Abstract
Despite 14-3-3 proteins being implicated in the control of the eukaryotic cell cycle, metabolism, cell signalling and survival, little is known about the global regulation or functions of the phosphorylation-dependent binding of 14-3-3s to diverse target proteins. We identified Arabidopsis cytosolic proteins that bound 14-3-3s in competition with a 14-3-3-binding phosphopeptide, including nitrate reductase, glyceraldehyde- 3-phosphate dehydrogenase, a calcium-dependent protein kinase, sucrose-phosphate synthase (SPS) and glutamyl-tRNA synthetase. Remarkably, in cells starved of sugars or fed with non-metabolizable glucose analogues, all 14-3-3 binding was lost and the target proteins were selectively cleaved into proteolytic fragments. 14-3-3 binding reappeared after several hours of re-feeding with sugars. Starvation-induced degradation was blocked by 5-amino imidazole-4-carboxamide riboside (which is converted to an AMP-mimetic) or the protease inhibitor MG132 (Cbz-leu-leu-leucinal). Extracts of sugar-starved (but not sugar-fed) Arabidopsis cells contained an ATP-independent, MG132-sensitive, neutral protease that cleaved Arabidopsis SPS, and the mammalian 14-3-3-regulated transcription factor, FKHR. Cleavage of SPS and phosphorylated FKHR in vitro was blocked by binding to 14-3-3s. The finding that 14-3-3s participate in a nutrient-sensing pathway controlling cleavage of many targets may underlie the effects of these proteins on plant development.
Keywords: Arabidopsis/protein phosphorylation/proteolysis/sugar-sensing/14-3-3 proteins
Introduction
14-3-3 proteins are an unusually highly conserved protein family with central regulatory roles in plant, fungal and mammalian cells (Aitken, 1996). 14-3-3s bind to phosphopeptide motifs in diverse target proteins (Muslin et al., 1996; Yaffe et al., 1997; Moorhead et al., 1999), and through these interactions, 14-3-3s have been reported to modulate the activities of enzymes (Toroser et al., 1998; Moorhead et al., 1999), regulate subcellular location of targets (Brunet et al., 1999; Lopez-Girona et al., 1999; May and Soll, 2000) and act as ‘adaptor proteins’ that mediate interactions between components of signal transduction pathways (for example, Van Der Hoeven et al., 2000).
In plants, 14-3-3s have been discovered primarily as regulators of enzymes of cytosolic metabolism and ion pumps (Baunsgaard et al., 1998; Toroser et al., 1998; Moorhead et al., 1999), while in mammalian cells they are best known as regulators of many components of oncogenic signalling pathways (Aitken, 1996; Van Der Hoeven et al., 2000) and of cell cycle checkpoints (Lopez-Girona et al., 1999). Perhaps this apparent disparity between the two Kingdoms is an artificial bias due to the different emphases of molecular studies in plant and mammalian cells. Indeed, it is striking that for the well-defined cases, 14-3-3 binding to individual plant and mammalian targets contributes in different ways to cell protection and survival (MacKintosh, 1998; Xing et al., 2000). For example, by binding to and inhibiting phosphorylated plant nitrate reductase (NR) in leaves in the dark (Bachmann et al., 1996; Moorhead et al., 1996), 14-3-3s ensure that toxic nitrite is not produced under conditions where it cannot be reduced further; while binding of 14-3-3s to the mammalian protein BAD prevents BAD from binding to Bcl-XL and thus blocks apoptosis (Zha et al., 1996). However, although 14-3-3s are repeatedly being implicated in the regulation of defined points that control cell growth, metabolism and development, little is known of how the global phosphorylation-dependent interactions of diverse targets with 14-3-3s are actually regulated within a single cell.
In order to tackle the question of global control by 14-3-3s, we have developed a combination of sensitive affinity-binding procedures and reagents (Moorhead et al., 1999). Here, we have used these methods to identify 14-3-3-regulated proteins in Arabidopsis cell extracts, and to monitor how their phosphorylation and 14-3-3 binding status change in response to hormones and nutrients. We discovered that sugar starvation has a dramatic effect on the interactions of 14-3-3s with all their target proteins in the cells.
Sugar starvation is a normal part of a plant’s lifestyle. The supply of sugars generated by photosynthesis is not continuous and stops partially or completely in the dark, during winter and when leaves are shed. Starvation is in fact one of the essential signals for plant development, and ‘sugar-sensing’ pathways regulate processes from germination through photomorphogenesis, flowering and senescence (Dijkwel et al., 1997; Pego et al., 1999). For example, sugars inhibit the red-light-triggered developmental transition that converts a starving etiolated seedling into a vegetative plant that is fully capable of photosynthesis (Dijkwel et al., 1997). Many plant genes whose expression is up- or down-regulated by sugars have been identified (Koch, 1996), and studies of Arabidopsis mutants that are altered in their sugar-sensing responses are starting to give clues about the relevant signal transduction pathways (Smeekens, 1998; Zhou et al., 1998). Some effects of sugar feeding can be mimicked by sugar analogues that can be phosphorylated by hexokinase, but not by sugars that are not substrates for this enzyme (Jang et al., 1997; Zhou et al., 1998). However, whether hexokinase acts as a ‘sugar receptor’ in addition to performing its metabolic function is controversial (Jang et al., 1997; Wiese et al., 1999). Other ideas about plant sugar-sensing come largely from analogies with sugar-sensing pathways in yeasts and other organisms.
In contrast to these uncertainties, here we demonstrate a sugar-sensing pathway in a plant cell culture system, with a clear role for 14-3-3s in controlling the cleavage of their diverse targets involved in plant metabolism and signalling.
Results
Identification of 14-3-3-binding proteins in Arabidopsis cell extracts
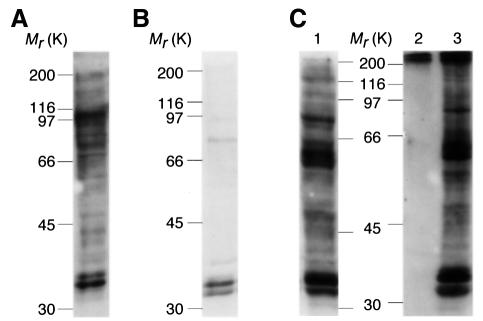
Digoxygenin (DIG)-14-3-3 overlays (Moorhead et al., 1999) of extracts of Arabidopsis cells grown in 10–90 mM sucrose or 20 mM glucose revealed many proteins (Figures 1A and 3A, lanes 2 and 6) whose binding to 14-3-3 proteins was blocked by competition with a 14-3-3-binding phosphopeptide (Muslin et al., 1996; Yaffe et al., 1997; Moorhead et al., 1999) (Figure 1B). The proteins detected on the overlays were co-immunoprecipitated with an anti-14-3-3 antibody, confirming that the native proteins bind to endogenous 14-3-3s in Arabidopsis cell extracts (Figure 1C). 14-3-3-binding proteins were purified by specific phosphopeptide elution from a 14-3-3 affinity column (Moorhead et al., 1999), and identified by comparing MALDI-TOF profiles of tryptic digests with the sequence databases (Figure 2). These analyses indicated that there was a high probability that the purified proteins included the cytosolic proteins NR [already known to be inhibited by 14-3-3 proteins (Bachmann et al., 1996; Moorhead et al., 1996)], glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and glutamyl-tRNA synthetase (GluRS) (Figure 2 and Table I). The previously identified plant 14-3-3-binding proteins, sucrose-phosphate synthase (SPS) and a calcium-dependent protein kinase (CDPK) capable of phosphorylating the 14-3-3-binding site (Ser543) of NR (Toroser et al., 1998; Moorhead et al., 1999), were also identified in the phosphopeptide elution pool from the 14-3-3 column by western blotting and activity (Moorhead et al., 1999; data not shown).
Fig. 1. DIG-14-3-3-binding proteins identified in extracts of Arabidopsis cells. (A) DIG-14-3-3 overlay of crude extract from mid-log-phase cells grown on standard medium containing sucrose, auxin and cytokinin. (B) DIG-14-3-3 overlay of a duplicate of the blot shown in (A) where the DIG-14-3-3s had been pre-incubated with a synthetic phosphopeptide carrying the canonical 14-3-3-binding sequence, ARAApSAPA (1 mM). (C) DIG-14-3-3 overlay of crude extract from cells grown on standard medium (lane 1), immunoprecipitate with pre-immune IgG (lane 2) or anti-14-3-3 immunoprecipitate of crude extract (lane 3). Note that the two proteins of ∼30 kDa whose binding to DIG-14-3-3s is not blocked by the phosphopeptide (B) are Arabidopsis 14-3-3s that can form dimers with the DIG-14-3-3 probe (not shown).
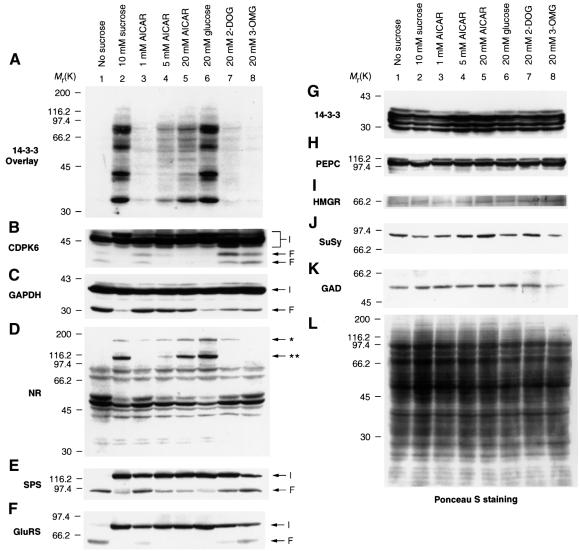
Fig. 3. Extracts of Arabidopsis cells assayed by DIG-14-3-3 overlay and western blotting. (A) DIG-14-3-3 overlay of extracts (40 µg) of Arabidopsis cells 24 h after transfer from a standard medium to a medium with no sugar (lane 1); 10 mM sucrose (lane 2); no sugar + 1 mM AICAR (lane 3); no sugar + 5 mM AICAR (lane 4); no sugar + 20 mM AICAR (lane 5); 20 mM glucose (lane 6); 20 mM 2-deoxyglucose (lane 7); 20 mM 3-O-methyl glucose (lane 8). (B–K) Western blots of the same samples as in (A) but probed with an anti-CDPK6 antibody (B), anti-GAPDH (C), anti-NR (D), anti-SPS (E), anti-GluRS (F), anti-14-3-3 (G), anti-PEPC (H), anti-HMGR (I), anti-SuSy (J), anti-GAD (K). Arrows labelled I indicate the positions of intact proteins; arrows labelled F show positions of proteolytic fragments. (L) Ponceau S protein stain of the same samples. Similar results were seen in experiments with three different sets of cells. In (D) the double asterisk indicates intact NR and the single asterisk an unknown protein that is detected by the anti-NR antibody.
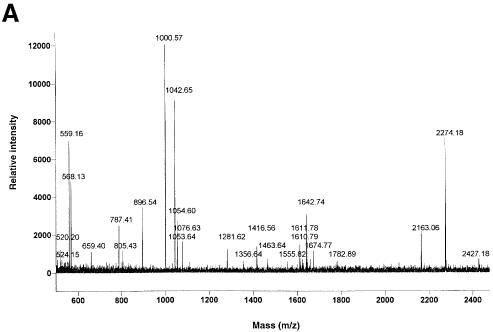
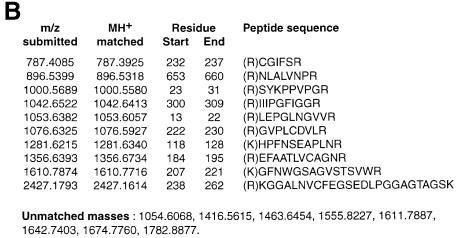
Fig. 2. Example of MALDI-TOF analysis of proteins eluted by phosphopeptide from a 14-3-3 column. A protein band of ∼100 kDa was cut from a blot of purified proteins (a repeat of the experiment shown in Table I), digested with trypsin and subjected to MALDI-TOF analysis (Materials and methods). (A) MALDI-TOF mass spectrum of the tryptic digest. (B) The spectrum shown in (A) was compared with the sequence databases using the MS-fit program (http://falcon.ludwig.ucl.ac.uk/mshome3.2.htm). The best fit for the data, with 55% of peptide masses finding a match, was Arabidopsis NR (NCBI accession No. 128188). All matched peptides had an Arg or Lys immediately N-terminal to the matched sequences in the intact protein, as expected for a tryptic digest.
Table I. Criteria for identifying proteins purified by 14-3-3 affinity chromatography.
| Name | Accession No. (NCBI) | Molecular mass predicted/actual in kDa | % masses matcheda | Protein coincided with a protein that binds 14-3-3s on overlays |
|---|---|---|---|---|
| Nitrate reductase (NR) | 128188 | 102.8/109 | 46% (18/39) | ? |
| 102.8/105 | 40% (23/57) | yes | ||
| 102.8/101 | 58% (14/24) | yes | ||
| Glutamyl-tRNA synthetase (GluRS) | 3435196 | 81/82 | 40% (4/10) | yes |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | 120667 | 37/39 | 22% (6/27) | yes |
aMatched peptides were restricted to those preceded by R or K as expected for a tryptic digest.
Selective loss of 14-3-3-binding signals in extracts of sugar-starved cells
The DIG-14-3-3 overlay assay was used to monitor changes in the global 14-3-3-binding status of target proteins. Extracts of cells harvested at different densities or grown under different hormone regimes showed changes in the relative intensities of particular DIG-14-3-3-binding proteins (data not shown). Most striking, however, was the complete disappearance of 14-3-3-binding protein bands in extracts of cells that were starved of sugars (Figure 3A), or grown in the presence of the non-metabolized sugars 3-O-methyl glucose (Figure 3A, lane 8) or mannitol (not shown) for 24 h. The sugar 2-deoxyglucose, which is a substrate for hexokinase but not metabolized further, had only a very marginal effect in preventing the loss of 14-3-3 binding (Figure 3A, lane 7). 14-3-3 binding bands were recovered when cells were re-fed with sucrose for several hours (data not shown). The samples were well-matched by general protein staining (Figure 3L). These results indicated that metabolism of sugars was essential for the binding of 14-3-3s to their target proteins and prompted us to test the effects of drugs that affect energy metabolism. 5-aminoimidazole-4-carboxamide riboside (AICAR), a drug that is phosphorylated by adenosine kinase to give the AMP-mimetic ZMP, largely blocked the starvation-induced loss of 14-3-3-binding protein bands (Figure 3A, compare lane 1 with lanes 3, 4 and 5).
As expected for a nitrate-inducible enzyme, NR activity was undetectable in cells starved of nitrogen for 24 h. However, in contrast to the global effect of sugar starvation, nitrogen starvation affected the intensity of 14-3-3 binding to only a few proteins in 14-3-3 overlay assays (Figure 4, compare lanes 1 and 3).
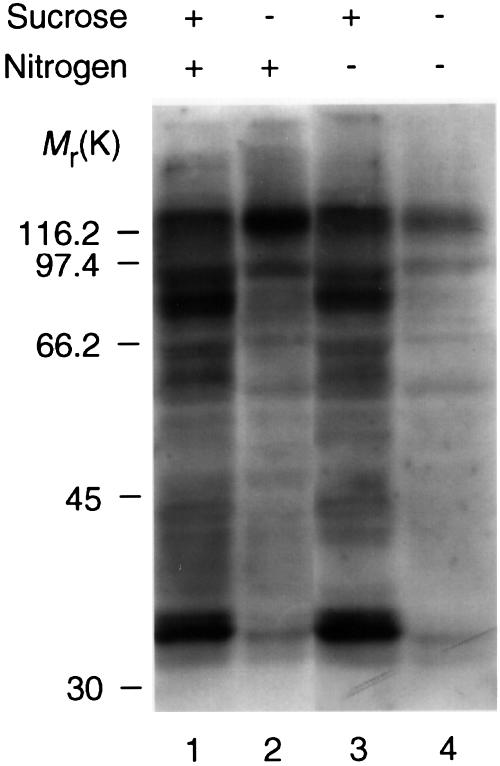
Fig. 4. Extracts of Arabidopsis cells assayed by DIG-14-3-3 overlay: comparison of the effects of nitrogen and sugar starvation. DIG-14-3-3 overlay of extracts (45 µg) of Arabidopsis cells 24 h after transfer from a standard medium to one with 10 mM sucrose (lane 1) or no sucrose (lane 2) and extracts of cells 24 h after transfer to a medium containing 10 mM sucrose and no nitrogen (lane 3) or no sucrose and no nitrogen (lane 4). See Materials and methods for media compositions.
Loss of 14-3-3 binding coincides with sugar-induced cleavage of target proteins
14-3-3 proteins generally bind to phosphorylated sites on their targets (Muslin et al., 1996; Yaffe et al., 1997; Moorhead et al., 1999), suggesting that the complete lack of interactions in sugar-starved cells may have been caused by dephosphorylation of the target proteins. However, western blotting revealed that the loss of 14-3-3-binding proteins in the overlays of sugar-starved cell extracts coincided with the selective degradation of proteins that had been bound to 14-3-3s in fed-cell extracts (Figure 3). Moreover, the ability of AICAR to preserve the 14-3-3 binding capability of target proteins in starved cells was mirrored by the protection of 14-3-3-binding targets from degradation (Figure 3). In every case investigated, degradation products could be seen in lanes where the intact protein was depleted. For example, an antibody that recognizes three CDPK bands showed that a CDPK of 52 kDa was depleted, and faster-migrating immunoreactive bands were present on the blots, in extracts of cells grown in the absence of metabolizable sugars or AICAR (Figure 3B). GAPDH is commonly assayed as a control abundant and ‘constitutively expressed’ protein, and indeed the amount of intact GAPDH protein looked similar in all samples (Figure 3C). However, a GAPDH degradation product of ∼30 kDa was observed in extracts where 14-3-3 binding was absent (Figure 3C), consistent with the existence of a sub-population that is degraded in starved cells. Similarly, the intact NR, SPS and GluRS proteins were depleted and distinct degradation products of these proteins were seen in sugar-starved cell extracts (Figure 3D, E and F). Degradation fragments were also observed when the cells were extracted directly into boiling SDS sample buffer, indicating that degradation had occurred in vivo, and not during the preparation of cell extracts. While in Figure 3 there appears to be some variation in the sensitivities of different target proteins to AICAR, 2-deoxyglucose and 3-O-methyl glucose, examination of blots with different exposure times to developing reagents for several different experiments suggests that these apparent differences may simply reflect the normal limitations of western blotting for quantitative analysis.
Consistent with a loss of intact protein, sugar-starved cell extracts contained <5% [0.115 ± 0.087 (for four experiments) nmol/min/mg] of the total NR activity present in sugar-fed cell extracts [3.2 ± 0.6(4) nmol/min/mg]. However, the specific activity of SPS was similar in all extracts [3.8 ± 0.3(4) nmol/min/mg for sugar-fed extracts and 3.5 ± 0.4(4) nmol/min/mg for sugar-starved extracts], suggesting that the large SPS fragment in starved cell extracts is active, although it may have altered kinetic properties (Sonnewald et al., 1993).
‘Control’ proteins are not degraded in sugar-starved cells
In contrast to the 14-3-3-binding partners, the abundance of total 14-3-3s was similar in all extracts (Figure 3G). Of several other protein ‘controls’ that are regulated by phosphorylation but not known to bind 14-3-3s, one form of phosphoenolpyruvate carboxylase (PEPC) (Nimmo et al., 1990) was actually less abundant in sucrose-fed cell extracts compared with cells that had been fed with glucose or non-metabolizable sugars (Figure 3H), indicating that PEPC protein displays regulation by sugars but by a signalling mechanism that is distinct from those that operate on the 14-3-3-binding targets. The relative abundance of HMGCoA reductase (HMGR) (Dale et al., 1995; Douglas et al., 1997) (Figure 3I), sucrose synthase (SuSy; Figure 3J) and glutamate decarboxylase (GAD; Figure 3K) also varied in a different way from the 14-3-3-binding proteins.
Sugar-starved cells contain a protease whose activity towards its substrates is blocked by their binding to 14-3-3s
The results presented above did not reveal whether or not the proteins were bound to 14-3-3s at the actual moment of degradation. 14-3-3 proteins might protect their binding partners from degradation, target them for degradation, or neither. These results also raised questions regarding the identity of the relevant protease and whether it is constitutively active or is itself regulated by sugars. In order to address these questions, Arabidopsis cell extracts were assayed for in vitro proteolysis of 14-3-3-binding partners. Extracts from starved cells were prepared that still contained some intact SPS and, when incubated at 30°C, the intact SPS was degraded (Figure 5A, compare lanes 1 and 2), and degradation was blocked by the protease inhibitor MG132 (Figure 5A, lane 3). Added 14-3-3 proteins also protected SPS from degradation (Figure 5A, lane 4), while a phosphopeptide containing the canonical 14-3-3-binding sequence (ARAA phosphoSAPA) enhanced the rate of SPS proteolysis by the starved-cell protease, indicating that direct binding of 14-3-3s was protecting this protein from proteolysis (Figure 5A, lane 5). In contrast to the starved cell extract, SPS in a fed cell extract was not degraded upon incubation of the extract at 30°C, even in the presence of 14-3-3-binding phosphopeptide (Figure 5B, lanes 3 and 4). Furthermore, mixing experiments showed that the extract of sugar-starved cells did not degrade the 14-3-3-binding proteins in sugar-fed cell extracts in the presence or absence of a synthetic 14-3-3-binding phosphopeptide (to block binding of 14-3-3s to their targets) (data not shown). The starved cell extracts, therefore, appeared to contain a protease that was absent and/or blocked by an interfering factor in the sugar-fed cell extract. However, an alternative possibility was that even after dissociating 14-3-3 proteins from SPS with the phosphopeptide, the SPS in the fed-cell extracts had been modified in some way that made it inaccessible to proteolysis. Therefore, in order to check whether the protease was only active in sugar-starved cells, it was necessary to use an exogenous substrate.
Fig. 5. In vitro degradation of Arabidopsis SPS in sugar-starved cell extracts by an MG132-inhibited protease and enhanced proteolysis stimulated by a 14-3-3-binding phosphopeptide. (A) A sugar-starved extract (25 µg) that still contained some intact 14-3-3-binding proteins was incubated at 30°C for 15 min in a total volume of 20 µl, and the products were analysed by western blotting with anti-SPS. No incubation (lane 1); incubation with no additions (lane 2); incubation with 50 µM MG132 (lane 3); incubation with 10 µg of 14-3-3s (lane 4); incubation with 10 µg of 14-3-3s and 1 mM 14-3-3-binding phosphopeptide ARAApSAPA (lane 5). (B) A sugar-starved extract (25 µg) (lanes 1 and 2) and a sugar-fed extract (12.5 µg) (lanes 3 and 4) were incubated at 30°C for 10 min in a total volume of 20 µl and the products analysed by western blotting with anti-SPS. No additions (lanes 1 and 3); incubation with 1 mM 14-3-3-binding phosphopeptide (lanes 2 and 4).
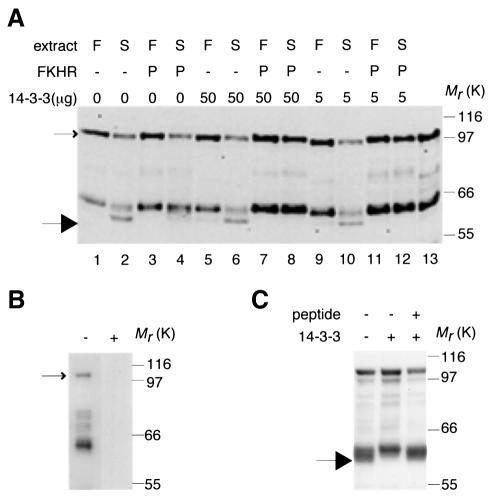
We found that the plant protease also cleaved a mammalian 14-3-3-binding protein, namely the forkhead transcription factor, FKHR (Brunet et al., 1999) (Figure 6). Unphosphorylated, bacterially-expressed glutathione S-transferase (GST)–FKHR was cleaved to a distinct fragment (large arrow in Figure 6) by starved plant cell extracts in the presence or absence of added 14-3-3s. Extracts of sugar-fed cells contained little or no FKHR protease activity (Figure 6A). FKHR that was phosphorylated with protein kinase B (PKB) (Brunet et al., 1999; Rena et al., 1999) and capable of binding to 14-3-3s in overlay assays (Figure 6B) was slightly protected from degradation (Figure 6A, lane 4), presumably due to binding to Arabidopsis 14-3-3s in the extract, and degradation of phosphorylated FKHR was completely blocked by adding 14-3-3 proteins before adding the extracts (Figure 6A, lanes 8 and 12). 14-3-3s had no effect on degradation of unphosphorylated FKHR (Figure 6A, lanes 6 and 10). Both binding of 14-3-3s to FKHR (Figure 6B) and the ability of 14-3-3s to protect phosphorylated FKHR from degradation by the starved-cell extract (Figure 6C) were blocked by the synthetic 14-3-3-binding peptide, ARAAphosphoSAPA.
Fig. 6. Phosphorylation and 14-3-3 binding protects GST–FKHR from degradation by a protease present in extracts from sugar-starved Arabidopsis cells. GST–FKHR was expressed in Escherichia coli and purified on glutathione-Sepharose. GST–FKHR was detected by western blotting with anti-GST. Intact GST–FKHR is indicated by a small arrow. The large arrow indicates the position of a new FKHR cleavage product that was generated during the incubation, and that runs below a fragment of FKHR that was present in the original preparation (lane 13). (A) Unphosphorylated FKHR (–) or FKHR phosphorylated with PKB (P) (2 µg of each in 25 µl) was incubated with starved (S) or fed (F) cell extract (10 µg of each) in the absence (0) or presence of 14-3-3 (50 µg in lanes 5–8, or 5 µg in lanes 9–12). Note that phosphorylation by PKB was stoichiometric as witnessed by the slight upwards band-shift of intact FKHR. (B) 14-3-3-binding phosphopeptide blocks binding of DIG-14-3-3s to FKHR. Blots of phosphorylated GST–FKHR were probed with DIG-14-3-3s alone (–), or DIG-14-3-3s pre-incubated with 1 mM 14-3-3-binding phosphopeptide, ARAApSAPA (+). (C) 14-3-3-binding phosphopeptide blocks the ability of 14-3-3 to prevent FKHR degradation. Phosphorylated GST–FKHR was incubated with protease from starved cell extracts in the presence (+) or absence (–) of 14-3-3s (25 µg), with (+) or without (–) 1 mM 14-3-3-binding phosphopeptide, ARAApSAPA.
The protease is blocked by MG132 in vitro and in vivo
The starved-cell protease was blocked completely by the cysteine protease inhibitors MG132, E64 and leupeptin, while phenylmethylsulfonyl fluoride (PMSF), EDTA, pepstatin, calpastatin CS peptide and clasto-lactacystin β-lactone had no effect (data not shown). This protease is, therefore, distinct from a previously-identified, PMSF-inhibited NR protease (Weiner and Kaiser, 1999). The protease identified here was ATP-independent, which rules out the proteasome, and was not active below pH 6.5, suggesting that it is a cytosolic, rather than a lysosomal enzyme. Consistent with the MG132-sensitive SPS- and FKHR-protease being responsible for the cleavage of Arabidopsis 14-3-3-binding proteins in starved cells, the in vivo proteolysis of NR, SPS, CDPK and GAPDH (Figure 7C–F) was blocked by MG132. Intriguingly, MG132 also promoted 14-3-3 binding of the intact proteins in vivo (Figure 7B), suggesting that proteolysis may in turn control the phosphorylation of 14-3-3-binding sites.
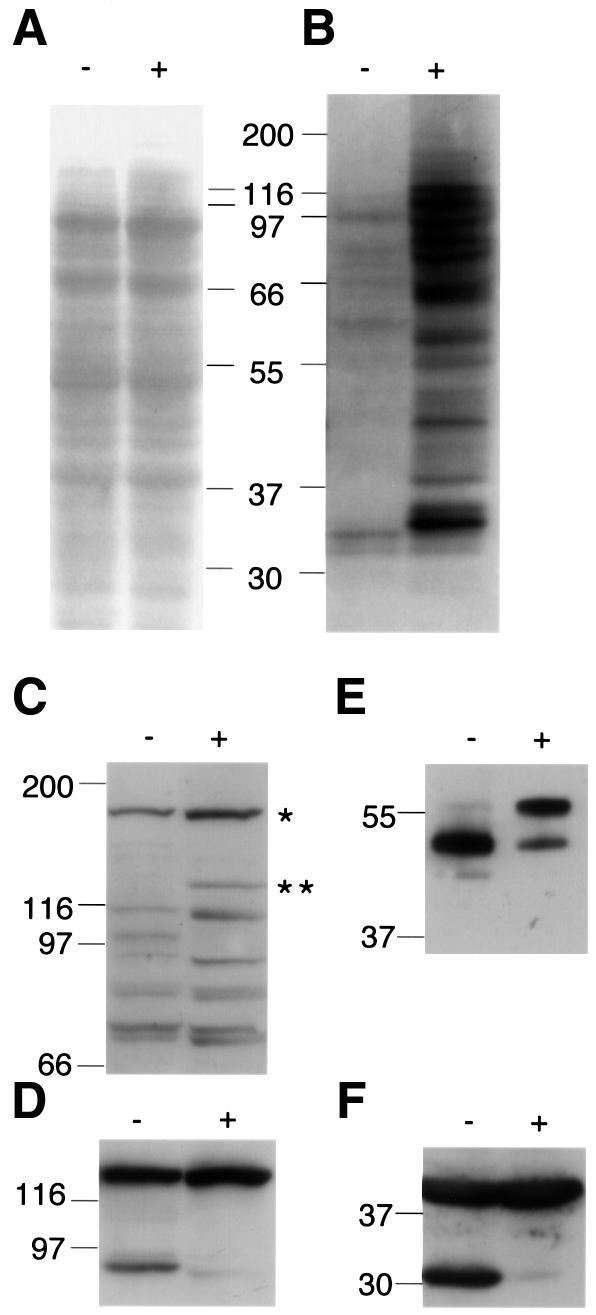
Fig. 7. MG132 blocks the cleavage of 14-3-3-binding proteins and prevents the loss of their ability to bind to DIG-14-3-3s in response to starvation in intact cells. (A) Ponceau S protein stain of blot of extracts (45 µg of each) of cells that had been starved of sugars for 24 h in the presence (+) or absence (–) of the protease inhibitor, MG132 (100 µM). Parallel blots were probed with DIG-14-3-3s (B); anti-NR antibody (C); anti-SPS (D); anti-CDPK6 (E); and anti-GAPDH (F). In (C), the double asterisk indicates intact NR and the single asterisk is an unknown protein that is detected by the anti-NR antibody.
Discussion
In this study, using sensitive, specific binding procedures we have distinguished ∼30 Arabidopsis cell proteins that bind 14-3-3s in competition with 14-3-3-binding phosphopeptides (Figure 1; Moorhead et al., 1999), and whose phosphorylation and 14-3-3 binding status changes in response to hormones (S.E.M.Meek and C.MacKintosh, unpublished results) and nutrients (Figure 3). The real strength of the affinity-binding approach is that 14-3-3-binding proteins that are regulated by extracellular stimuli can be purified after elution with phosphopeptides from a 14-3-3 affinity column, and identified by mass spectrometry and amino acid sequencing. Among the new targets that have been defined here are the cytosolic-specific isoforms of enzymes of plant metabolism and signalling. The discovery that 14-3-3s interact with several cytosolic enzymes in primary nitrogen and carbon assimilation and protein synthesis (Moorhead et al., 1999; this paper) suggests that 14-3-3s have the potential for a major impact on metabolite partitioning in plants. Arabidopsis proteins purified by 14-3-3 affinity chromatography, but whose specificity for binding to 14-3-3s is still being tested, include further cytosolic enzymes of metabolism, vesicle trafficking and protein kinases (V.Cotelle and C.MacKintosh, unpublished results).
While we have observed changes in the 14-3-3-binding status of individual target proteins in response to hormones and growth factors in both plant and mammalian cells (S.E.M.Meek, J.Harthill and C.MacKintosh, unpublished results), the most dramatic event occurred in response to sugar starvation in Arabidopsis plant cells. In sugar-fed cells, many proteins were phosphorylated (Moorhead et al., 1999) and bound to 14-3-3 proteins (Figures 1 and 3). In sugar-starved cells, however, all 14-3-3 binding was lost, and the target proteins that we identified were all cleaved to give distinct proteolytic cleavage fragments. A protease whose properties are consistent with it being the enzyme responsible for the cleavage events seen in vivo was identified in extracts of sugar-starved cells but not in extracts of sugar-fed cells. This protease was completely inactive below pH 6.5, which is suggestive of a cytosolic enzyme rather than a lysosomal or vacuolar protease. It is well-known that starvation of eukaryotic cells triggers a non-selective autophagic process that delivers cellular proteins and organelles to lysosomes for complete degradation and recycling (Aubert et al., 1996; Klionsky and Ohsumi, 1999). Autophagy is also involved in remodelling intracellular structures for cell differentiation. The starvation-induced, selective and limited cleavage of 14-3-3-binding proteins by a neutral protease is clearly not autophagy. However, it is possible that there is a mechanistic link between the two processes. Perhaps cleavage of key 14-3-3-binding partners in the cytoplasm acts as a trigger to initiate autophagy.
Because of the diversity of plant 14-3-3-binding target proteins, we would expect numerous cellular processes to be altered depending on the phosphorylation, the 14-3-3-binding status, and the state of cleavage of these targets. We propose that the dramatic and selective cleavage of 14-3-3-regulated proteins in sugar-starved plant cells explains the major shifts in metabolism, including reduced nitrate assimilation and sugar synthesis, which have been detailed in many physiological studies of sugar-starved plants (Koch, 1996; Matt et al., 1998; Morcuende et al., 1998). Cleavage of 14-3-3-binding partners may not necessarily lead to loss of function. Starved-cell extracts that contained the 95 kDa SPS cleavage fragment, but very little detectable intact SPS, contained just as much SPS activity as fed-cell extracts, although it is possible that the regulatory properties may be altered. In contrast, NR activity in starved-cell extracts was very low.
14-3-3 binding blocked proteolysis of target proteins in vitro (Figures 5 and 6). Presumably, therefore, 14-3-3s must be dissociated from their targets before proteolysis occurs in vivo. Dissociation of 14-3-3–target complexes might be triggered by starvation-induced dephosphorylation of the target proteins (we showed previously that plant proteins that have been dephosphorylated cannot bind to 14-3-3 proteins; Moorhead et al., 1999). Another possibility is that the 14-3-3s themselves are modified in sugar-starved cells to lower their affinity for target binding. If the latter 14-3-3-centred mechanism for dissociation were correct, we would have to propose that the target proteins and their fragments in sugar-starved cells subsequently lost their phosphorylated binding motifs to render them incapable of binding 14-3-3s in overlays (Figure 3). In this scenario, possible mechanisms include dephosphorylation, loss of the binding site by cleavage or an inhibitory phosphorylation close to the 14-3-3-binding site on the target proteins.
While the precise mechanisms that render the target proteins susceptible to proteolysis are not clear, other findings may have provided two clues. First, AICAR, a drug that is phosphorylated by adenosine kinase to give the AMP-mimetic ZMP, largely blocked the starvation-induced loss of 14-3-3-binding protein bands (Figure 3A, compare lane 1 with lanes 3, 4 and 5). While it does seem counterintuitive that AICAR/AMP should mimic the effects of feeding, this finding suggests that AMP blocks some critical aspect of the starvation response. In mammalian cells, AICAR activates several enzymes, including the AMP-activated protein kinase (AMPK) (Corton et al., 1995). AMP also activates the homologous plant kinase, SnRK1, by inhibiting its dephosphorylation (Sugden et al., 1999). Moreover, an SnRK1-like protein kinase phosphorylates NR (Douglas et al., 1997) and other 14-3-3-binding proteins (C.MacKintosh, unpublished results), and activation of an SnRK1-like protein kinase by sugars has been reported in plants (Bhalerao et al., 1999). One hypothesis, therefore, is that AICAR blocks loss of 14-3-3 binding (Figure 3A) by activating a plant SnRK1 leading to the phosphorylation of plant proteins that can then bind 14-3-3s. Secondly, the intriguing observation that the protease inhibitor MG132 also promoted 14-3-3 binding of the intact proteins in vivo (Figure 7B) suggests that proteolysis may in turn control the phosphorylation of 14-3-3-binding sites. This is perhaps not surprising since at least one Arabidopsis protein kinase, a CDPK, binds 14-3-3s and the degradation of a CDPK in starved cells is blocked by MG132 (Figure 7E). However, an alternative possibility is that the starvation-induced protease cleaves off the 14-3-3-binding phosphorylation sites from the target proteins. There may, therefore, be a complex regulatory relationship between control of the phosphorylation and cleavage status of 14-3-3-regulated proteins.
Consistent with previous observations that plant cells can survive several days of sucrose starvation (Aubert et al., 1996), the population of cells used in our studies survived starvation for 24 h, and responded to re-feeding with sucrose. At the whole-plant level, overexpression of 14-3-3 proteins has been found to promote phenotypes associated with a general enhancement of cell survival. For example, transgenic potatoes overexpressing 14-3-3s display a prolonged stay-green phenotype with delayed leaf senescence (Markiewicz et al., 1996), and antisense 14-3-3 plants show early senescence (Wilczynski et al., 1998). It is tempting to speculate that our finding that 14-3-3s protect their targets from proteolysis may underlie the phenotypes of these transgenic plants.
Materials and methods
Cell culture
Arabidopsis suspension cells were routinely cultured in standard media containing sucrose (90 mM), auxin and cytokinin, as described by Moorhead et al. (1999). The standard medium contained (for 1 l) 4.43 g of Murashige and Skoog basal salts with minimal organics (MSMO from Sigma), 30 g of sucrose, 1-naphthaleneacetic acid (0.5 mg) and kinetin (0.05 mg). The nitrogen sources in the standard medium were ammonium nitrate (1650 mg/l) and potassium nitrate (1900 mg/l). For nitrogen starvation experiments, these two components were omitted, and other salts and vitamins were mixed individually to the MSMO recipe supplied by Sigma.
Peptides, antibodies and assays
Peptides were synthesized and purified by Graham Bloomberg (University of Bristol, Bristol, UK) using N-9-fluorenylmethoxycarbonyl chemistry on an Applied Biosystems 430A peptide synthesizer. Antibodies against Arabidopsis cytosolic GluRS were raised in sheep against the synthetic peptide KPIVLFSIPDGRAA that was linked using glutaraldehyde to a mixture of BSA and keyhole limpet haemocyanin (10 mg of each protein and 5 mg of peptide). The antibody was affinity purified on a column of peptide immobilized to activated CH-Sepharose (Pharmacia Biotech Europe, Belgium) according to the manufacturer’s instructions. Assays of NR (in the presence of EDTA) and SPS (under Vmax conditions) were as described by Moorhead et al. (1999).
14-3-3 overlays, 14-3-3 affinity chromatography and western blotting
Cell-free extract preparation, 14-3-3 proteins, DIG-14-3-3 overlays, 14-3-3 affinity chromatography, immunoprecipitation and western blotting with anti-spinach 14-3-3 antibodies were as described by Moorhead et al. (1999). To check whether purified proteins bound to DIG-14-3-3s, overlays could also be performed on Fluorotrans membranes that had been stained with sulforhodamine B (see below) and destained in 30% methanol in 0.1 M ammonium bicarbonate.
Identification of 14-3-3-binding proteins
Proteins were electroblotted from SDS gels to PVM (Fluorotrans) membranes (0.2 µm; Pall Europe Limited, Portsmouth, UK), dried under vacuum and stained with 0.005% (w/v) sulforhodamine B, 30% (v/v) methanol and 0.2% (v/v) acetic acid. Protein bands were excised, and the stain removed by washing twice in 50% (v/v) methanol in 50 mM ammonium bicarbonate, and once in 50 mM ammonium bicarbonate. Protein was digested overnight at 30°C on a shaking platform with 50 ng/µl trypsin in 20 mM ammonium bicarbonate in 1% (w/v) octyl glucoside (∼5 µl per band). The supernatant was kept and membrane washed with shaking at 30°C in 5 µl of 50% (v/v) acetonitrile, 0.1% (v/v) trifluoroacetic acid. Both supernatants were combined and masses of peptides were determined by MALDI-TOF spectrometry on a PerSeptive Biosystems Voyager DE STR system. The masses of intact protein and tryptic peptides (with Arg or Lys immediately N-terminal to the matched sequence) were compared with the sequence databases using the MS-fit and MS-digest programs (http://falcon.ludwig.ucl.ac.uk/mshome3.2.htm). The Fluorotrans membrane was chosen after preliminary trials showed that while Immobilon or nitrocellulose membranes were better at binding proteins during blotting, Fluorotrans had the advantage of releasing a better yield of tryptic peptides.
Reproducibility
With the exception of mass spectrometry, results shown are typical of at least two, and more usually three or four separate experiments.
Acknowledgments
Acknowledgements
We are grateful to Graham Rena, Chris Armstrong, Andrew Paterson and Sue Hall of the MRC Unit for purified FKHR and active PKB, and to the following people for antibodies: Karen Koch, University of Gainesville, FL, for antibodies that recognize both sucrose synthase proteins from maize (SUS1 and SH1); Cathrine Lillo, Stavanger College, Norway, for anti-NR; Mark Stitt, University of Heidelberg, for anti-SPS; Albert Ferrer, University of Barcelona, Spain, for anti-HMGR; Hugh Nimmo, University of Glasgow, UK, for anti-PEPC; Ming-Che Shih, University of Iowa, IA, for anti-GAPDH; Hillel Fromm, Weizmann Institute of Science, Rehovot, Israel, for anti-GAD. We thank Steve Millam, Scottish Crop Research Institute, for help in setting up the cell culture. This work was supported by the European Community (B104 CT97 2231), the UK BBSRC and UK MRC.
References
- Aitken A. (1996) 14-3-3 and its possible role in co-ordinating multiple signalling pathways. Trends Cell Biol., 6, 341–347. [DOI] [PubMed] [Google Scholar]
- Aubert S., Gout,E., Bligny,R., Marty-Mazars,D., Barrieu,F., Alabouvette,J., Marty,F. and Douce,R. (1996) Ultrastructural and biochemical characterization of autophagy in higher plant cells subjected to carbon deprivation: control by the supply of mitochondria with respiratory substrates. J. Cell Biol., 133, 1251–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann M., Huber,J.L., Liao,P.C., Gage,D.A. and Huber,S.C. (1996) The inhibitor protein of phosphorylated nitrate reductase from spinach (Spinacia oleracea) leaves is a 14-3-3 protein. FEBS Lett., 387, 127–131. [DOI] [PubMed] [Google Scholar]
- Baunsgaard L., Fuglsang,A.T., Jahn,T., Korthout,H.A., de Boer,A.H. and Palmgren,M.G. (1998) The 14-3-3 proteins associate with the plant plasma membrane H+-ATPase to generate a fusicoccin binding complex and a fusicoccin responsive system. Plant J., 13, 661–671. [DOI] [PubMed] [Google Scholar]
- Bhalerao R.P., Salchert,K., Bako,L., Okresz,L., Szabados,L., Muranaka,T., Machida,Y., Schell,J. and Koncz,C. (1999) Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc. Natl Acad. Sci. USA, 96, 5322–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A. et al. (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell, 96, 857–868. [DOI] [PubMed] [Google Scholar]
- Corton J.M., Gillespie,J.G., Hawley,S.A. and Hardie,D.G. (1995) 5-aminoimidazole-4-carboxamide ribonucleoside. A specific method for activating AMP-activated protein kinase in intact cells? Eur. J. Biochem., 229, 558–565. [DOI] [PubMed] [Google Scholar]
- Dale S., Arro,M., Becerra,B., Morrice,N.G., Boronat,A., Hardie,D.G. and Ferrer,A. (1995) Bacterial expression of the catalytic domain of 3-hydroxy-3-methylglutaryl-CoA reductase (isoform HMGR1) from Arabidopsis thaliana, and its inactivation by phosphorylation at Ser577 by Brassica oleracea 3-hydroxy-3-methylglutaryl-CoA reductase kinase. Eur. J. Biochem., 233, 506–513. [DOI] [PubMed] [Google Scholar]
- Dijkwel P.P., Huijser,C., Weisbeek,P.J., Chua,N.H. and Smeekens,S.C. (1997) Sucrose control of phytochrome A signaling in Arabidopsis. Plant Cell, 9, 583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P., Pigaglio,E., Ferrer,A., Halford,N.G. and MacKintosh,C. (1997) Three spinach leaf nitrate reductase-3-hydroxy-3-methylglutaryl-CoA reductase kinases that are regulated by reversible phosphorylation and/or Ca2+ ions. Biochem. J., 325, 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J.C., Leon,P., Zhou,L. and Sheen,J. (1997) Hexokinase as a sugar sensor in higher plants. Plant Cell, 9, 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J. and Ohsumi,Y. (1999) Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol., 15, 1–32. [DOI] [PubMed] [Google Scholar]
- Koch K.E. (1996) Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol., 47, 509–540. [DOI] [PubMed] [Google Scholar]
- Lopez-Girona A., Furnari,B., Mondesert,O. and Russell,P. (1999) Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature, 397, 172–175. [DOI] [PubMed] [Google Scholar]
- MacKintosh C. (1998) Regulation of cytosolic enzymes in primary metabolism by reversible protein phosphorylation. Curr. Opin. Plant Biol., 1, 224–229. [DOI] [PubMed] [Google Scholar]
- Markiewicz E., Wilczynski,G., Rzepecki,R., Kulma,A. and Szopa,J. (1996) The 14-3-3 protein binds to the nuclear matrix endonuclease and has a possible function in the control of plant senescence. Cell. Mol. Biol. Lett., 1, 391–415. [Google Scholar]
- Matt P., Schurr,U., Klein,D., Krapp,A. and Stitt,M. (1998) Growth of tobacco in short-day conditions leads to high starch, low sugars, altered diurnal changes in the Nia transcript and low nitrate reductase activity, and inhibition of amino acid synthesis. Planta, 207, 27–41. [DOI] [PubMed] [Google Scholar]
- May T. and Soll,J. (2000) 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell, 12, 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead G., Douglas,P., Morrice,N., Scarabel,M., Aitken,A. and MacKintosh,C. (1996) Phosphorylated nitrate reductase from spinach leaves is inhibited by 14-3-3 proteins and activated by fusicoccin. Curr. Biol., 6, 1104–1113. [DOI] [PubMed] [Google Scholar]
- Moorhead G. et al. (1999) Phosphorylation-dependent interactions between enzymes of plant metabolism and 14-3-3 proteins. Plant J., 18, 1–12. [DOI] [PubMed] [Google Scholar]
- Morcuende R., Krapp,A., Hurry,V. and Stitt,M. (1998) Sucrose-feeding leads to increased rates of nitrate assimilation, increased rates of α-oxoglutarate synthesis, and increased synthesis of a wide spectrum of amino acids in tobacco leaves. Planta, 206, 394–409. [Google Scholar]
- Muslin A.J., Tanner,J.W., Allen,P.M. and Shaw,A.S. (1996) Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell, 84, 889–897. [DOI] [PubMed] [Google Scholar]
- Nimmo H.G., Carter,P.J., Fewson,C.A., McNaughton,G.A., Nimmo,G.A. and Wilkins,M.B. (1990) Regulation of phosphoenolpyruvate carboxylase: an example of signal transduction via protein phosphorylation in higher plants. Adv. Enzyme Regul., 30, 121–131. [DOI] [PubMed] [Google Scholar]
- Pego J.V., Weisbeek,P.J. and Smeekens,S.C. (1999) Mannose inhibits Arabidopsis germination via a hexokinase-mediated step. Plant Physiol., 119, 1017–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rena G., Guo,S., Cichy,S.C., Unterman,T.G. and Cohen,P. (1999) Phosphorylation of the transcription factor forkhead family member FKHR by protein kinase B. J. Biol. Chem., 274, 17179–17183. [DOI] [PubMed] [Google Scholar]
- Smeekens S. (1998) Sugar regulation of gene expression in plants. Curr. Opin. Plant Biol., 1, 230–234. [DOI] [PubMed] [Google Scholar]
- Sonnewald U., Quick,W.P., MacRae,E., Krause,K.P. and Stitt,M. (1993) Purification, cloning and expression of spinach leaf sucrose-phosphate synthase in Escherichia coli. Planta, 189, 174–181. [DOI] [PubMed] [Google Scholar]
- Sugden C., Crawford,R.M., Halford,N.G. and Hardie,D.G. (1999) Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5′-AMP. Plant J., 19, 433–439. [DOI] [PubMed] [Google Scholar]
- Toroser D., Athwal,G.S. and Huber,S.C. (1998) Site-specific regulatory interaction between spinach leaf sucrose-phosphate synthase and 14-3-3 proteins. FEBS Lett., 435, 110–114. [DOI] [PubMed] [Google Scholar]
- Van Der Hoeven P.C., Van Der Wal,J.C., Ruurs,P., Van Dijk,M.C. and Van Blitterswijk,J. (2000) 14-3-3 isotypes facilitate coupling of protein kinase C-ζ to Raf-1: negative regulation by 14-3-3 phosphorylation. Biochem. J., 345, 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner H. and Kaiser,W.M. (1999) 14-3-3 proteins control proteolysis of nitrate reductase in spinach leaves. FEBS Lett., 455, 75–78. [DOI] [PubMed] [Google Scholar]
- Wiese A., Groner,F., Sonnewald,U., Deppner,H., Lerchl,J., Hebbeker,U., Flugge,U. and Weber,A. (1999) Spinach hexokinase I is located in the outer envelope membrane of plastids. FEBS Lett., 461, 13–18. [DOI] [PubMed] [Google Scholar]
- Wilczynski G., Kulma,A. and Szopa,J. (1998) The expression of 14-3-3 isoforms in potato is developmentally regulated. J. Plant Physiol., 153, 118–126. [Google Scholar]
- Xing H., Zhang,S., Weinheimer,C., Kovacs,A. and Muslin,A.J. (2000) 14-3-3 proteins block apoptosis and differentially regulate MAPK cascades. EMBO J., 19, 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe M.B., Rittinger,K., Volinia,S., Caron,P.R., Aitken,A., Leffers,H., Gamblin,S.J., Smerdon,S.J. and Cantley,L.C. (1997) The structural basis for 14-3-3:phosphopeptide binding specificity. Cell, 91, 961–971. [DOI] [PubMed] [Google Scholar]
- Zha J., Harada,H., Yang,E., Jockel,J. and Korsmeyer,S.J. (1996) Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-XL. Cell, 87, 619–628. [DOI] [PubMed] [Google Scholar]
- Zhou L., Jang,J.C., Jones,T.L. and Sheen,J. (1998) Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc. Natl Acad. Sci. USA, 95, 10294–10299. [DOI] [PMC free article] [PubMed] [Google Scholar]