5 α- and β-receptor subunits require IL-5, adenovirus E1B 55-kDa requires the adenovirus E4 34-kDa, and Cdk2 requires cyclin E for nuclear import (Goodrum et al., 1996; Jans et al., 1997; Moore et al., 1999). Although IκBα is sufficiently small to diffuse passively through the NPC, a recent report suggests that it is also imported by a piggy-back mechanism through its association with an ankyrin repeat-binding protein (Turpin et al., 1999). The nuclear import of several small polypeptides such as histone H1 (Jakel et al., 1999) and ribosomal proteins (Jakel and Gorlich, 1998) is carrier-mediated, whereas rad24 (Lopez-Girona et al., 1999) and U1 snRNP-specific protein C (Klein Gunnewiek et al., 1997) localize to the nucleus by passive diffusion and subsequent retention in the nucleus.
Why does eIF4E require a carrier for nuclear import? Since eIF4E, which is the most limiting initiation factor (Duncan et al., 1987), interacts with several more abundant cytoplasmic proteins, it is probable that in the absence of 4E-T, eIF4E would be retained in the cytoplasm. This is in good agreement with our results showing that overexpression of 4E-T wt, but not the Y30A mutant, causes the nuclear accumulation of HA-eIF4E in cells treated with LMB (Figure 9). Also, the observations that eIF4E, which immunoprecipitates with 4E-T, is not associated with eIF4Gs or 4E-BPs (Figure 2B), and that eIF4E and 4E-T interact through conserved binding domains (Figure 3), demonstrate that the interaction of 4E-T, the eIF4Gs and the 4E-BPs with eIF4E is mutually exclusive. Indeed, eIF4G is not co-precipitated with the eIF4E–4E-T complex (Figure 2B). Consistent with this, overexpression of 4E-T wt, but not 4E-T Y30A, strongly inhibits cap-dependent translation of a reporter luciferase construct (J.Dostie, unpublished observations). Thus, the use of 4E-T in mediating the nuclear import of eIF4E provides an efficient mechanism specifically to recruit eIF4E to the import machinery by competing with eIF4E-binding proteins. Direct binding of eIF4E to importin α as an alternative means to import eIF4E to the nucleus would not be effective, since importin α is a general adaptor protein, which localizes primarily to the nucleus and at the nuclear membrane, and thus would not be an effective competitor with eIF4Gs and 4E-BPs for binding to eIF4E.
A database search identified 4E-T orthologs in mouse and rat but did not identify any putative 4E-T homolog in S.cerevisiae. The Drosophila cup protein (Keyes and Spradling, 1997) and CG11097 gene product (DDBJ/EMBL/GenBank accession No. AAF59398.1), and the Caenorhabditis elegans clone F26F3 (DDBJ/EMBL/GenBank accession No. S43585), share limited homology with a short segment of 4E-T (amino acids 201–261). It is possible that in yeast and less complex eukaryotes, eIF4E uses a different adaptor protein that shares little homology with 4E-T. It is also possible that in S.cerevisiae there is more free eIF4E and that, consequently, eIF4E localizes to the nucleus by passive diffusion.
The use of 4E-T might provide a means for the control of eIF4E nuclear import through the regulation of 4E-T. Since 4E-T shuttles between the nucleus and the cytoplasm, either the nuclear import or export of 4E-T could be modulated. Phosphorylation affects the nuclear import and/or export of numerous proteins. For example, nuclear import of nucleoplasmin (Vancurova et al., 1995) and c-rel (Gilmore and Temin, 1988; Mosialos et al., 1991) is enhanced by phosphorylation. Also, phosphorylation of Jak family members by STAT kinases (Leonard and O’Shea, 1998) triggers their nuclear import, whereas dephosphorylation of NF-AT4 by calcineurin is required for nuclear entry (Zhu et al., 1998). Additionally, nuclear export of MK2 (Engel et al., 1998) is activated by phosphorylation, whereas phosphorylation of cyclin B1 (Li et al., 1997a) inhibits its nuclear export. Since 4E-T is a phosphoprotein (J.Dostie, unpublished observations), the nuclear localization of 4E-T and, consequently, that of eIF4E may also be regulated by phosphorylation. eIF4E contains a putative NES (amino acids 126–137 in human eIF4E), but it is not known whether it is functional. Further studies with the heterokaryon assay using eIF4E mutants will be required in order to determine the nuclear export mechanism of eIF4E.
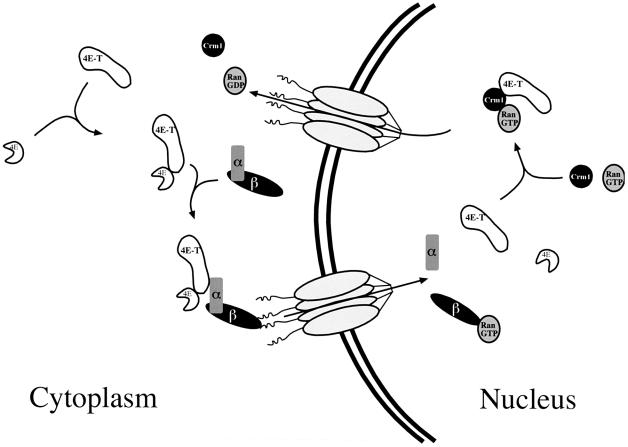
The results presented here support the conclusion that eIF4E is imported into the nucleus via the importin αβ pathway by a piggy-back mechanism through its association with 4E-T (see model in Figure 10). Upon nuclear entry, eIF4E and 4E-T would dissociate from importin α as a result of the interaction of Ran⋅GTP with importin β. This process probably occurs immediately and rapidly following the transport, because 4E-T resides most of the time in the cytoplasm. Although the relative affinity of 4E-T for eIF4E in the presence or absence of importin α is not known, it is possible that dissociation of 4E-T from importin α also results in the release of eIF4E. 4E-T and eIF4E may dissociate because of a decrease in affinity or because of their interaction with nuclear proteins. Following the release of eIF4E, 4E-T shuttles back to the cytoplasm via the CRM1/exportin 1 pathway. Since nuclear import or export of 4E-T does not require interaction with eIF4E, it is possible that 4E-T also functions as an import or export adaptor for other proteins. Therefore, the identification of 4E-T-binding proteins is of interest.
Fig. 10. 4E-T mediates the nuclear import of eIF4E. See the text for details.
The nuclear function of eIF4E is not known. The cap is involved in several aspects of RNA metabolism in the nucleus, including 3′ end mRNA processing (Hart et al., 1985), pre-mRNA splicing (Konarska et al., 1984; Izaurralde et al., 1994) and nuclear export of RNA polymerase II transcripts (Hamm and Mattaj, 1990; Jarmolowski et al., 1994). Several nuclear cap-binding proteins mediate the nuclear function of the cap structure (Patzelt et al., 1983; Rozen and Sonenberg, 1987; Izaurralde et al., 1992). For example, the nuclear cap-binding complex (CBC), which is composed of CBP20 and CBP80, functions in pre-mRNA splicing (Izaurralde et al., 1994; Lewis et al., 1996) and mRNA 3′ end processing (Flaherty et al., 1997). CBC is also required for U snRNA export (Izaurralde et al., 1995), and although it does not seem to facilitate mRNA export dramatically, a study on the Chironomus tentans Balbiani ring mRNA export demonstrates that CBC binds co-transcriptionally to the cap and accompanies the ribonucleoprotein particle during nuclear export (Visa et al., 1996).
We have shown recently that nuclear eIF4E co-localizes with splicing factors in speckles (Dostie et al., 2000). The pattern of eIF4E nuclear localization is modulated by inhibitors of RNA polymerase II, and is regulated by the dual specificity kinase Clk/Sty, suggesting that, in addition to its role in translation, eIF4E may also be involved in nuclear mRNA metabolism. Another intriguing, but more remote possibility is that eIF4E is involved in mRNA surveillance by nuclear translation to target mRNAs that contain premature nonsense codons for degradation (Li et al., 1997b).
Materials and methods
Isolation of 4E-T cDNA clones and DNA sequence analysis
Partial 4E-T cDNA clones were isolated from a λgt11 human placenta cDNA library (kindly provided by M.Park, McGill University) following far-western screening using a 32P-labeled eIF4E probe as previously described (Pause et al., 1994). An 809 bp DNA fragment was generated by PCR using a partial cDNA clone as template and the following primers: 5′-GCCTTCATGCCTTTCTG-3′ and 5′-GGCACCTTATCAAGGTTAAAG-3′. The amplified DNA was 32P-labeled by random priming using [α-32P]dCTP, random hexamers and the Klenow fragment of DNA polymerase (Feinberg and Vogelstein, 1984). A λ Uni-ZAP-XR human fetal brain cDNA library (kindly provided by G.Rouleau, Université de Montréal) was screened with this probe to obtain a longer cDNA clone. Phages (1 × 106) displayed on duplicate sets of filters (Hybond-N+; Amersham) were screened. Isolation of the positive clones was performed as described in the Stratagene ZAP Express instruction manual. Oligonucleotides used for sequencing were derived from either pBluescript or from the 4E-T DNA sequence. Nucleotide sequence was determined from both strands using the dideoxy chain termination method (Sanger et al., 1977) and the T7 polymerase sequencing kit (Amersham Pharmacia Biotech). Regions of compression were re-sequenced using 7-deaza-dGTP.
Northern blotting
Total RNA from HeLa cells was isolated using Trizol (Life Technology, Inc.) and purified twice in batch mode with oligo(dT) beads (Amersham Pharmacia Biotech). Poly(A)+ RNA (5 µg) was fractionated by electrophoresis on a 1% agarose/formaldehyde gel, transferred to a Hybond-N+ membrane and cross-linked to the membrane using UV light (Stratalinker). The membrane was pre-hybridized in 5× SSPE (0.9 M sodium chloride, 50 mM sodium phosphate, 5 mM EDTA pH 7.7), 5× Denhardt’s solution, 50% formamide, 1% SDS, 10% dextran sulfate, 100 µg/ml denatured salmon sperm for 4 h at 42°C. The probe was generated as described above, and hybridization was performed in the same buffer containing the 4E-T probe at 1 × 106 c.p.m./ml for 16 h at 42°C. The membrane was washed to a final stringency of 0.1% SSPE, 0.1% SDS at 68°C, and exposed against a Kodak XAR film for 48 h with intensifying screens.
Antibodies
A GST–4E-T fusion protein comprising amino acids 1–471 of 4E-T was purified on glutathione–Sepharose according to the manufacturer’s instructions (Amersham Pharmacia Biotech). Two New Zealand White rabbits were immunized with GST–4E-T as previously described (Gradi et al., 1998). Anti-GST- and anti-4E-T-specific antibodies were purified from the resulting serum by affinity chromatography using GST and GST–4E-T (1–471) columns. Anti-eIF4E no. 5853 (Frederickson et al., 1991), and anti-eIF4GI C (Imataka et al., 1997) were described previously. Anti-eIF4A and anti-myc 9E10 antibody were kindly provided by H.Trachsel (University of Berne) and M.Tremblay (McGill University), respectively. Monoclonal anti-eIF4E and anti-actin antibodies were purchased from Transduction Laboratories. The anti-HA 12CA5 was purchased from BabCo. Peroxidase-coupled anti-mouse and anti-rabbit IgG were purchased from Amersham Corp. Texas red- and fluorescein-coupled secondary antibodies were purchased from Molecular Probes.
Cell culture, cell extracts and western blotting
Cells were plated on 100 mm dishes and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). At 80% density, the cells were washed twice in phosphate-buffered saline (PBS), scraped and pelleted by slow centrifugation. Cell pellets were resuspended in either buffer B [50 mM Tris–HCl pH 7.4, 150 mM potassium chloride, 20% glycerol, 1 mM EDTA, 1 mM dithiothreitol (DTT), 0.2% Tween-20 and Complete™ (Boehringer Mannheim)] or RIPA buffer (50 mM Tris–HCl pH 7.4, 150 mM sodium chloride, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate and Complete™) and incubated for 30 min at 4°C. Alternatively, cells were resuspended in buffer A (20 mM Tris–HCl pH 7.5, 100 mM potassium chloride, 2 mM EDTA, 2 mM DTT and Complete™) and lysed by three freeze–thaw cycles. Cell debris was removed by centrifugation at 10 000 g for 10 min at 4°C, and protein concentration was determined with the Bio-Rad assay. Polypeptides were resolved by SDS–PAGE and transferred onto 0.45 µm nitrocellulose membranes. Membranes were blocked for 16 h at 4°C with 5% skim milk in PBS containing 0.2% Tween-20 (PBST). Primary antibodies were incubated for 2 h at room temperature followed by four washes of 15 min in PBST. Membranes were incubated with either peroxidase- or 125I-coupled secondary antibodies for 30 min at room temperature and washed four times for 15 min in PBST. Detection of peroxidase-coupled secondary antibodies was performed with Enhanced-ECL (Amersham Corp.).
Far-western analysis and Coomassie Blue staining
Partially purified mouse Flag-HMK-eIF4E fusion protein (2.5 µg) was 32P-labeled with heart muscle kinase (Sigma) as described (Blanar and Rutter, 1992). Membranes were incubated with the probe as previously described (Methot et al., 1997). HeLa cell extract (100 µg) was resolved by SDS–PAGE, incubated for 30 min at room temperature in Coomassie Blue staining solution [50% methanol, 0.05% Coomassie Brilliant Blue R-250 (Bio-Rad), 10% acetic acid], and unbound dye was removed with destaining solution (5% methanol, 7% acetic acid).
Vaccinia virus infection, immunoprecipitation and m7GDP resin precipitation
HeLa cells (70% confluency) were infected with recombinant vaccinia virus vTF7-3 for 1 h at 37°C (Fuerst et al., 1986). Cells were washed with serum-free medium and transfected with plasmid DNA (5 µg) using Lipofectin (Life Technologies, Inc.) according to the manufacturer’s recommendations. Cell extract (200 µg) prepared with buffer B as described above was incubated with 2 µg of anti-HA 12CA5 antibody for 2 h at 4°C. Protein G–Sepharose (50 µl of a 50% slurry) was added and the mixture was incubated for an additional hour at 4°C (Amersham Corp.). Immunoprecipitates were collected by slow centrifugation (3000 g), washed three times with buffer B, boiled for 6 min in Laemmli buffer and resolved by SDS–PAGE. For co-immunoprecipitation of total cell extract, 1 mg of HeLa extract in buffer B was incubated with 1 µg of anti-4E-T and processed as described above. For m7GDP resin precipitation, 1 mg of HeLa extract in buffer A was incubated with 25 µl of m7GDP resin (Edery et al., 1988) for 2 h at 4°C. The resin was washed three times with 1 ml of buffer A, boiled for 6 min in Laemmli buffer and resolved by SDS–PAGE.
Cell fractionation
HeLa cell fractionation into cytoplasmic, post-nuclear, nucleoplasmic and nuclear fractions was performed as previously described (Lejbkowicz et al., 1992) with minor modifications. Briefly, cells (5 × 106) were washed twice with ice-cold PBS, scraped and pelleted by slow centrifugation. Cell pellets were washed with isotonic buffer 1 [10 mM HEPES–KOH pH 7.4, 250 mM sucrose, 1 mM phenylmethylsulfonyl fluoride (PMSF) and Complete™], resuspended in 1 ml of buffer 2 (10 mM HEPES–KOH pH 7.4, 250 mM sucrose, 2 mM EDTA, 0.1 mM DTT, 1 mM PMSF and Complete™) and disrupted by 50 strokes with a pestle B homogenizer. Lysates were centrifuged at 1000 g for 10 min at 4°C. This step was repeated twice, and the supernatant was regarded as the cytoplasmic fraction. Nuclei were resuspended in 1 ml of buffer 3 (10 mM HEPES–KOH pH 7.4, 250 mM sucrose, 2 mM EDTA, 0.1 mM DTT, 2 mM magnesium chloride, 0.1% Triton X-114, 1 mM PMSF and Complete™), incubated on ice for 15 min and centrifuged at 1000 g for 10 min at 4°C. The supernatant was collected and regarded as the post-nuclear fraction. Nuclei were resuspended in 0.5 ml of buffer 4 (10 mM HEPES–KOH pH 7.4, 250 mM sucrose, 2 mM EDTA, 0.1 mM DTT, 0.5% NP-40, 50 mM sodium chloride, 1 mM PMSF and Complete™), incubated on ice for 30 min and centrifuged at 1000 g for 10 min at 4°C. This step was repeated twice, and the combined supernatants were regarded as the nucleoplasmic fraction. The NP-40-extracted nuclear pellet was resuspended in 0.2 ml of buffer 5 (20 mM HEPES–KOH pH 7.9, 25% glycerol, 420 mM sodium chloride, 1.5 mM magnesium chloride, 0.2 mM EDTA, 0.5 mM DTT, 1 mM PMSF and Complete™), incubated on ice for 20 min and centrifuged at 14 000 g for 2 min at 4°C. The supernatant was collected and regarded as the nuclear fraction.
Construction of plasmids
pcDNA3-4E-T was constructed by cloning the full-length 4E-T cDNA into pcDNA3 (Invitrogen). HA-4E-T was generated by subcloning the coding region of 4E-T from pcDNA3-4E-T into PACTAG2 (kindly provided by M.Tremblay, McGill University). All 4E-T mutants were constructed by PCR using either pcDNA3-4E-T or HA-4E-T as template. The plasmids myc-PK and myc-NPc (kindly provided by G.Dreyfuss, University of Pennsylvania School of Medicine) were used to generate myc-PK- and myc-NPc-4E-T constructs. To subclone 4E-T into myc-PK, primers were designed to include a KpnI site and a NotI site in the forward and reverse oligonucleotide, respectively (Siomi and Dreyfuss, 1995). To generate myc-NPc-4E-T constructs, primers were designed to include an EcoRI site and an XhoI site in the forward and reverse oligonucleotide, respectively (Michael et al., 1997). HA-eIF4E wt and HA-eIF4E W73A were described previously (Pyronnet et al., 1999). For the purification of baculovirus-expressed recombinant Flag-HMK-4E-T, the coding region of 4E-T was subcloned into the baculovirus transfer vector pVL1392-FlagHMK (Pharmingen) (Haghighat et al., 1996).
Transient transfections and immunofluorescence
HeLa cells (50% confluency) were transfected with 10 µg of plasmid DNA by the calcium phosphate transfection method (Graham and Van Der Eb, 1973). After 24 h, cells were trypsinized and plated at 2 × 104 per chamber on Lab-Tek chamber slides (Nunc). At 36 h post-transfection, cells were fixed for 30 min with 4% formaldehyde in PBS and permeabilized for 30 min with 4% formaldehyde/0.2% Tween-20 in PBS at room temperature. Cells were rehydrated briefly with 0.2% Tween-20 in PBS before blocking for 16 h in a solution containing 50% FBS, 6% skim milk, 3% bovine serum albumin (BSA), 0.2% Tween-20 and 0.02% sodium azide. Cells were incubated with primary antibodies for 2 h at room temperature and washed extensively with 0.2% Tween-20/PBS before and after incubation with secondary antibodies for 30 min at room temperature. Cells were mounted in 30% glycerol in PBS and analyzed with a Kontron IBAS confocal imaging system. Images were taken from a 63× objective of a Zeiss LSM 410 microscope. For drug treatment, LMB (kindly provided by M.Yoshida, University of Tokyo) was added to the media at a final concentration of 2 ng/ml, and incubated for the time period indicated in the figure legends.
Pull-down assay
S-Tag-importin α and GST–importin β were purified as previously described (Chi et al., 1996). Baculovirus Flag-HMK-4E-T was purified from Spodoptera frugiperda (Sf9) cells as previously described (Haghighat et al., 1995), except that cells were disrupted by a Dounce homogenizer (50 strokes, pestle B) in transport buffer (20 mM HEPES–KOH pH 7.0, 150 mM potassium acetate, 2 mM magnesium acetate, 2 mM DTT, 0.1% casamino acids, 0.1% Tween-20 and Complete™) and purification was performed in transport buffer. Purification of untagged eIF4E was performed as previously described (Edery et al., 1988), except that the protein was eluted in transport buffer. Approximately 3 µg of S-Tag-importin α, 375 ng of GST–importin β, 1 µg of Flag-HMK-4E-T and 1 µg of eIF4E were used in the assay. Proteins were mixed and pre-incubated on ice for 30 min in a final volume of 30 µl. Transport buffer (250 µl) and 15 µl of glutathione–Sepharose resin (Amersham Pharmacia Biotech) were then added to the samples. The samples were incubated for 40 min at room temperature end-over-end and washed three times with transport buffer. The precipitates were eluted in sample buffer, resolved by SDS–PAGE and analyzed by western blotting.
Nucleotide sequence accession number
The nucleotide and protein sequence of 4E-T have been submitted to the DDBJ/EMBL/GenBank database under accession No. AF240775.
Acknowledgments
Acknowledgements
We thank M.Park and G.Rouleau for human placenta and fetal brain cDNA libraries, respectively, M.Tremblay for PACTAG plasmid and anti-HA 12CA5 antibody, H.Trachsel for anti-eIF4A antibody, G.Dreyfuss for myc-PK and myc-NPc plasmids, and M.Yoshida for LMB. We thank H.Imataka, S.Pyronnet and E.Izaurralde for comments on the manuscript. We are indebted to Colin Lister and Chantal Binda for excellent technical assistance and to H.Dilhuidy for technical support with the confocal microscopy. J.D. was supported by a studentship from the Medical Research Council of Canada. This work was supported by a grant from the Medical Research Council of Canada to N.S. N.S. is a Distinguished Scientist of the Medical Research Council of Canada, and a Howard Hughes Medical Institute International Scholar.
References
- Adam E.J. and Adam,S.A. (1994) Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J. Cell Biol., 125, 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanar M.A. and Rutter,W.J. (1992) Interaction cloning: identification of a helix–loop–helix zipper protein that interacts with c-Fos. Science, 256, 1014–1018. [DOI] [PubMed] [Google Scholar]
- Chi N.C., Adam,E.J., Visser,G.D. and Adam,S.A. (1996) RanBP1 stabilizes the interaction of Ran with p97 nuclear protein import. J. Cell Biol., 135, 559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J., Lejbkowicz,F. and Sonenberg,N. (2000) Nuclear eukaryotic initiation factor 4E (eIF4E) colocalizes with splicing factors in speckles. J. Cell Biol., 148, 239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan R., Milburn,S.C. and Hershey,J.W. (1987) Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J. Biol. Chem., 262, 380–388. [PubMed] [Google Scholar]
- Edery I., Altmann,M. and Sonenberg,N. (1988) High-level synthesis in Escherichia coli of functional cap-binding eukaryotic initiation factor eIF-4E and affinity purification using a simplified cap-analog resin. Gene, 74, 517–525. [DOI] [PubMed] [Google Scholar]
- Engel K., Kotlyarov,A. and Gaestel,M. (1998) Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation. EMBO J., 17, 3363–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A.P. and Vogelstein,B. (1984) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem., 137, 266–267. [DOI] [PubMed] [Google Scholar]
- Fischer U., Huber,J., Boelens,W.C., Mattaj,I.W. and Luhrmann,R. (1995) The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell, 82, 475–483. [DOI] [PubMed] [Google Scholar]
- Flaherty S.M., Fortes,P., Izaurralde,E., Mattaj,I.W. and Gilmartin,G.M. (1997) Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc. Natl Acad. Sci. USA, 94, 11893–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M., Ohno,M., Yoshida,M. and Mattaj,I.W. (1997) CRM1 is an export receptor for leucine-rich nuclear export signals. Cell, 90, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Frederickson R.M., Montine,K.S. and Sonenberg,N. (1991) Phosphorylation of eukaryotic translation initiation factor 4E is increased in Src-transformed cell lines. Mol. Cell. Biol., 11, 2896–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst T.R., Niles,E.G., Studier,F.W. and Moss,B. (1986) Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl Acad. Sci. USA, 83, 8122–8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore T.D. and Temin,H.M. (1988) v-rel oncoproteins in the nucleus and in the cytoplasm transform chicken spleen cells. J. Virol., 62, 703–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.C., Raught,B. and Sonenberg,N. (1999) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem., 68, 913–963. [DOI] [PubMed] [Google Scholar]
- Goodrum F.D., Shenk,T. and Ornelles,D.A. (1996) Adenovirus early region 4 34-kilodalton protein directs the nuclear localization of the early region 1B 55-kilodalton protein in primate cells. J. Virol., 70, 6323–6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D., Kostka,S., Kraft,R., Dingwall,C., Laskey,R.A., Hartmann,E. and Prehn,S. (1995) Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol., 5, 383–392. [DOI] [PubMed] [Google Scholar]
- Gorlich D., Pante,N., Kutay,U., Aebi,U. and Bischoff,F.R. (1996) Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J., 15, 5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Gradi A., Imataka,H., Svitkin,Y.V., Rom,E., Raught,B., Morino,S. and Sonenberg,N. (1998) A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol., 18, 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F.L. and Van Der Eb,A.J. (1973) A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology, 52, 456–467. [DOI] [PubMed] [Google Scholar]
- Haghighat A., Mader,S., Pause,A. and Sonenberg,N. (1995) Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J., 14, 5701–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighat A., Svitkin,Y., Novoa,I., Kuechler,E., Skern,T. and Sonenberg,N. (1996) The eIF4G–eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J. Virol., 70, 8444–8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm J. and Mattaj,I.W. (1990) Monomethylated cap structures facilitate RNA export from the nucleus. Cell, 63, 109–118. [DOI] [PubMed] [Google Scholar]
- Hart R.P., McDevitt,M.A. and Nevins,J.R. (1985) Poly(A) site cleavage in a HeLa nuclear extract is dependent on downstream sequences. Cell, 43, 677–683. [DOI] [PubMed] [Google Scholar]
- Henkel T., Zabel,U., van Zee,K., Muller,J.M., Fanning,E. and Baeuerle,P.A. (1992) Intramolecular masking of the nuclear location signal and dimerization domain in the precursor for the p50 NF-κB subunit. Cell, 68, 1121–1133. [DOI] [PubMed] [Google Scholar]
- Imataka H. and Sonenberg,N. (1997) Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol., 17, 6940–6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H., Olsen,H.S. and Sonenberg,N. (1997) A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J., 16, 817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E., Stepinski,J., Darzynkiewicz,E. and Mattaj,I.W. (1992) A cap binding protein that may mediate nuclear export of RNA polymerase II-transcribed RNAs. J. Cell Biol., 118, 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E., Lewis,J., McGuigan,C., Jankowska,M., Darzynkiewicz,E. and Mattaj,I.W. (1994) A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell, 78, 657–668. [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Lewis,J., Gamberi,C., Jarmolowski,A., McGuigan,C. and Mattaj,I.W. (1995) A cap-binding protein complex mediating U snRNA export. Nature, 376, 709–712. [DOI] [PubMed] [Google Scholar]
- Jakel S. and Gorlich,D. (1998) Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J., 17, 4491–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel S., Albig,W., Kutay,U., Bischoff,F.R., Schwamborn,K., Doenecke,D. and Gorlich,D. (1999) The importin β/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J., 18, 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans D.A., Briggs,L.J., Gustin,S.E., Jans,P., Ford,S. and Young,I.G. (1997) The cytokine interleukin-5 (IL-5) effects cotransport of its receptor subunits to the nucleus in vitro. FEBS Lett., 410, 368–372. [DOI] [PubMed] [Google Scholar]
- Jarmolowski A., Boelens,W.C., Izaurralde,E. and Mattaj,I.W. (1994) Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol., 124, 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D., Roberts,B.L., Richardson,W.D. and Smith,A.E. (1984) A short amino acid sequence able to specify nuclear location. Cell, 39, 499–509. [DOI] [PubMed] [Google Scholar]
- Keyes L.N. and Spradling,A.C. (1997) The Drosophila gene fs(2)cup interacts with otu to define a cytoplasmic pathway required for the structure and function of germ-line chromosomes. Development, 124, 1419–1431. [DOI] [PubMed] [Google Scholar]
- Klein Gunnewiek J.M., van Aarssen,Y., van der Kemp,A., Nelissen,R., Pruijn,G.J. and van Venrooij,W.J. (1997) Nuclear accumulation of the U1 snRNP-specific protein C is due to diffusion and retention in the nucleus. Exp. Cell Res., 235, 265–273. [DOI] [PubMed] [Google Scholar]
- Konarska M.M., Padgett,R.A. and Sharp,P.A. (1984) Recognition of cap structure in splicing in vitro of mRNA precursors. Cell, 38, 731–736. [DOI] [PubMed] [Google Scholar]
- Kudo N., Wolff,B., Sekimoto,T., Schreiner,E.P., Yoneda,Y., Yanagida,M., Horinouchi,S. and Yoshida,M. (1998) Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res., 242, 540–547. [DOI] [PubMed] [Google Scholar]
- Kumagai A. and Dunphy,W.G. (1999) Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes Dev., 13, 1067–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamphear B.J., Kirchweger,R., Skern,T. and Rhoads,R.E. (1995) Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem., 270, 21975–21983. [DOI] [PubMed] [Google Scholar]
- Lang V., Zanchin,N.I., Lunsdorf,H., Tuite,M. and McCarthy,J.E. (1994) Initiation factor eIF-4E of Saccharomyces cerevisiae. Distribution within the cell, binding to mRNA and consequences of its overproduction. J. Biol. Chem., 269, 6117–6123. [PubMed] [Google Scholar]
- Lazaris-Karatzas A., Montine,K.S. and Sonenberg,N. (1990) Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature, 345, 544–547. [DOI] [PubMed] [Google Scholar]
- Lejbkowicz F., Goyer,C., Darveau,A., Neron,S., Lemieux,R. and Sonenberg,N. (1992) A fraction of the mRNA 5′ cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc. Natl Acad. Sci. USA, 89, 9612–9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard W.J. and O’Shea,J.J. (1998) Jaks and STATs: biological implications. Annu. Rev. Immunol., 16, 293–322. [DOI] [PubMed] [Google Scholar]
- Lewis J.D., Gorlich,D. and Mattaj,I.W. (1996) A yeast cap binding protein complex (yCBC) acts at an early step in pre-mRNA splicing. Nucleic Acids Res., 24, 3332–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Meyer,A.N. and Donoghue,D.J. (1997a) Nuclear localization of cyclin B1 mediates its biological activity and is regulated by phosphorylation. Proc. Natl Acad. Sci. USA, 94, 502–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Leonard,D. and Wilkinson,M.F. (1997b) T cell receptor (TCR) mini-gene mRNA expression regulated by nonsense codons: a nuclear-associated translation-like mechanism [published erratum appears in J. Exp. Med. (1997) 185, 1883]. J. Exp. Med., 185, 985–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Girona A., Furnari,B., Mondesert,O. and Russell,P. (1999) Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature, 397, 172–175. [DOI] [PubMed] [Google Scholar]
- Mader S. and Sonenberg,N. (1995) Cap binding complexes and cellular growth control. Biochimie, 77, 40–44. [DOI] [PubMed] [Google Scholar]
- Mader S., Lee,H., Pause,A. and Sonenberg,N. (1995) The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol. Cell. Biol., 15, 4990–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J., Gingras,A.C., Sonenberg,N. and Burley,S.K. (1997) Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell, 89, 951–961. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J., Gingras,A.C., Sonenberg,N. and Burley,S.K. (1999) Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell, 3, 707–716. [DOI] [PubMed] [Google Scholar]
- Mathews M.B., Sonenberg,N. and Hershey,J.W.B. (1996) Origins and targets of translational control. In Hershey,J.W.B., Mathews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 1–29. [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- Methot N., Rom,E., Olsen,H. and Sonenberg,N. (1997) The human homologue of the yeast Prt1 protein is an integral part of the eukaryotic initiation factor 3 complex and interacts with p170. J. Biol. Chem., 272, 1110–1116. [DOI] [PubMed] [Google Scholar]
- Michael W.M., Eder,P.S. and Dreyfuss,G. (1997) The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J., 16, 3587–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minich W.B., Balasta,M.L., Goss,D.J. and Rhoads,R.E. (1994) Chromatographic resolution of in vivo phosphorylated and nonphosphorylated eukaryotic translation initiation factor eIF-4E: increased cap affinity of the phosphorylated form. Proc. Natl Acad. Sci. USA, 91, 7668–7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.D., Yang,J., Truant,R. and Kornbluth,S. (1999) Nuclear import of Cdk/cyclin complexes: identification of distinct mechanisms for import of Cdk2/cyclin E and Cdc2/cyclin B1. J. Cell Biol., 144, 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.S. (1998) Ran and nuclear transport. J. Biol. Chem., 273, 22857–22860. [DOI] [PubMed] [Google Scholar]
- Mosialos G., Hamer,P., Capobianco,A.J., Laursen,R.A. and Gilmore,T.D. (1991) A protein kinase-A recognition sequence is structurally linked to transformation by p59v-rel and cytoplasmic retention of p68c-rel. Mol. Cell. Biol., 11, 5867–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S. and Dreyfuss,G. (1999) Transport of proteins and RNAs in and out of the nucleus. Cell, 99, 677–690. [DOI] [PubMed] [Google Scholar]
- Patzelt E., Blaas,D. and Kuechler,E. (1983) CAP binding proteins associated with the nucleus. Nucleic Acids Res., 11, 5821–5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A., Belsham,G.J., Gingras,A.C., Donze,O., Lin,T.A., Lawrence,J.C.,Jr and Sonenberg,N. (1994) Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′ cap function. Nature, 371, 762–767. [DOI] [PubMed] [Google Scholar]
- Poulin F., Gingras,A.C., Olsen,H., Chevalier,S. and Sonenberg,N. (1998) 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J. Biol. Chem., 273, 14002–14007. [DOI] [PubMed] [Google Scholar]
- Pyronnet S., Imataka,H., Gingras,A.C., Fukunaga,R., Hunter,T. and Sonenberg,N. (1999) Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J., 18, 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinker-Schaeffer C.W., Graff,J.R., De Benedetti,A., Zimmer,S.G. and Rhoads,R.E. (1993) Decreasing the level of translation initiation factor 4E with antisense RNA causes reversal of ras-mediated transformation and tumorigenesis of cloned rat embryo fibroblasts. Int. J. Cancer, 55, 841–847. [DOI] [PubMed] [Google Scholar]
- Robbins J., Dilworth,S.M., Laskey,R.A. and Dingwall,C. (1991) Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell, 64, 615–623. [DOI] [PubMed] [Google Scholar]
- Rozen F. and Sonenberg,N. (1987) Identification of nuclear cap specific proteins in HeLa cells. Nucleic Acids Res., 15, 6489–6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen,S. and Coulson,A.R. (1977) DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See Y. and Jakowski,G. (1989) Estimating molecular weights of polypeptides by SDS gel electrophoresis. In Creighton,T.E. (ed.), Protein Structure: A Practical Approach. IRL Press, Oxford, UK, pp. 1–21. [Google Scholar]
- Shatkin A.J. (1976) Capping of eucaryotic mRNAs. Cell, 9, 645–653. [DOI] [PubMed] [Google Scholar]
- Siomi H. and Dreyfuss,G. (1995) A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol., 129, 551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N., Rupprecht,K.M., Hecht,S.M. and Shatkin,A.J. (1979) Eukaryotic mRNA cap binding protein: purification by affinity chromatography on sepharose-coupled m7GDP. Proc. Natl Acad. Sci. USA, 76, 4345–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K., Ford,C.S., Guthrie,C. and Weis,K. (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell, 90, 1041–1050. [DOI] [PubMed] [Google Scholar]
- Turpin P., Hay,R.T. and Dargemont,C. (1999) Characterization of IκBα nuclear import pathway. J. Biol. Chem., 274, 6804–6812. [DOI] [PubMed] [Google Scholar]
- Vancurova I., Paine,T.M., Lou,W. and Paine,P.L. (1995) Nucleoplasmin associates with and is phosphorylated by casein kinase II. J. Cell Sci., 108, 779–787. [DOI] [PubMed] [Google Scholar]
- Visa N., Izaurralde,E., Ferreira,J., Daneholt,B. and Mattaj,I.W. (1996) A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J. Cell Biol., 133, 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz A.J., Flynn,A., Proud,C.G. and Cooper,J.A. (1997) Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J., 16, 1909–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz A.J., Johnson,J.C., Penn,B., Mahalingam,M., Kimball,S.R. and Cooper,J.A. (1999) Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol. Cell. Biol., 19, 1871–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W., Meinkoth,J.L., Tsien,R.Y. and Taylor,S.S. (1995) Identification of a signal for rapid export of proteins from the nucleus. Cell, 82, 463–473. [DOI] [PubMed] [Google Scholar]
- Zhu J., Shibasaki,F., Price,R., Guillemot,J.C., Yano,T., Dotsch,V., Wagner,G., Ferrara,P. and McKeon,F. (1998) Intramolecular masking of nuclear import signal on NF-AT4 by casein kinase I and MEKK1. Cell, 93, 851–861. [DOI] [PubMed] [Google Scholar]