Abstract
The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η (pol η), which is involved in the replication of damaged DNA. Pol η catalyzes efficient and accurate translesion synthesis past cis-syn cyclobutane di-thymine lesions. Here we show that human pol η can catalyze translesion synthesis past an abasic (AP) site analog, N-2-acetylaminofluorene (AAF)-modified guanine, and a cisplatin-induced intrastrand cross-link between two guanines. Pol η preferentially incorporated dAMP and dGMP opposite AP, and dCMP opposite AAF-G and cisplatin-GG, but other nucleotides were also incorporated opposite these lesions. However, after incorporating an incorrect nucleotide opposite a lesion, pol η could not continue chain elongation. In contrast, after incorporating the correct nucleotide opposite a lesion, pol η could continue chain elongation, whereas pol α could not. Thus, the fidelity of translesion synthesis by human pol η relies not only on the ability of this enzyme to incorporate the correct nucleotide opposite a lesion, but also on its ability to elongate only DNA chains that have a correctly incorporated nucleotide opposite a lesion.
Keywords: DNA polymerase η/translesion synthesis/xeroderma pigmentosum variant (XP-V)
Introduction
A myriad of lesions in DNA are caused by ubiquitous environmental and endogenous genotoxic agents. These lesions can interfere with normal DNA metabolism including DNA replication, eventually resulting in mutations that lead to carcinogenesis and/or cell death. To maintain the integrity of the genetic material, cells possess multiple pathways to repair various types of DNA damage, such as nucleotide excision and base excision repair pathways (Friedberg et al., 1995). However, not all lesions on the genome can be repaired efficiently by these processes in time for DNA replication, and some types of lesion are repaired very inefficiently. To prevent acute cell death through arrested DNA replication at unrepaired lesions, cells have a mechanism, referred to as translesion synthesis, which allows DNA synthesis to proceed past lesions. Recently, a novel family of DNA polymerases that can catalyze translesion synthesis was identified (Cordonnier and Fuchs, 1999; Friedberg and Gerlach, 1999; Johnson et al., 1999c; Woodgate, 1999). Genetic evidence revealed that there are two subpathways of mutagenic and relatively accurate translesion syntheses. In Escherichia coli, two DNA polymerases named Pol IV (dinB) (Wagner et al., 1999) and Pol V (umuD′2C) (Reuven et al., 1999; Tang et al., 1999) were identified. The products of these two genes are involved in mutagenesis, suggesting that these polymerases catalyze error-prone translesion synthesis. In the yeast Saccharomyces cerevisiae, Rev3 was identified as DNA polymerase ζ (Nelson et al., 1996b). Genetic studies also indicated that this gene took part in the mutagenic pathway. The human counterparts of dinB (Gerlach et al., 1999; Ogi et al., 1999) and REV3 (Gibbs et al., 1998; Lin et al., 1999) have also been identified and found to have mutagenic properties.
In addition to these mutagenic DNA polymerases, human and yeast cells have another DNA polymerase named pol η. Human pol η is the product of the XPV gene, which is mutated in a cancer-prone genetic disorder, xeroderma pigmentosum variant (XP-V) (Johnson et al., 1999a; Masutani et al., 1999b). XP-V cells are defective in the replication of damaged DNA (Lehmann et al., 1975) and show hypermutability after exposure to UV irradiation or DNA-damaging agents (Maher et al., 1976; Wang et al., 1991, 1993; Misra and Vos, 1993; Waters et al., 1993; Raha et al., 1996). These in vivo observations imply that few errors are introduced when pol η carries out translesion synthesis inside the cell. Mutations in the RAD30 gene of S.cerevisiae inactivate the accurate translesion pathway (McDonald et al., 1997; Roush et al., 1998), indicating that the RAD30 gene product, yeast pol η, is also involved in this pathway. These observations are well explained by the fact that both human and yeast pol η can catalyze translesion synthesis past cis-syn di-thymine dimers by incorporating dAMP efficiently (Johnson et al., 1999b; Masutani et al., 1999a,b). Paradoxically, we have recently found that DNA synthesis by pol η using undamaged templates in vitro was highly error-prone (Matsuda et al., 2000). In order to try and resolve this paradox, we have examined the substrate range and mechanisms of translesion synthesis of the XPV gene product, human pol η.
Results
Processivity of human DNA polymerase η
To examine the biological activities of human pol η, a recombinant protein tagged with His6 at its C-terminal side was expressed in insect cells and purified as described in Materials and methods. The histidine-tagged protein corrected the defective translesion DNA synthesis in XP-V cell extracts, and as such is biologically active (Masutani et al., 1999b). The processivity of the recombinant pol η was examined on singly primed phagemids under conditions of excess molar amounts of template–primer over enzyme. From a labeled primer of 20 nucleotides, pol η elongated DNA chains by 1–8 nucleotides as shown by enzyme titration and time course experiments (Figure 1). Thus, human pol η is a distributive polymerase like other translesion polymerases such as E.coli pol IV (dinB) (Wagner et al., 1999), pol V (umuD′2C) (Reuven et al., 1999; Tang et al., 1999) and yeast pol η (Washington et al., 1999), suggesting that the role of human pol η is restricted to incorporating a few nucleotides opposite and/or past a lesion. Under the same conditions, pol α elongated DNA chains by 1–11 nucleotides, in good agreement with previous data (Copeland and Wang, 1991).
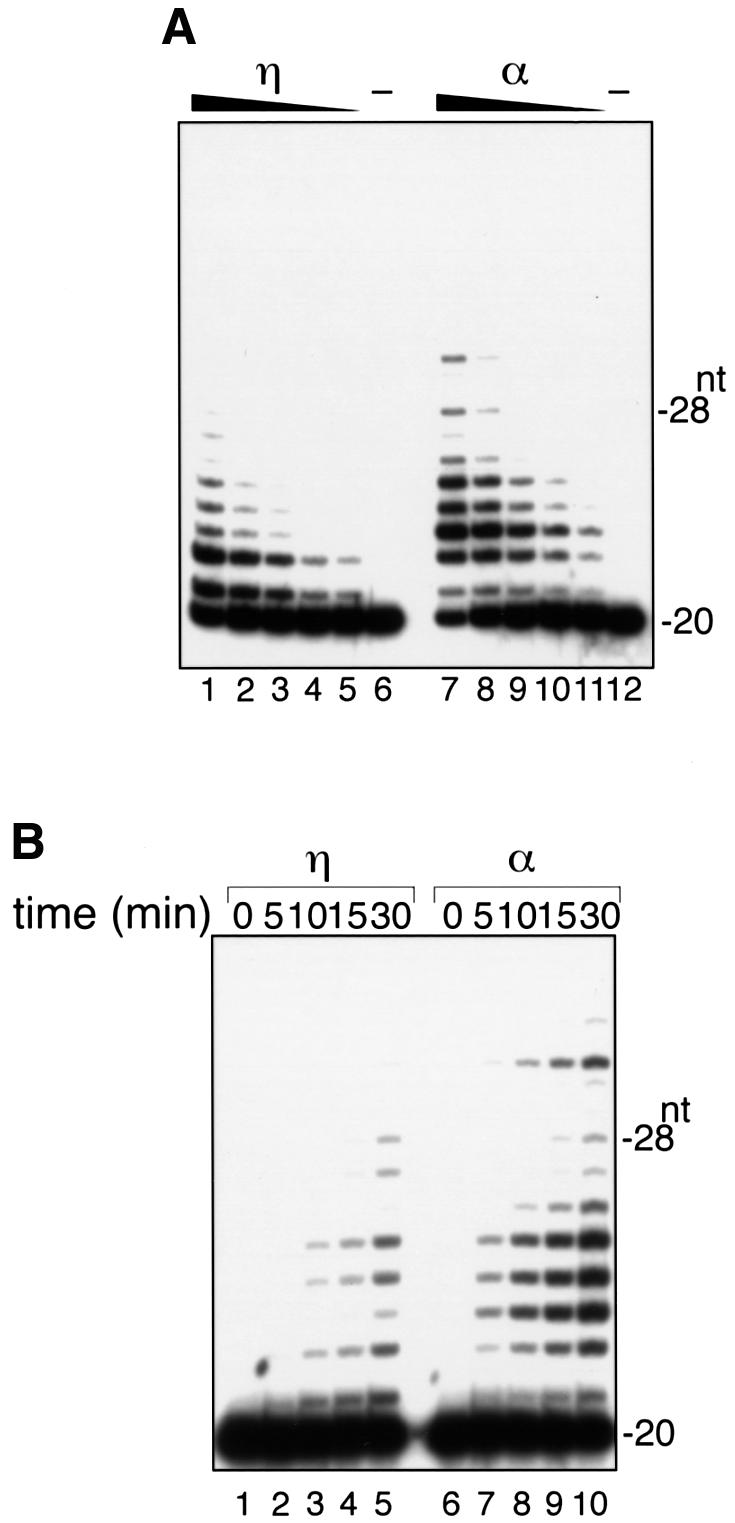
Fig. 1. Processivity of human pol η. (A) Enzyme titration. Decreasing amounts of pol η (3.2, 1.6, 0.8, 0.4 and 0.2 fmol in lanes 1, 2, 3, 4 and 5, respectively) or pol α (0.3, 0.15, 0.07, 0.04 and 0.02 fmol in lanes 7, 8, 9, 10 and 11, respectively) were incubated with 500 fmol of the single-stranded phagemid template annealed to 5′-32P-labeled primer for 15 min in the reaction mixture. Lanes 6 and 12 contained no enzyme. The products were subjected to polyacrylamide gel electrophoresis under denaturing conditions. An autoradiogram of the gel is shown. (B) Time course. Pol η (0.8 fmol in lanes 1–5) or pol α (0.15 fmol in lanes 6–10) was incubated with 500 fmol of 5′-32P- labeled primer–templates for the indicated time periods. An autoradiogram of the gel is shown.
Translesion synthesis by human DNA polymerase η
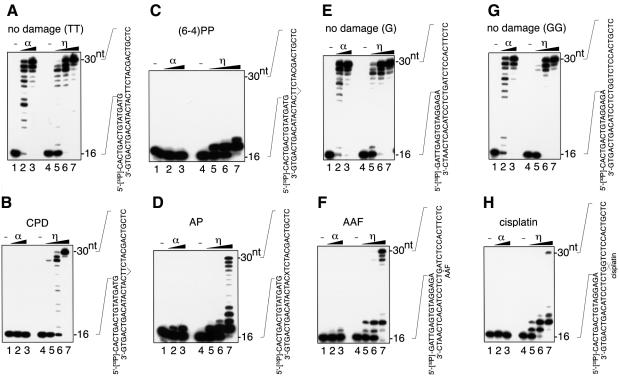
To examine the translesion activity of human pol η, a 30mer DNA oligomer containing a single lesion was synthesized, annealed to a 5′-32P-labeled 16mer DNA oligomer and used as template (Figure 2). The primer on this template was positioned so that the first nucleotide was always incorporated opposite an abasic (AP) site analog or an N-2-acetylaminofluorene (AAF)-modified guanine. On templates containing a cis-syn cyclobutane pyrimidine dimer (CPD) or the (6–4) photoproduct, cis-diamminedichloroplatinum(II) (cisplatin), which induces an intrastrand cross-link between two guanines, the positioning of the primer ensured that the first two nucleotides were incorporated opposite these lesions. DNA polymerase α and pol η could synthesize DNA products of up to 30mer in length equally well on the undamaged template, if enough enzyme was added (Figure 2A, E and G). As reported previously (Masutani et al., 1999a,b), pol η, but not pol α, was able to bypass the CPD on the template efficiently (Figure 2B). When the DNA template contained a (6–4) photoproduct, pol η incorporated one nucleotide opposite the first thymine and another nucleotide opposite the second thymine of the lesion in reactions containing excess enzyme, but bypass product was hardly detected (Figure 2C). Long exposure of the gel revealed that pol η gave a small amount of bypass products on the template but the reaction was much less efficient than that on any other templates used in this study (data not shown). Pol α did not incorporate any nucleotide opposite the (6–4) photoproduct. On the AP template, which contained a stable analog of the abasic site, pol α was able to incorporate one nucleotide opposite the lesion, but could not continue DNA synthesis. Pol η was also inhibited, and stopped after incorporating one nucleotide opposite the lesion, but bypass products could be obtained by adding excess amounts of the enzyme (Figure 2D). Similar results were obtained with templates containing an AAF-modified guanine and an intrastrand cross-link between two guanines created by cisplatin (Figure 2F and H). On both templates, pol α hardly incorporated any nucleotide opposite the lesions, and no bypass products were detected even in the presence of excess enzyme. In contrast, pol η gave bypass products on the AAF-G template, but DNA synthesis stopped mainly after the incorporation of two nucleotides, the second of which was incorporated opposite the A next to the lesion (Figure 2F). On the cisplatin-GG template, pol η also gave some bypass products, although most products contained only one nucleotide opposite the first G of the cross-link and especially the second nucleotide opposite the second G of the lesion (Figure 2H). From these results, we concluded that human pol η had the ability to bypass the AP analogs, AAF-G and cisplatin-GG. However, it must be mentioned that while the human pol η incorporated nucleotides opposite these lesions efficiently, only a small proportion of the incorporated nucleotides were elongated, even in the presence of an excess amount of pol η (Figure 2D, F and H, lanes 7). On the other hand, low but detectable amounts of elongated products were observed when limiting amounts of enzyme were added to the reactions (Figure 2D, F and H, lanes 5 and 6), although the products were <30 nucleotides in length, perhaps reflecting the low processivity of pol η. These results suggest that some of the lesions were bypassed in one processive reaction by pol η, but that others were never bypassed by the polymerase.
Fig. 2. Translesion synthesis by human pol η. Increasing amounts of pol α (4.7 and 23.6 fmol in lanes 2 and 3, respectively) or pol η (0.25, 1.4 and 7.1 fmol in lanes 5, 6 and 7, respectively) were incubated with the 5′-32P-labeled primer–templates indicated beside each panel at 37°C for 15 min in the standard reaction mixture. Lanes 1 and 4 contained no enzyme. The products were subjected to polyacrylamide gel electrophoresis under denaturing conditions. The autoradiograms of the gels are shown. (A) Undamaged control for CPD and (6–4) photoproduct [(6–4)PP]. (B) CPD at the bridged TT. (C) The (6–4) photoproduct at the bridged TT. (D) The abasic analog at X. (E) Undamaged control for AAF. (F) AAF-modified guanine at the indicated site. (G) Undamaged control for cisplatin. (H) Intrastrand cross-link of two guanines by cisplatin at the indicated site.
Nucleotide selectivity of human DNA polymerase η during incorporation opposite lesions
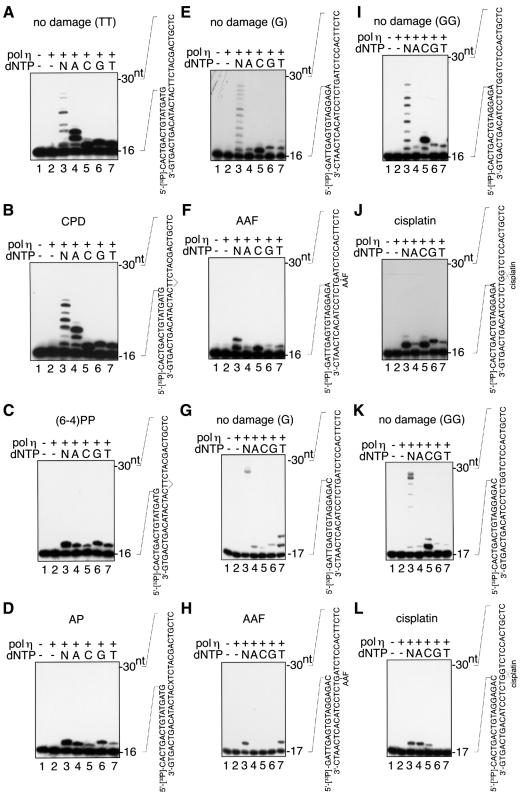
To examine which kind of nucleotide could be incorporated opposite a lesion by pol η, polymerization reactions were performed in the presence of only one kind of deoxyribonucleotide (Figure 3). For these experiments, limiting amounts of pol η were used to facilitate the identification of the most efficiently incorporated nucleotide. As noted in our previous reports (Masutani et al., 1999a,b), human pol η has deoxynucleotidyl transferase activity and stops synthesis after incorporating an incorrect nucleotide even on undamaged templates (Figure 3A, E, G, I and K). This activity was more obvious when T was the template nucleotide. Under these conditions, dAMP dCMP, dGMP and dTMP were almost equally incorporated opposite T (Figure 3A), suggesting that the pol η tends to misincorporate nucleotides opposite T preferentially. The misincorporation observed in reactions containing only one kind of nucleotide would be suppressed in the presence of all four nucleotides. These observations are consistent with our recent finding that human pol η replicates undamaged DNA with low fidelity, especially when the sequence on the template nucleotide is T (Matsuda et al., 2000). On a template containing the cis-syn cyclobutane di-thymine dimer, pol η only bypassed the lesion when dAMP was incorporated as the first and second nucleotide. DNA synthesis stopped after the incorporation of another dAMP opposite the C next to the lesion (Figure 3B). As well as on undamaged template, pol η incorporated dCMP, dGMP or dTMP opposite the first T of the lesion in the reactions where only one nucleotide was included. On the template containing the (6–4) photoproduct, which pol η hardly bypasses, all four nucleotides were incorporated opposite the first thymine of the lesion, but synthesis stopped thereafter (Figure 3C).
Fig. 3. Nucleotide selectivity of pol η incorporation opposite lesions. Pol η (0.25 fmol in lanes 2–7) was incubated with 30mer DNA annealed to a 5′-32P-labeled 16mer (A–F, I and J) or 17mer (G, H, K and L) DNA in the presence of all four dNTPs (lane 3) or with one of the indicated dNTPs (lanes 4–7), or in the absence of dNTPs (lane 2). The autoradiograms of the gels are shown. (A) Undamaged control for the CPD and (6–4)PP templates with a 16mer primer. (B) CPD at the bridged TT with a 16mer primer. (C) The (6–4) photoproduct at the bridged TT with a 16mer primer. (D) The AP analog at X with a 16mer primer. (E) Undamaged control for the AAF-G template with a 16mer primer. (F) AAF-G at the indicated site with a 16mer primer. (G) Undamaged control for AAF-G template with a 17mer primer. (H) AAF-G at the indicated site with a 17mer primer. (I) Undamaged control for the cisplatin-GG template with a 16mer primer. (J) Cisplatin-GG at the indicated site with a 16mer primer. (K) Undamaged control for the cisplatin-GG template with a 17mer primer. (L) Cisplatin-GG at the indicated site with a 17mer primer.
Similar experiments were performed with AP-, AAF-G- and cisplatin-GG-containing templates. On the AP template, the polymerase preferentially incorporated dAMP or dGMP opposite the lesion (Figure 3D). However, hardly any nucleotide was incorporated opposite T next to the lesion, even in the presence of dATP, by the enzyme present in limiting amounts. This observation is in agreement with the result that pol η mainly stopped DNA synthesis after incorporating one nucleotide opposite the AP analog site (Figure 2D). Human pol η preferentially incorporated dCMP opposite AAF-G, although the other three nucleotides were also incorporated to some extent (Figure 3F). As human pol η mainly stopped synthesis after incorporating a nucleotide opposite the A next to AAF-G (Figure 2F), we examined which nucleotide was incorporated opposite the A on this template. In contrast to reactions containing undamaged templates, where all four nucleotides were incorporated (Figure 3G), pol η preferentially incorporated dTMP opposite A on the AAF-modified template (Figure 3H). Preferential incorporation of dCMP was observed opposite the first G of cisplatin-GG, but it was not the only nucleotide incorporated (Figure 3J). On this template, the second dCMP was hardly incorporated opposite the second G of the cross-link, whereas two dCMPs were incorporated opposite GG on the undamaged template (Figure 3I). In fact, dAMP was incorporated more efficiently than dCMP using this template (Figure 3L). However, we found that dCMP was incorporated preferentially opposite the second G of cisplatin-GG in a different sequence (Vaisman et al., 2000), suggesting that nucleotide selection by pol η is influenced by the sequence context of the lesion. From these results, we concluded that although pol η could incorporate the correct nucleotides preferentially opposite the above lesions, the misincorporation rate was much higher than that expected of the XPV gene product in vivo, which carries out relatively accurate translesion synthesis (Maher et al., 1976; Wang et al., 1991, 1993).
Ability of human DNA polymerase η to elongate DNA chains past lesions
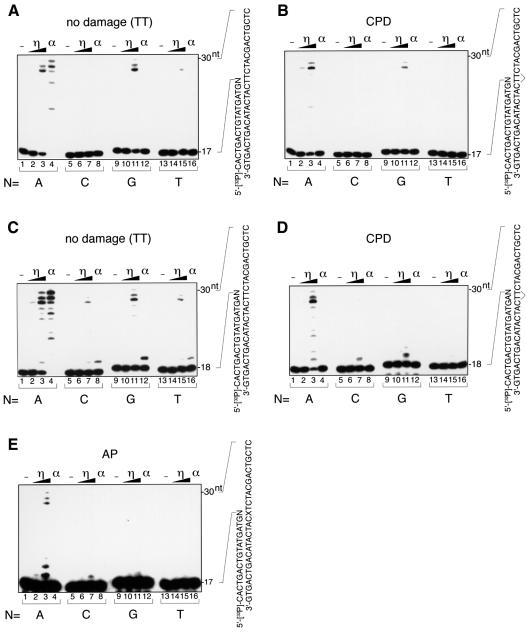
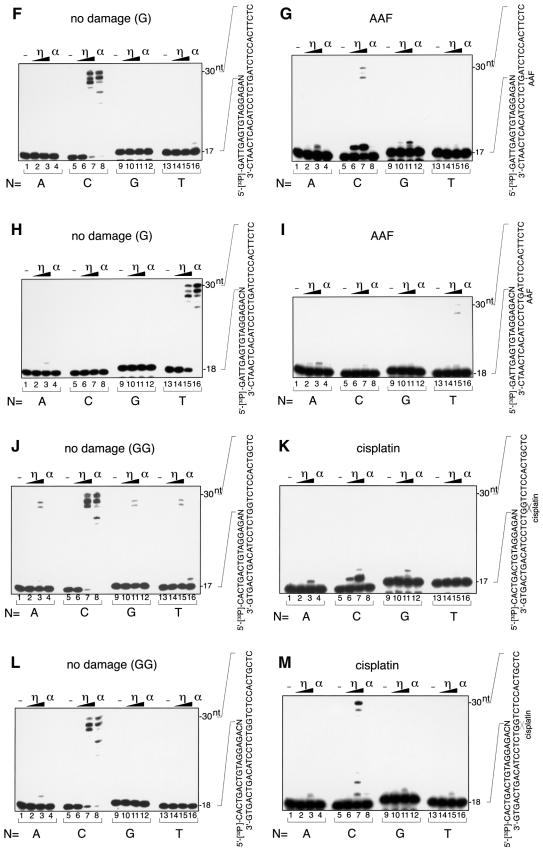
For lesion bypass, it is important that nucleotides are incorporated beyond the lesion after the initial incorporation of a nucleotide opposite the lesion. To test whether human pol η had this activity, we designed sets of 17mer and 18mer primers that had different sequences at their 3′ ends and examined which sequence allowed pol η to elongate past the lesion (Figure 4). The annealing of these primers to a 30mer DNA template containing a lesion placed the 3′ nucleotide of the primer opposite the lesion. When the template contained no lesion, pol η and pol α could elongate DNA more efficiently from a paired primer terminus than from a mispaired primer terminus (Figure 4A, C, F, H, J and L). However, pol η could synthesize DNA chains from a mispaired primer terminus when excess amounts of enzyme were added to the reaction. In particular, G–T mismatches at the primer terminus were elongated more efficiently than other mismatches (Figure 4A and C). When the template contained the CPD, pol η could synthesize DNA chains more efficiently from primers that had the sequence AA opposite the two thymines of the dimer (Figure 4B and D). Pol η could also elongate DNA chains from mispaired primers on the CPD template, and especially from primer termini having G opposite the dimer. However, the elongation of mispaired primers was less efficient on the CPD template than on undamaged templates (compare Figure 4B and D with A and C). As DNA synthesis by pol η was inhibited when the template contained AP, AAF-G or cisplatin-GG, the amounts of enzyme were increased 2-fold in experiments with templates containing these lesions. As for AP, pol η could elongate DNA from primers more efficiently when A rather than G was located opposite the lesion (Figure 4E). On the AAF-G template, pol η could continue DNA synthesis much more efficiently when the nucleotide opposite the lesion was C, although most of the incorporation stopped after the incorporation of one more nucleotide (Figure 4G). This is consistent with the major arrest of DNA synthesis that occurred after the incorporation of a nucleotide opposite A next to the lesion (Figure 2F). We also performed the experiment against the A next to AAF-G on the template. In this case, the primer with T opposite A was elongated, but other primers were not (Figure 4I). When the template contained cisplatin-GG, the primer containing C opposite the first G of the cross-link was used efficiently for DNA synthesis by pol η, but most of the incorporation stopped after the insertion of another nucleotide opposite the second G of the lesion (Figure 4K). This is also in agreement with the results shown in Figure 2H, which indicated that DNA synthesis mainly stopped after the incorporation of a nucleotide opposite the second G of the cross-link. Although dAMP was incorporated preferentially opposite the second G (Figure 3L), only the primer containing C opposite the second G of the cross-link was used efficiently to elongate DNA (Figure 4M). Thus, translesion synthesis past cisplatin-GG occurred only when two dCMPs were incorporated opposite two guanines at the cross-link. Pol α was unable to elongate DNA chains from any of these primer–templates containing the lesion, indicating that the critical bypass activity of pol η relies on its ability to continue elongation when the correct nucleotide has been incorporated opposite a lesion.
Fig. 4. Ability of pol η to elongate DNA chains past lesions. Sets of 5′-32P-labeled 17mer (A, B, E, F, G, J and K) or 18mer (C, D, H, I, L and M) primers, which contained different sequences at their 3′ ends (indicated by N, where N was A, C, G and T in lanes 1–4, 5–8, 9–12 and 13–16, respectively), were annealed to 30mer templates. Increasing amounts of pol η or pol α were incubated with these primed templates. The auto-radiograms of the gels are shown. The amounts of pol η were 0.12 fmol in lanes 2, 6, 10 and 14, 0.7 fmol in lanes 3, 7, 11 and 15 (A–D, F, H, J and L), 0.25 fmol in lanes 2, 6, 10 and 14, and 1.4 fmol in lanes 3, 7, 11 and 15 (E, G, I, K and M). Pol α (lanes 4, 8, 12 and 16) was present at 4.7 fmol. (A) Undamaged control for the CPD template with a 17mer primer. (B) CPD at the bridged TT with a 17mer primer. (C) Undamaged control for the CPD template with an 18mer primer. (D) CPD at the bridged TT with an 18mer primer. (E) The AP analog at X with a 17mer primer. (F) Undamaged control for the AAF-G template with a 17mer primer. (G) AAF-G at the indicated site with a 17mer primer. (H) Undamaged control for the AAF-G template with an 18mer primer. (I) AAF-G at the indicated site with an 18mer primer. (J) Undamaged control for the cisplatin-GG template with a 17mer primer. (K) Cisplatin-GG at the indicated site with a 17mer primer. (L) Undamaged control for the cisplatin-GG template with an 18mer primer. (M) Cisplatin-GG at the indicated site with an 18mer primer.
Discussion
Substrate range for bypass replication by human pol η
XP-V cells are known to show hypermutability after exposure not only to UV light but also to various DNA-damaging agents (Friedberg et al., 1995). In addition, extracts from XP-V cells are reported to be defective in replicating DNA containing UV-induced CPD and AAF-modified bases (Cordeiro-Stone et al., 1997; Ensch-Simon et al., 1998; Svoboda et al., 1998; Cordonnier et al., 1999; Masutani et al., 1999a). These observations indicate that the XPV gene product, human DNA polymerase η, is involved in accurate translesion synthesis past lesions induced by chemical agents and UV-induced CPD.
CPD was certainly the best substrate for bypass replication by pol η, while the (6–4) photoproduct was hardly replicated at all. We report here that pol η could replicate an AP analog, AAF-G, and cisplatin-GG, but less efficiently than CPD. These observations are consistent with the assumption that pol η evolved mainly to overcome UV radiation, a natural cause of DNA damage from ancient times. Among the major types of damage induced by UV light, the (6–4) photoproduct is repaired much more quickly than the CPD lesion by the nucleotide excision repair pathway (Friedberg et al., 1995). This explains well why pol η can deal efficiently with the CPD but hardly at all with the (6–4) photoproduct. AAF-G and cisplatin-GG are caused by chemical agents and can be repaired by the nucleotide excision repair pathway. AP sites, which can result from the depurination or depyrimidination of DNA by spontaneous hydrolysis of N-glycosylic bonds, are the most frequent type of lesion in cells and are repaired efficiently by the base excision repair pathway (Friedberg et al., 1995). Although these repair pathways mainly contribute to the removal of these lesions, it would be surprising if they were able to repair all lesions before DNA replication. Thus, human pol η may contribute to the general damage tolerance of cells by allowing replication past these lesions as well.
Recent findings indicate that human cells have a set of genes encoding DNA polymerases involved in translesion synthesis, including REV3 (pol ζ) (Xiao et al., 1998; Lin et al., 1999), DINB1 (Gerlach et al., 1999; Ogi et al., 1999) and hRAD30B, which is the second homolog of yeast RAD30 (hRAD30A is another name for XPV) (McDonald et al., 1999). In yeast, pol ζ was shown to be involved in translesion synthesis past AAF-modified bases (Baynton et al., 1998) and AP sites in cooperation with Rev1 (Nelson et al., 1996a). Although the substrate specificities of novel human polymerases have not been fully determined yet, it is possible that these human gene products also contribute to the damage tolerance of cells by allowing replication bypass of certain specific lesions.
Low fidelity DNA synthesis by pol η
We have recently found that DNA synthesis by the human pol η on undamaged DNA is highly mutagenic (Matsuda et al., 2000). This could be due to its loose requirement for correct Watson–Crick base pairing, a property that would allow pol η to catalyze translesion synthesis past many types of lesion. In contrast, other replicative DNA polymerases strictly require correct Watson–Crick base pairing for DNA synthesis and cannot catalyze translesion synthesis efficiently (Kunkel and Bebenek, 2000).
Here we have shown that human pol η is essentially an error-prone DNA polymerase. Pol η misincorporation was particularly evident when the template sequence was a T. More importantly, pol η could elongate DNA chains from mispaired template–primers especially when the 3′ nucleotide of the primer was G and located opposite T. This finding agrees well with the observation that the most frequent mutation caused by in vitro DNA replication by human pol η is a base substitution of T for C (Matsuda et al., 2000). Pol η could also elongate DNA chains from other mispaired template–primers. It must be mentioned, however, that these reactions were only observed efficiently under conditions where excess amounts of pol η were added to the reactions. This may mean that repeated attacks of pol η on the mispaired 3′ terminus of the primer are required for the elongation of DNA chains containing a mispaired template–primer. As cells possess a wide range of replicative DNA polymerases, such as δ and ε, that are capable of rapidly removing mispaired primer ends through their 3′ to 5′ proofreading exonuclease activities (Bambara et al., 1997; Waga and Stillman, 1998), it seems unlikely that the primer ends would be available for repeated elongation by pol η. Thus, our results suggest rather that the action of pol η may be restricted to the synthesis of a few nucleotides opposite and past the lesion to prevent mutations. The low processivity of the enzyme would restrict DNA synthesis to short patches only. In addition, cells might have some mechanism to regulate pol η. It is also possible that a protein factor exists in cells, which increases the fidelity of pol η. These possibilities should be clarified before we can understand why the translesion synthesis defective in XP-V patients is accurate.
Molecular mechanisms of translesion synthesis by pol η
In this study, we demonstrated that the fidelity of translesion synthesis by human pol η relies on the enzyme’s ability to select the correct nucleotide to be incorporated opposite a lesion and to elongate only DNA chains that have a correctly base-paired primer terminus. Although neither one of these activities was completely efficient, together they allowed DNA polymerase η to bypass lesions with a low mutation frequency.
Human pol η catalyzed DNA synthesis past a CPD much more efficiently than other lesions. Owing to the efficient deoxynucleotidyl transferase activity of pol η on this template, we could not determine the identity of the major nucleotide incorporated opposite the lesion in reactions containing only one kind of nucleotide. Recently, however, human pol η was shown to incorporate dAMP more frequently than any other nucleotide opposite thymines of a CPD by steady-state kinetic experiments (Johnson et al., 2000) (R.Kusumoto, C.Masutani, S.Iwai and F.Hanaoka, unpublished observation). Importantly, pol η could elongate primers having an A opposite the T of a dimer more preferentially than other primers, and the preference was greater on the CPD-containing template than on undamaged templates (Figure 4A–D), although elongation from a mispaired primer on a CPD-containing template was also observed. Thus, the preferential incorporation of a correct nucleotide opposite a lesion and the preferential elongation of DNA chains having a correct nucleotide opposite a lesion could explain the considerable accuracy of translesion synthesis by pol η.
On AP analog-, AAF-G- and cisplatin-GG-containing templates, translesion syntheses by pol η were less efficient than on CPD templates, but detectable translesion products were still observed. Because of the low processivity of the enzyme, excess enzyme was required to detect fully elongated products of 30 nucleotides. However, small amounts of bypass products were observed in reactions containing lower concentrations of enzyme, and a portion of the arrested replication products opposite and around the lesions was never elongated even in the presence of excess enzyme (Figure 2). These observations suggest that certain portions of the lesions were bypassed in one processive reaction and that others were hardly ever bypassed even in the presence of excess enzyme. Opposite the AP analog, pol η incorporated dAMP and dGMP with almost equal efficiency. After incorporating a nucleotide, pol η more efficiently elongated a product having an A rather than a G opposite the lesion. This might be because the T residue next to the AP site on the template facilitated the incorporation of dAMP and/or further elongation by pol η. However, pol η bypassed the lesion by incorporating a nucleotide opposite the AP site and did not do so through slippage of the template–primer because products of 30 nucleotides were obtained (Figure 2D). Thus, human pol η follows the A-rule (Strauss, 1991). Purines might be incorporated more efficiently than pyrimidines owing to the stronger stacking interaction of purine bases with the 3′ nucleotide of the primer (Saenger, 1984). On the cisplatin-GG template, human pol η incorporated dCMP opposite the first G of the cross-link, but dAMP was incorporated more efficiently than dCMP opposite the second G of the lesion in our sequence context. However, only the incorporation of dCMP opposite the lesion allowed further elongation of DNA chains past the lesion, resulting in accurate translesion synthesis of the cisplatin-induced cross-link. As for the AAF-G-containing template, pol η also elongated DNA chains when the correct nucleotide was incorporated opposite the lesion. Interestingly, nucleotide selectivity by pol η opposite the A next to AAF-G was more accurate on the lesion-containing template than on the undamaged template (Figure 3G and H), suggesting that human pol η can catalyze more accurate DNA synthesis on damage-containing templates than on undamaged templates.
Very recently, Yuan et al. (2000) reported that yeast pol η could incorporate dGMP or dAMP opposite an AP site and dCMP opposite an AAF-G, but that the enzyme was unable to extend DNA synthesis past these lesions. They also showed that yeast pol ζ could extend DNA synthesis past the AP site following pol η action. Although human pol η can catalyze translesion synthesis past these lesions by itself, it seems likely that more efficient translesion synthesis occurrs through the combined actions of pol η and other DNA polymerases, including replicative polymerases in human cells.
Materials and methods
Proteins
Recombinant human pol η tagged with His6 at its C-terminal side was expressed in Sf9 insect cells using the baculovirus expression system. Cells were collected by low-speed centrifugation and washed twice with ice-cold phosphate-buffered saline. The cell pellets were suspended in 8 vols of ice-cold buffer A (20 mM sodium phosphate pH 7.3, 10% glycerol, 10 mM β-mercaptoethanol) containing 0.3 M NaCl, 1% NP-40, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 50 µM EGTA, 0.4 µg/ml antipain, 0.4 µg/ml aprotinin, 0.1 µg/ml leupeptin and 80 ng/ml pepstain A, and then incubated on ice for 30 min with occasional agitation. The suspension was centrifuged at 12 000 g for 1 h. The supernatant was adjusted to 20 mM imidazole and loaded onto a HiTrap Q (Amersham Pharmacia) column equilibrated with buffer B (buffer A containing 0.3 M NaCl, 20 mM imidazole and 0.25 mM PMSF). After washing the column with the same buffer, flow-through fractions were loaded onto a nickel-NTA agarose (Qiagen) column equilibrated with buffer B. The column was washed with buffer B, and bound materials were eluted by a linear gradient of 20–250 mM imidazole in buffer B. The peak fractions were dialyzed against buffer C (buffer A containing 0.1 M NaCl, 1 mM EDTA, 0.25 mM PMSF) and loaded onto a MonoS PC1.6/5 (Amersham Pharmacia) column equilibrated with buffer C. The column was washed with buffer C, and proteins were eluted by a linear gradient of 0.1–0.5 M NaCl in buffer C. Peak fractions were stored at –80°C.
DNA polymerase α was purified from mouse FM3A cells as described previously (Eki et al., 1991).
Processivity assays
The singly primed phagemid DNA templates were prepared by mixing a T7 primer (5′-TAATACGACTCACTATAGGG-3′), which was labeled at its 5′ end using T4 polynucleotide kinase and [γ-32P]ATP, and single-stranded pBS-KS(+) DNA (Stratagene) at a molar ratio of 2:1.
Pol η or pol α was incubated with 500 fmol of the singly primed template at 37°C in 10 µl reactions containing 40 mM Tris–HCl pH 8.0, 1 mM MgCl2, 500 µM each of the four dNTPs, 10 mM dithiothreitol (DTT), 250 µg/ml bovine serum albumin (BSA), 60 mM KCl and 2.5% glycerol. Reaction times and amounts of enzymes are indicated in the figure legends. Reactions were terminated by adding 10 µl of formamide and boiling. Products were electrophoresed on a 20% polyacrylamide–7 M urea gel and autoradiographed.
Translesion synthesis assays
The 5′-32P-labeled primer–template DNA was prepared by mixing the 16mer, 17mer or 18mer primer labeled at its 5′ end with the 30mer DNA at a molar ratio of 1:1. The 30mer oligomers containing the CPD (Murata et al., 1990), (6–4) photoproduct (Iwai et al., 1996) and AP site analog (Fujiwara et al., 1999) were synthesized as described. The AAF-modified template was prepared by treating the 30mer oligonucleotide with N-acetoxy-AAF as described (van Vuuren et al., 1993). The cisplatin-modified 30mer oligonucleotide was prepared as described previously (Fujiwara et al., 1999).
Standard reactions of 10 µl contained 40 mM Tris–HCl pH 8.0, 1 mM MgCl2, 100 µM each of the four dNTPs, 10 mM DTT, 250 µg/ml BSA, 60 mM KCl, 2.5% glycerol and 40 nM of the 5′-32P-labeled primer–template DNA. In the experiments shown in Figure 3I and J, 100 mM KCl was included to strengthen annealing and avoid misannealing of the templates with the primers. Reactions were performed at 37°C for 15 min and terminated by adding 10 µl of formamide followed by boiling. Products were electrophoresed on a 20% polyacrylamide–7 M urea gel and autoradiographed.
Acknowledgments
Acknowledgements
We are grateful to Marito Araki, Ayumi Yamada, Mayumi Yuasa, Tomokazu Nogimori and other members of Dr Hanaoka’s laboratory at Osaka University for helpful discussions. This work was supported by grants from the Ministry of Education, Science, Sports and Culture of Japan, from CREST and from the Biodesign Research Program of RIKEN.
References
- Bambara R.A., Murante,R.S. and Henricksen,L.A. (1997) Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem., 272, 4647–4650. [DOI] [PubMed] [Google Scholar]
- Baynton K., Bresson-Roy,A. and Fuchs,R.P. (1998) Analysis of damage tolerance pathways in Saccharomyces cerevisiae: a requirement for Rev3 DNA polymerase in translesion synthesis. Mol. Cell. Biol., 18, 960–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland W.C. and Wang,T.S. (1991) Catalytic subunit of human DNA polymerase α overproduced from baculovirus-infected insect cells. Structural and enzymological characterization. J. Biol. Chem., 266, 22739–22748. [PubMed] [Google Scholar]
- Cordeiro-Stone M., Zaritskaya,L.S., Price,L.K. and Kaufmann,W.K. (1997) Replication fork bypass of a pyrimidine dimer blocking leading strand DNA synthesis. J. Biol. Chem., 272, 13945–13954. [DOI] [PubMed] [Google Scholar]
- Cordonnier A.M. and Fuchs,R.P. (1999) Replication of damaged DNA: molecular defect in xeroderma pigmentosum variant cells. Mutat. Res., 435, 111–119. [DOI] [PubMed] [Google Scholar]
- Cordonnier A.M., Lehmann,A.R. and Fuchs,R.P. (1999) Impaired translesion synthesis in xeroderma pigmentosum variant extracts. Mol. Cell. Biol., 19, 2206–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eki T., Enomoto,T., Masutani,C., Miyajima,A., Takada,R., Murakami,Y., Ohno,T., Hanaoka,F. and Ui,M. (1991) Mouse DNA primase plays the principal role in determination of permissiveness for polyomavirus DNA replication. J. Virol., 65, 4874–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensch-Simon I., Burgers,P.M.J. and Taylor,J.-S. (1998) Bypass of a site-specific cis-syn thymine dimer in an SV40 vector during in vitro replication by HeLa and XPV cell-free extracts. Biochemistry, 37, 8218–8226. [DOI] [PubMed] [Google Scholar]
- Friedberg E.C. and Gerlach,V.L. (1999) Novel DNA polymerases offer clues to the molecular basis of mutagenesis. Cell, 98, 413–416. [DOI] [PubMed] [Google Scholar]
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC. [Google Scholar]
- Fujiwara Y., Masutani,C., Mizukoshi,T., Kondo,J., Hanaoka,F. and Iwai,S. (1999) Characterization of DNA recognition by the human UV-damaged-DNA binding protein. J. Biol. Chem., 274, 20027–20033. [DOI] [PubMed] [Google Scholar]
- Gerlach V.L., Aravind,L., Gotway,G., Schultz,R.A., Koonin,E.V. and Friedberg,E.C. (1999) Human and mouse homologs of Escherichia coli DinB (DNA polymerase IV), members of the UmuC/DinB superfamily. Proc. Natl Acad. Sci. USA, 96, 11922–11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs P.E., McGregor,W.G., Maher,V.M., Nisson,P. and Lawrence,C.W. (1998) A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase ζ. Proc. Natl Acad. Sci. USA, 95, 6876–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai S., Shimizu,M., Kamiya,H. and Ohtsuka,E. (1996) Synthesis of a phosphoramidite coupling unit of the pyrimidine (6–4) pyrimidone photoproduct and its incorporation into oligodeoxynucleotides. J. Am. Chem. Soc., 118, 7642–7643. [Google Scholar]
- Johnson R.E., Kondratick,C.M., Prakash,S. and Prakash,L. (1999a) hRAD30 mutations in the variant form of xeroderma pigmentosum. Science, 285, 263–265. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Prakash,S. and Prakash,L. (1999b) Efficient bypass of a thymine–thymine dimer by yeast DNA polymerase, pol η. Science, 283, 1001–1004. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Washington,M.T., Prakash,S. and Prakash,L. (1999c) Bridging the gap: a family of novel DNA polymerases that replicate faulty DNA. Proc. Natl Acad. Sci. USA, 96, 12224–12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.E., Washington,M.T., Prakash,S. and Prakash,L. (2000) Fidelity of human DNA polymerase η. J. Biol. Chem., 275, 7447–7450. [DOI] [PubMed] [Google Scholar]
- Kunkel T.A. and Bebenek,K. (2000) DNA replication fidelity. Annu. Rev. Biochem., 69, in press. [DOI] [PubMed] [Google Scholar]
- Lehmann A.R., Kirk-Bell,S., Arlett,C.F., Paterson,M.C., Lohman,P.H., de Weerd-Kastelein,E.A. and Bootsma,D. (1975) Xeroderma pigmentosum cells with normal levels of excision repair have a defect in DNA synthesis after UV-irradiation. Proc. Natl Acad. Sci. USA, 72, 219–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Wu,X. and Wang,Z. (1999) A full-length cDNA of hREV3 is predicted to encode DNA polymerase ζ for damage-induced mutagenesis in humans. Mutat. Res., 433, 89–98. [DOI] [PubMed] [Google Scholar]
- Maher V.M., Ouellette,L.M., Curren,R.D. and McCormick,J.J. (1976) Frequency of ultraviolet light-induced mutations is higher in xeroderma pigmentosum variant cells than in normal human cells. Nature, 261, 593–595. [DOI] [PubMed] [Google Scholar]
- Masutani C., Araki,M., Yamada,A., Kusumoto,R., Nogimori,T., Maekawa,T., Iwai,S. and Hanaoka,F. (1999a) Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J., 18, 3491–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C. et al. (1999b) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature, 399, 700–704. [DOI] [PubMed] [Google Scholar]
- Matsuda T., Bebenek,K., Masutani,C., Hanaoka,F. and Kunkel,T.A. (2000) Low fidelity DNA synthesis by human DNA polymerase η. Nature, 404, 1011–1013. [DOI] [PubMed] [Google Scholar]
- McDonald J.P., Levine,A.S. and Woodgate,R. (1997) The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics, 147, 1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J.P., Rapic-Otrin,V., Epstein,J.A., Broughton,B.C., Wang,X., Lehmann,A.R., Wolgemuth,D.J. and Woodgate,R. (1999) Novel human and mouse homologs of Saccharomyces cerevisiae DNA polymerase η. Genomics, 60, 20–30. [DOI] [PubMed] [Google Scholar]
- Misra R.R. and Vos,J.M. (1993) Defective replication of psoralen adducts detected at the gene-specific level in xeroderma pigmentosum variant cells. Mol. Cell. Biol., 13, 1002–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T., Iwai,S. and Ohtsuka,E. (1990) Synthesis and characterization of a substrate for T4 endonuclease V containing a phosphorodithioate linkage at the thymine dimer site. Nucleic Acids Res., 18, 7279–7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996a) Deoxycytidyl transferase activity of yeast REV1 protein. Nature, 382, 729–731. [DOI] [PubMed] [Google Scholar]
- Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996b) Thymine–thymine dimer bypass by yeast DNA polymerase ζ. Science, 272, 1646–1649. [DOI] [PubMed] [Google Scholar]
- Ogi T., Kato,T.J., Kato,T. and Ohmori,H. (1999) Mutation enhancement by DINB1, a mammalian homologue of the Escherichia coli mutagenesis protein DinB. Genes Cells, 4, 607–618. [DOI] [PubMed] [Google Scholar]
- Raha M., Wang,G., Seidman,M.M. and Glazer,P.M. (1996) Mutagenesis by third-strand-directed psoralen adducts in repair-deficient human cells: high frequency and altered spectrum in a xeroderma pigmentosum variant. Proc. Natl Acad. Sci. USA, 93, 2941–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuven N.B., Arad,G., Maor-Shoshani,A. and Livneh,Z. (1999) The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA and SSB and is specialized for translesion replication. J. Biol. Chem., 274, 31763–31766. [DOI] [PubMed] [Google Scholar]
- Roush A.A., Suarez,M., Friedberg,E.C., Radman,M. and Siede,W. (1998) Deletion of the Saccharomyces cerevisiae gene RAD30 encoding an Escherichia coli DinB homolog confers UV radiation sensitivity and altered mutability. Mol. Gen. Genet., 257, 686–692. [DOI] [PubMed] [Google Scholar]
- Saenger W. (1984) Principles of Nucleic Acid Structure. Springer Verlag, Berlin, Germany. [Google Scholar]
- Strauss B.S. (1991) The ‘A rule’ of mutagen specificity: a consequence of DNA polymerase bypass of non-instructional lesions? BioEssays, 13, 79–84. [DOI] [PubMed] [Google Scholar]
- Svoboda D.L., Briley,L.P. and Vos,J.-M.H. (1998) Defective bypass replication of a leading strand cyclobutane thymine dimer in xeroderma pigmentosum variant cell extracts. Cancer Res., 58, 2445–2448. [PubMed] [Google Scholar]
- Tang M., Shen,X., Frank,E.G., O’Donnell,M., Woodgate,R. and Goodman,M.F. (1999) UmuD′2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl Acad. Sci. USA, 96, 8919–8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman A., Masutani,C., Hanaoka,F. and Chaney,S.G. (2000) Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase η. Biochemistry, 39, 4575–4580. [DOI] [PubMed] [Google Scholar]
- van Vuuren A.J., Appeldoorn,E., Odijk,H., Yasui,A., Jaspers,N.G., Bootsma,D. and Hoeijmakers,J.H. (1993) Evidence for a repair enzyme complex involving ERCC1 and complementing activities of ERCC4, ERCC11 and xeroderma pigmentosum group F. EMBO. J., 12, 3693–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waga S. and Stillman,B. (1998) The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem., 67, 721–751. [DOI] [PubMed] [Google Scholar]
- Wagner J., Gruz,P., Kim,S.R., Yamada,M., Matsui,K., Fuchs,R.P. and Nohmi,T. (1999) The dinB gene encodes a novel E.coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol. Cell, 4, 281–286. [DOI] [PubMed] [Google Scholar]
- Wang Y.C., Maher,V.M. and McCormick,J.J. (1991) Xeroderma pigmentosum variant cells are less likely than normal cells to incorporate dAMP opposite photoproducts during replication of UV-irradiated plasmids. Proc. Natl Acad. Sci. USA, 88, 7810–7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-C., Maher,V.M., Mitchell,D.L. and McCormick,J.J. (1993) Evidence from mutation spectra that the UV hypermutability of xeroderma pigmentosum variant cells reflects abnormal, error-prone replication on a template containing photoproducts. Mol. Cell. Biol., 13, 4276–7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington M.T., Johnson,R.E., Prakash,S. and Prakash,L. (1999) Fidelity and processivity of Saccharomyces cerevisiae DNA polymerase η. J. Biol. Chem., 274, 36835–36838. [DOI] [PubMed] [Google Scholar]
- Waters H.L., Seetharam,S., Seidman,M.M. and Kraemer,K.H. (1993) Ultraviolet hypermutability of a shuttle vector propagated in xeroderma pigmentosum variant cells. J. Invest. Dermatol., 101, 744–748. [DOI] [PubMed] [Google Scholar]
- Woodgate R. (1999) A plethora of lesion-replicating DNA polymerases. Genes Dev., 13, 2191–2195. [DOI] [PubMed] [Google Scholar]
- Xiao W., Lechler,T., Chow,B.L., Fontanie,T., Agustus,M., Carter,K.C. and Wei,Y.F. (1998) Identification, chromosomal mapping and tissue-specific expression of hREV3 encoding a putative human DNA polymerase ζ. Carcinogenesis, 19, 945–949. [DOI] [PubMed] [Google Scholar]
- Yuan F., Zhang,Y., Rajpal,D.K., Wu,X., Guo,D., Wang,M., Taylor,J.-S. and Wang,Z. (2000) Specificity of DNA lesion bypass by the yeast DNA polymerase η. J. Biol. Chem., 275, 8233–8239. [DOI] [PubMed] [Google Scholar]