Abstract
Elp3 and Gcn5 are histone acetyltransferases (HATs) that function in transcription as subunits of Elongator and SAGA/ADA, respectively. Here we show that mutations that impair the in vitro HAT activity of Elp3 confer typical elp phenotypes such as temperature sensitivity. Combining an elp3Δ mutation with histone H3 or H4 tail mutations confers lethality or sickness, supporting a role for Elongator in chromatin remodelling in vivo. gcn5Δelp3Δ double mutants display a number of severe phenotypes, and similar phenotypes result from combining the elp mutation with mutation in a gene encoding a SAGA-specific, but not an ADA-specific subunit, indicating that Elongator functionally overlaps with SAGA. Because concomitant active site alterations in Elp3 and Gcn5 are sufficient to confer severe phenotypes, the redundancy must be specifically related to the HAT activity of these complexes. In support of this conclusion, gcn5Δelp3Δ phenotypes are suppressed by concomitant mutation of the HDA1 and HOS2 histone deacetylases. Our results demonstrate functional redundancy among transcription-associated HAT and deacetylase activities, and indicate the importance of a fine-tuned acetylation–deacetylation balance during transcription in vivo.
Keywords: Elongator/ELP3/GCN5/histone acetyltransferase/histone deacetylase
Introduction
In order to facilitate transcription in a repressive chromatin environment, the RNA polymerase II (RNAPII) transcription machinery can enlist chromatin-remodelling complexes (Workman and Kingston, 1998). Conversely, transcriptional repression often entails recruitment of factors stabilizing chromatin, such as histone deacetylases (HDACs) (Ayer, 1999). Remodelling factors that allow transcriptional activators and RNAPII transcription factors to work in chromatin have been divided into two major groups: ATP-dependent chromatin-remodelling machines (the SWI/SNF group of proteins) (Cairns, 1998) and histone acetyltransferases (HATs) (Mizzen and Allis, 1998; Struhl, 1998; Brown et al., 2000). The discovery that a well known transcriptional co-activator, Gcn5, also possesses intrinsic HAT activity provided the first direct link between histone acetylation and gene activation (Brownell et al., 1996). Since then, many studies have confirmed and extended the initial findings in different eukaryotic systems (Wade et al., 1997; Mizzen and Allis, 1998; Workman and Kingston, 1998). In yeast, Gcn5 is the catalytic component of at least two multi-protein complexes, SAGA and ADA (Grant et al., 1997; Eberharter et al., 1999). Consistent with a role for Gcn5 in chromatin remodelling of promoters, recruitment of SAGA by a transcriptional activator leads to histone acetylation and concomitant activation of transcription in vitro (Utley et al., 1998). Moreover, mutation of sequences encoding residues critical for Gcn5 HAT activity in vitro is sufficient to significantly affect the function of the protein in transcriptional activation in vivo (Kuo et al., 1998; Wang et al., 1998), and to generate localized alterations in the chromatin structure of target gene promoters (Gregory et al., 1998; Kuo et al., 1998).
Recently, we isolated the form of RNAPII that is responsible for transcriptional elongation (Otero et al., 1999). This form of RNAPII is distinct from previously characterized RNAPII holoenzymes in that it lacks Mediator/Srb components, but instead carries a novel multi-subunit complex, termed Elongator. One of the subunits of Elongator was identified as being a novel HAT (Otero et al., 1999; Wittschieben et al., 1999). This protein, Elp3, is highly conserved among eukaryotes and can acetylate all four histones in vitro. The function of Elp3 HAT might be to confer to elongating RNAPII holoenzyme the ability to act as a ‘chromatin conditioner’ in generally establishing and maintaining active chromatin in eukaryotes (Travers, 1999; Wittschieben et al., 1999).
The genome of the yeast Saccharomyces cerevisiae also encodes other HATs, such as Esa1, Sas2, Sas3, Hpa2 and Hat1 (Kleff et al., 1995; Reifsnyder et al., 1996; Neuwald and Landsman, 1997; Smith et al., 1998; Angus-Hill et al., 1999; Brown et al., 2000). Among these HATs, only Esa1 is encoded by an essential gene (Smith et al., 1998). Indeed, the phenotypes resulting from the deletion of any of the others are rather mild. For example, cells lacking ELP3 and GCN5, the only HATs that have been connected directly to transcription both in vitro and in vivo, have doubling times comparable to those of wild type, and the strains only fail to grow under very specific conditions that are distinct for each mutation. While gcn5 mutants grow poorly during amino acid starvation (Georgakopoulos and Thireos, 1992) and fail to undergo high-frequency mating-type inter-conversion (switching-defective) (Breeden and Nasmyth, 1987; Cosma et al., 1999), elp3 mutants display a peculiar ‘slow-start’ phenotype, manifested as a pronounced delay in adaptation to new growth conditions. Elongator mutants are also salt- and temperature-sensitive (Otero et al., 1999; Wittschieben et al., 1999). The molecular basis for these growth phenotypes appears to be problems with activation of a few, specific genes. gcn5 mutant strains fail to activate the HIS4 gene in response to low nitrogen (Lucchini et al., 1984), and also show reduced activation of HO and some additional genes (Pollard and Peterson, 1997; Cosma et al., 1999; Krebs et al., 1999). Likewise, elp strains show significant delays in the activation of genes whose expression is required for the cells to grow under the relevant conditions. For example, activation of the genes GAL1–10, ENA1 and PHO5 is delayed and often reduced in elp3 mutants (Wittschieben et al., 1999).
The rather limited phenotypic consequences of mutating the transcription-related HATs described above could be explained by the existence of functional redundancy. As a consequence, cellular functions such as transcriptional activation might still proceed normally when one HAT complex is non-functional, but not when two (or more) are non-functional. Similar redundancies might underlie the limited consequence of mutating genes encoding HDACs (Rundlett et al., 1996). Here we show that single-site alterations in the HAT domain of Elp3 confer typical elp phenotypes such as slow growth adaptation and temperature sensitivity. A role for Elongator in chromatin modification and remodelling in vivo is supported by the finding that the elp3 mutation in combination with histone tail alterations confers sickness or lethality. In support of the existence of functional overlaps between HATs, Elongator and SAGA, or, specifically, the acetyltransferase activities of Gcn5 and Elp3, can indeed not be abrogated at the same time without severe consequences for cell growth. The severe consequences of concomitant gcn5Δelp3Δ mutation can, however, be counteracted by also mutating the genes encoding the HDACs, HDA1 and HOS2. Deletion of these genes individually fails to suppress the gcn5Δelp3Δ phenotypes, indicating functional redundancy among HDACs as well.
Results
The HAT activity of Elp3 is essential for its function in vivo
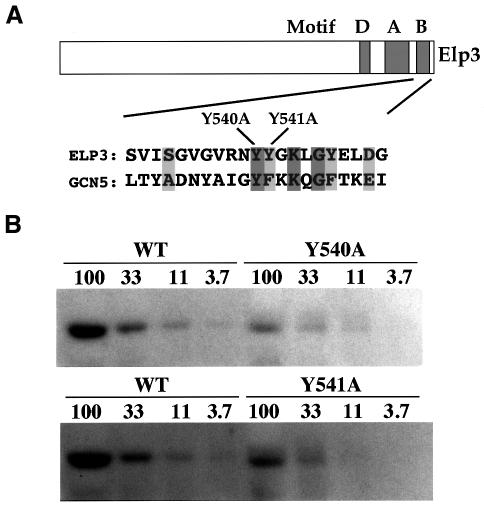
The presence of Elp3 protein in elongating RNAPII holoenzyme could potentially serve to couple histone modification to a DNA-tracking engine, leading to more or less genome-wide potentiation of transcriptional activation in yeast, and domain-specific changes in higher eukaryotes (Travers, 1999; Wittschieben et al., 1999). In order to investigate the structure–function relationship of Elp3 HAT in vitro, mutations were made in ELP3 that changed two conserved tyrosine residues to alanine in the B motif of the putative catalytic HAT domain (Y540A and Y541A, respectively; Figure 1A). These residues are likely to be directly involved in acetyl-CoA binding (Dutnall et al., 1998; Rojas et al., 1999). Similar mutations in yeast Gcn5 (Y220A and F221A, respectively) have previously been shown to reduce the HAT activity of this protein to <10% of wild-type levels in vitro, and to significantly affect its function in transcription in vivo (Kuo et al., 1998; Wang et al., 1998). Baculoviruses that enable expression of mutated Elp3 protein in insect cells were generated. As reported previously by Wittschieben et al. (1999), Elp3 protein was insoluble, making it necessary to employ an in-gel HAT assay for measuring enzyme activity (Brownell and Allis, 1995; Wittschieben et al., 1999). As expected, the mutant proteins had a significantly reduced activity compared with wild type: a reduction to <25% for Y540A and ∼35% for Y541A (Figure 1B).
Fig. 1. Elp3 mutants have reduced HAT activity in vitro. (A) Alignment of Elp3 and Gcn5 HAT motif B based on Neuwald and Landsman (1997) with the positions of the changes introduced in Elp3 indicated. (B) The HAT activity of Elp3 mutants is significantly reduced. Titrations of equal amounts of wild-type and mutant proteins were fractionated in 10% SDS–PAGE gels containing either BSA (not shown) or 1 mg/ml histone. After renaturation of separated proteins, the gels were incubated with tritium-labeled acetyl-CoA. Autoradiograms of dried gels are shown. Relative amounts of protein loaded are indicated above the lanes. Based on two independent titration experiments, the Y540A mutant had 22–25% activity, while Y541A had 34–38% activity compared with wild type.
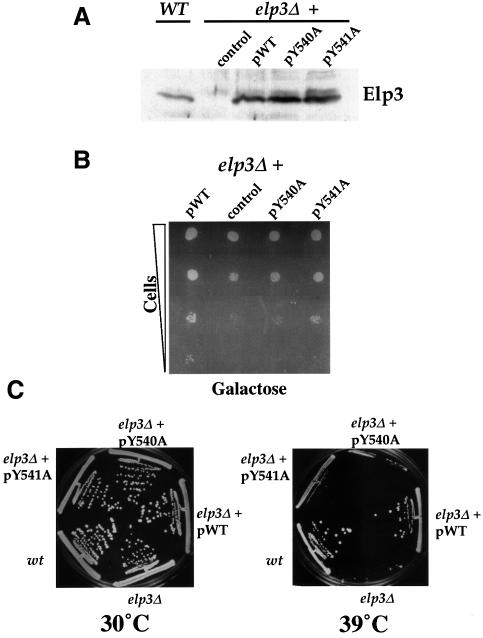
DNA fragments encoding the HAT domain mutations were exchanged for wild-type fragments in CEN plasmids carrying ELP3 expressed from its native promoter. In order to test their ability to complement elp phenotypes, these plasmids were then transformed into elp3Δ cells (Figure 2). Wild-type and mutant versions of Elp3 expressed to roughly the same level as endogenous Elp3 (Figure 2A), and could be co-immunoprecipitated with Elp1 and Elp2 from extracts with similar efficiency (data not shown), showing that they were incorporated into Elongator complexes. As expected, expression of wild-type Elp3 rescued the mutant phenotype. In contrast, Elp3 Y541A failed to fully complement both the slow adaptation to growth on galactose (Figure 2B) and the temperature-sensitive phenotype of the strain (Figure 2C), while cells expressing Elp3 Y540A displayed phenotypes that were indistinguishable from the strain that did not express Elp3. The in vivo effects of these mutations correlate with the lower level of in vitro HAT activity observed with Elp3 Y540A compared with Elp3 Y541A (Figure 1B). The data thus indicate that the acetyltransferase activity of Elp3 is essential for Elongator function in vivo.
Fig. 2. Elp3 HAT mutants display elp phenotypes. (A) Similar levels of wild-type and mutant Elp3 protein are detected in whole-cells extracts. Extracts from elp3Δ cells carrying CEN plasmids expressing wild-type Elp3 (pWT), no protein (control), Elp3 Y540A (pY540A) or Elp3 Y541A (pY541A) were compared with wild-type (ELP3) extracts. The result of a western blot probed with anti-Elp3 antibodies is shown. (B) Elp3 protein-carrying altered HAT domains are unable to complement the slow adaptation phenotype typical of elpΔ strains. Strains carrying the above CEN plasmids were grown in YPD and plated in a dilution series onto YPG (galactose) plates and incubated at 30°C for 2–3 days. (C) Elp3 protein-carrying altered HAT domains are unable to complement the temperature-sensitive phenotype of an elp3Δ strain. Strains were treated as in (B), but streaked on YPD plates and incubated for 3–5 days at 30 or 39°C.
Synthetic phenotype of strains lacking ELP3 and the tails of histones H3 and H4
gcn5 mutation is synthetic lethal with mutations encoding changes in the N-terminal tails of histones H3 and H4 (Zhang et al., 1998), indicating that histones are targets for the HAT activity of Gcn5 in vivo. To investigate whether histones could also be genetically linked to Elp3 in vivo, an elp3Δ strain was constructed in which the endogenous H3 and H4 genes (HHT1-HHF1 and HHT2-HHF2) were deleted so that survival was dependent on a plasmid borne copy of HHT2-HHF2. Plasmids carrying different mutated forms of the same histone genes were then introduced, and viability in the presence of these mutations was tested by plasmid shuffle experiments (Zhang et al., 1998) (Table I). Deletion of the N-terminal tail of histone H4 (Δ4–19) was lethal in the elp3Δ strain. An obvious interpretation of this result would be that Elp3 is involved in H3 acetylation in vivo. In the absence of H4 tails, this acetylation might become essential for viability. elp3Δ cells carrying a deletion of the N-terminal (Δ3–29) tail of histone H3 grew normally at 30°C, but were temperature-sensitive (37°C), pointing to a less important Elp3-mediated effect on histone H4 tails.
Table I. Plasmid shuffle of histone alleles.
| ELP3 | elp3Δ | |
|---|---|---|
| H4 Δ4–19 | viable | dead |
| H3 Δ3–29 | viable | ts– |
| H3 K14R | ||
| H4 K8,16R | viable | ts– |
| H3 K14Q | ||
| H4 K8,16Q | viable | viable |
| H3 K9R | ||
| H4 K5,12R | viable | viable |
The effects of mutated histone alleles were tested in ELP3 or elp3 cells carrying wild-type histones on a URA3-marked plasmid. After growth in URA+ media, the ability to grow in the absence of wild-type histones was tested by plating on 5-FOA-containing media. 5-FOA-resistant cells were plated on SD plates lacking uracil to confirm the loss of the URA3-marked plasmid, and were then plated on YPD plates and incubated at 30 and 37°C for 3–5 days to test for temperature sensitivity (ts–).
Distinct acetylation patterns typify nucleosome depositioning and transcription, respectively (Strahl and Allis, 2000), making synthetic phenotypes with mutations in sequences encoding specific lysine residues in the histone tails useful as tools to indicate the involvement of gene products in transcription-coupled chromatin modification (Zhang et al., 1998). We found that the ELP3 mutation combined with concomitant point mutations in sequences encoding the ‘transcription-related’ histone H3 lysine residue 14 and H4 lysine residues 8 and 16 to arginine (H3 K14R, H4 K8,16R) also conferred temperature sensitivity. In contrast, this phenotype was not observed when elp3Δ was combined with changes at these sites to glutamine (which might mimic constitutive acetylation of the sites), or when combined with changes in residues primarily thought to be related to nucleosome deposition (H3 K9R, H4 K5,12R). These results indicate that Elp3 is involved in transcription-associated chromatin modification and remodelling, and support the notion that histones are an important target for Elp3 activity in vivo.
Synthetic phenotype of strains lacking both GCN5 and ELP3
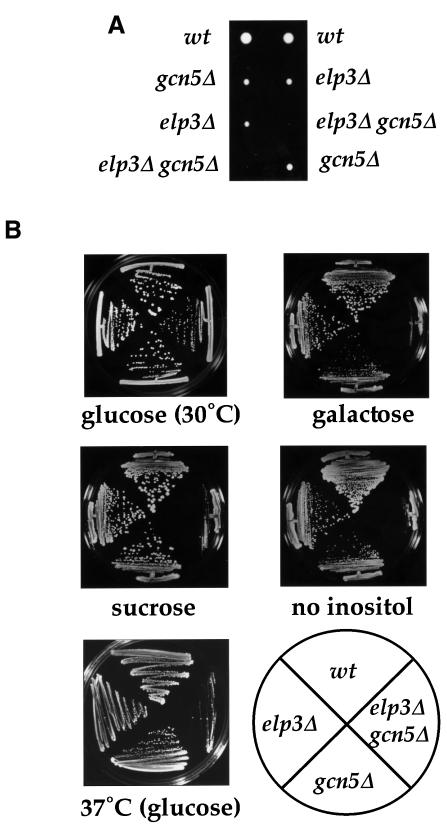
If Elp3 is involved in potentiation of transcriptional activation by histone acetylation, its in vivo function might at least partly overlap with that of the well characterized Gcn5 protein. To test this hypothesis, one copy of the GCN5 gene was deleted in ELP3/elp3Δ diploid yeast cells. The resulting strain was induced to sporulate and spores dissected. gcn5Δelp3Δ cells generated only very small, slow growing colonies (Figure 3A). Moreover, while gcn5Δ and elp3Δ single mutants grew on galactose, raffinose and sucrose as the sole carbon source, gcn5Δelp3Δ cells did not (Figure 3B and data not shown). Likewise, gcn5Δelp3Δ cells were unable to grow in the absence of inositol or at 37°C (Figure 3B). These results clearly demonstrate that GCN5 and ELP3 cannot be deleted at the same time without severe effects on cell growth, and thus indicate that the encoded proteins perform an overlapping function or co-operate during RNAPII transcription. The results gain further significance from the finding that ELP3 or GCN5 deletion in combination with deletion of HAT1 [encoding the only known B type (cytoplasmic) HAT (Kleff et al., 1995; Parthun et al., 1996)] does not lead to a new phenotype (data not shown; Ruiz-Garcia et al., 1998). Likewise, the ELP3 deletion in combination with deletion of genes encoding other HATs, such as SAS3 (Reifsnyder et al., 1996) and HPA2 (Angus-Hill et al., 1999), also does not result in new, dramatic phenotypes (data not shown). This indicates that the functional overlap is specifically related to transcription and not simply generally observed between HAT-encoding genes.
Fig. 3. Synthetic phenotype of the elp3Δgcn5Δ strain. (A) elp3Δgcn5Δ double mutants are slow growing. Tetrad dissections of gcn5Δ/GCN5 elp3Δ/ELP3 diploids are shown after 2 days of incubation at 30°C. (B) elp3Δgcn5Δ double mutants are unable to grow on a number of media. Cells of the indicated genotype were streaked on YPD [glucose (30°C)], YPG (galactose), YPS (sucrose) or inositol starvation (no inositol) plates, and incubated for 3–5 days at 30°C. To demonstrate temperature sensitivity, cells of the indicated genotype were streaked on YPD [37°C (glucose)] and incubated for 3–5 days at 37°C.
The phenotype of gcn5 elp3 is linked to the functions of SAGA and Elongator
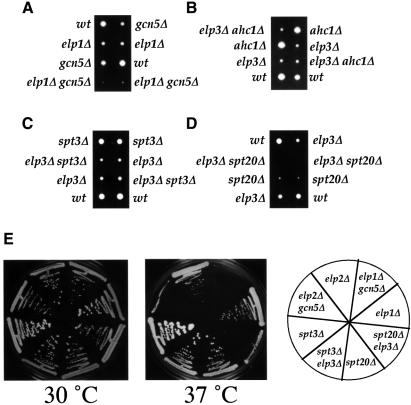
While Gcn5 is the catalytic component of at least two different HAT complexes, SAGA and ADA (Grant et al., 1997; Eberharter et al., 1999), Elp3 appears to exert its function mostly, if not exclusively, through the Elongator complex (Wittschieben et al., 1999). In agreement with this notion, mutation of the genes encoding the other subunits of Elongator (ELP1 and ELP2, respectively) in conjunction with a GCN5 mutation yielded phenotypes similar to those of the elp3Δgcn5Δ strain (Figure 4A and E, and data not shown). To investigate whether Elp3 (and therefore Elongator) functionally overlaps with Gcn5 in the context of the ADA or SAGA complex, we also tested whether combining mutations in ELP3 with mutations in the ADA-specific AHC1 gene, as well as in the SAGA-specific SPT20 and SPT3 genes, would confer phenotypes similar to those of elp3Δgcn5Δ. The ahc1 mutation disrupts the ADA complex, the spt20Δ mutation is believed to abolish all SAGA functions, and spt3Δ is only required for a HAT-independent function of SAGA (Grant et al., 1997; Roberts and Winston, 1997; Sterner et al., 1999). While elp3Δahc1Δ and elp3Δspt3Δ failed to confer a new phenotype (Figure 4B, C and E, and data not shown), elp3Δspt20Δ strains were, like gcn5Δelp3Δ, very slow growing and temperature-sensitive (Figure 4D and E). We conclude that the synthetic phenotype of the gcn5Δelp3Δ strain is due to functional overlap or redundancy between Elongator and SAGA. The finding that the spt3 elp3 mutation does not result in a new phenotype (these cells also grew on galactose, sucrose and in the absence of inositol; data not shown) furthermore suggests that the partial redundancy between the complexes is specifically related to the HAT functions of SAGA.
Fig. 4. Severe phenotypes of cells with concomitant disruptions of Elongator and SAGA genes. (A–D) elp1Δgcn5Δ and elp3Δspt20Δ, but not elp3Δahc1Δ and elp3Δspt3Δ double mutants are slow growing. Tetrad dissections of the respective diploids are shown after 2–3 days of incubation at 30°C. (E) elp3Δspt3Δ, but not elp1Δgcn5Δ, elp2Δgcn5Δ and elp3Δspt20Δ double mutants grow at 37°C. Cells of the indicated genotype were streaked on YPD and incubated for 3–5 days at 30 or 37°C. As expected, elp3Δahc1Δ cells are not temperature-sensitive either (data not shown).
The functional overlap between Gcn5 and Elp3 is due to their intrinsic HAT activity
The basis for the dramatic consequences of lacking both Elongator and SAGA function could be a disturbance of the integrity (or stability) of SAGA and Elongator resulting from the removal of one of their subunits, and/or an effect on the functions of Gcn5 and Elp3 that is distinct from their intrinsic HAT activity. Alternatively, the basis could be a specific requirement for Elp3- and Gcn5-catalyzed acetylation by these complexes. To distinguish between these possibilities, we investigated whether merely abrogating the HAT activity of Gcn5 and Elp3 would give rise to severe phenotypes. As expected, the gcn5Δelp3Δ strain was dependent on a plasmid-borne copy of ELP3 or GCN5 for normal growth on a number of media. To investigate the effect of concomitant HAT mutation in Gcn5 and Elp3, plasmids carrying different mutated forms of the GCN5 and ELP3 genes were introduced, and the consequences for growth were tested by plasmid shuffle experiments (see Materials and methods for details) (Figure 5). The different versions of Elp3 expressed to similar levels and incorporated into Elongator when expressed from such constructs (Figure 2A), and the same was true for Gcn5 and SAGA (Kuo et al., 1998). As expected, expression of just one wild-type version of either Gcn5 or Elp3 partially rescued the slow-growth phenotype of the gcn5Δelp3Δ strain, while expression of both led to wild-type-like growth. However, if the HAT activity of both Gcn5 and Elp3 was impaired by a point mutation in the sequence encoding the respective catalytic domains (Kuo et al., 1998), cells continued to grow very poorly. These data strongly indicate that the severe phenotypes of the gcn5Δelp3Δ strain are specifically due to the concomitant absence of the enzymatic activities of Gcn5 and Elp3.
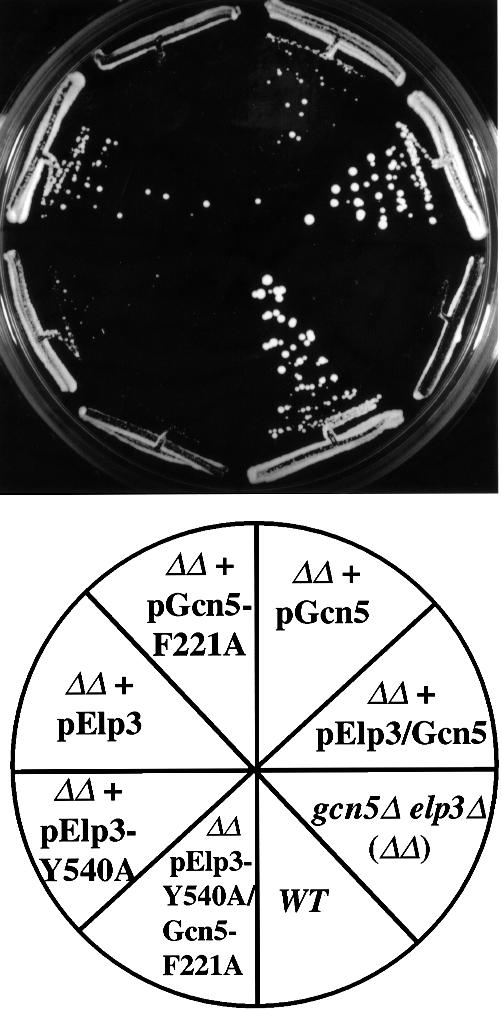
Fig. 5. Severe phenotypes of cells with concomitant disruption of Elp3 and Gcn5 HAT activity. The effects of altered HAT domains in GCN5 and ELP3 were tested in gcn5Δelp3Δ cells carrying wild-type ELP3 on a URA3-marked plasmid. Cells capable of losing the URA3 plasmid were grown on 5-FOA-containing synthetic media for 4–6 days at 30°C. See Materials and methods for details. The slight growth of gcn5Δelp3Δ cells carrying pELP3-Y540A was not observed in other similar experiments. gcn5Δelp3Δ cells expressing altered versions of the HAT domains were also temperature-sensitive and unable to grow on alternative carbon sources (data not shown).
Mutation of specific deacetylases can suppress the gcn5Δelp3Δ phenotype
Mutation of the gene encoding the HDAC Rpd3 has previously been shown to suppress phenotypes resulting from the gcn5 mutation (Perez-Martin and Johnson, 1998). The rpd3 mutation did not suppress elp3 phenotypes and rpd3Δgcn5Δelp3Δ cells had an even more severe growth defect than gcn5Δelp3Δ cells (data not shown). We also found that mutation of genes encoding other deacetylases, HDA1 and/or HOS2 (Rundlett et al., 1996), failed to suppress phenotypes resulting from single elp3Δ or gcn5Δ mutations (data not shown). In contrast, gcn5Δelp3Δ phenotypes could be partially suppressed by the mutation of HDA1 and almost entirely by further mutating HOS2 (Figure 6). These results support two conclusions: first, that HDACs are also functionally redundant; and secondly, that there is a fine-tuned, enzyme-regulated balance between acetylation and deacetylation in living cells, which can be shifted by mutating genes encoding the relevant enzymes.
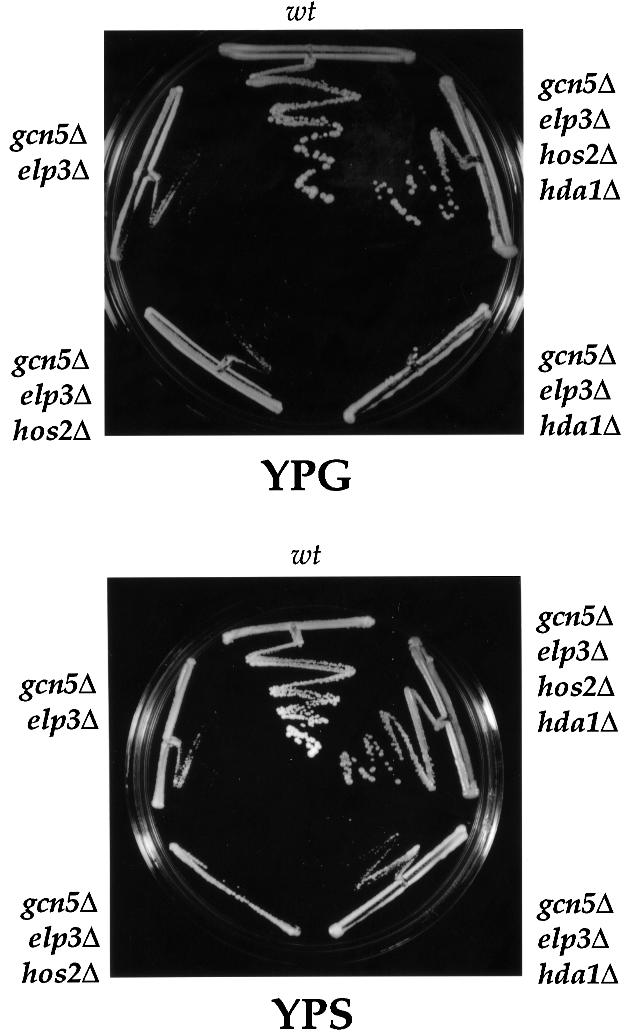
Fig. 6. The severe consequences of the gcn5Δelp3Δ mutation can be suppressed by mutation of specific HDACs. Mutation of HDA1 and HDA1 HOS2 suppresses the severe consequences of the gcn5Δelp3Δ mutation. Cells of the indicated genotype were streaked on YPG or YPS and incubated for 3–4 days at 30°C. Mutation of HDA1 and HOS2 also suppresses the temperature sensitivity (37°C) as well as the slow growth on YPD (30°C) of gcn5Δelp3Δ cells (data not shown).
Discussion
The findings reported here add to the understanding of the roles of HAT and HDACs in transcription in the following ways. First, they demonstrate a role for Elp3 acetyltransferase activity in chromatin modification and transcription in vivo. Secondly, they show that Gcn5 and Elp3 cannot be deleted simultaneously without severe effects on yeast growth and transcriptional activation. The partially redundant function of Elp3 and Gcn5 is exerted through the SAGA and Elongator complexes, as combinations of mutations in genes encoding other subunits of these complexes confer similar phenotypes. Moreover, it is the acetyltransferase activities of Gcn5 and Elp3 that are the determining factors for the phenotypes observed. The concomitant absence of these activities confers phenotypes similar to those observed after disruption of the entire open reading frames of the genes. Finally, the consequences of mutating both HATs can be counteracted by mutation of two, but not one, specific HDACs. This indicates the importance of a fine-tuned balance between acetylation and deacetylation in vivo, and points to the existence of functional redundancy among HDACs as well.
A role for Elongator in chromatin remodelling in vivo
Previously, we showed that Elp3 is an integral component of elongating RNAPII holoenzyme as a subunit of Elongator (Wittschieben et al., 1999). Motifs in Elp3 indicated that the protein is a HAT, and this was confirmed by an in vitro activity assay. The data presented here show that the acetyltransferase activity of Elp3 is essential for elongator function in vivo. Moreover, elp3 mutation is synthetically lethal with deletion of the tail of histone H4 (Δ4–19), and point mutation of sequences encoding transcription-related lysine residues results in temperature sensitivity in the elp3Δ background. A straightforward interpretation of the synthetic lethality resulting from ELP3 and H4 tail mutation would be that Elp3 primarily acetylates the H3 tail in vivo.
Our demonstration of a partially overlapping function for Elongator and SAGA might provide an attractive explanation as to why these complexes have remained highly conserved throughout evolution, yet are not encoded by essential genes in yeast: an important role for either may be at least partly substituted by that of the other. Cells lacking either GCN5 or ELP3 grow with a doubling time comparable to wild type (doubling times 90–110 min), but gcn5Δelp3Δ cells grow very slowly (doubling time >4 h). These cells are also unable to grow at elevated temperature, in the absence of inositol, or on galactose, sucrose and raffinose as the sole carbon source, indicating widespread transcription defects.
Functional redundancy among chromatin remodelling complexes: similarities and differences
The genetic interaction between Elongator and SAGA reported here provides evidence for redundancy specifically among HATs. A more widespread redundancy between chromatin remodellers has previously been suggested based on the results obtained by deleting SWI and ADA/GCN5 genes in the same cell (Pollard and Peterson, 1997; Roberts and Winston, 1997; Biggar and Crabtree, 1999). SWI/SNF and SAGA are likely to co-operate during promoter opening to allow activators and the basal transcription machinery access to DNA (Roberts and Winston, 1997; Pollard and Peterson, 1998; Biggar and Crabtree, 1999; Gregory et al., 1999; Sudarsanam et al., 1999). Several large transcription complexes, such as Mediator, Spt proteins, SWI/SNF, SAGA and Elongator, might well be partially redundant, or act in synergy, a concept previously articulated by Roberts and Winston (1997).
While the results of this study indicate that Gcn5 and Elp3 have a partially redundant function, there is also evidence to suggest that their normal role in transcription must be fundamentally different. First, the combination of swi/snf and gcn5/ada mutations gives rise to dramatic phenotypes (Pollard and Peterson, 1997; Roberts and Winston, 1997; Biggar and Crabtree, 1999), whereas the swi2ΔelpΔ strains display no clear, additional growth defect (data not shown). Secondly, while both swi and gcn5 phenotypes can be suppressed by deletion of one of the two sets of genes that encode histones H2A and H2B [(hta1-htb1)Δ] (Hirschhorn et al., 1992; Perez-Martin and Johnson, 1998), elp phenotypes cannot (data not shown). Thirdly, deletion of RPD3, encoding an HDAC that can be recruited to promoters by interactions with DNA-bound repressors (Kadosh and Struhl, 1997), leads to suppression of gcn5 phenotypes (Perez-Martin and Johnson, 1998), but does not result in suppression of the effects of the elp3 mutation. Taken together, these data, as well as functional data obtained in vitro (Cote et al., 1994; Imbalzano et al., 1994; Kwon et al., 1994; Utley et al., 1998; Otero et al., 1999), are consistent with the notion that the SWI/SNF complex and SAGA are primarily involved in assisting transcriptional activators and basal factors in their association with promoter sequences (Pollard and Peterson, 1998), while Elongator is likely to function mainly in elongation (Otero et al., 1999). It cannot be ruled out, however, that Elongator also plays a role during promoter remodelling, for example during its loading onto RNAPII at the initiation–elongation transition. Likewise, it is also possible that SAGA might play an as yet undefined role during transcript elongation through chromatin.
An acetylation–deacetylation balance and its importance for transcription in living cells
Our data indicate that the most likely explanation for the severe consequences of concomitantly disrupting GCN5 and ELP3 is the reduced acetylation of transcription-related lysine residues in histone tails. First, severe phenotypes are also observed in cells concomitantly expressing Gcn5 and Elp3 with single residue alterations in their HAT domains. Secondly, mutation of the deacetylases HDA1 and HOS2 in combination is sufficient to suppress gcn5Δelp3Δ phenotypes. These data suggest a significance of overall acetylation levels for transcription proficiency. It might indeed be relevant to think of the effects of deleting both Gcn5 and Elp3 in the light of the proposed existence of an enzyme-maintained acetylation–deacetylation equilibrium. In gcn5Δelp3Δ cells, this equilibrium may have become shifted too far towards deacetylation at certain genes. A shift towards deacetylation might create a local chromatin structure, a ‘chromatin micro-environment’, which is detrimental to the efficient opening of the chromatin structure required for access by transcription factors, as well as for efficient progress of the transcription machinery through the coding region at these genes.
Materials and methods
Yeast strains, media and DNA constructs
All the S.cerevisiae strains used for genetic analysis (Table II) were congenic with strain W303 (ade2 can1 his3 leu2 trp1 ura3) (Thomas and Rothstein, 1989), and were grown and manipulated essentially as described previously (Otero et al., 1999). Deletion of the open reading frame of yeast genes was performed as described (Lorenz et al., 1995). ELP2 is described elsewhere (Fellows et al., 2000). Plates used to investigate the ability of strains to ferment galactose, raffinose and sucrose contained 1 µg/ml antimycin A (Sigma).
Table II. Yeast strains.
| Strain | Genotype | Reference/source |
|---|---|---|
| JSY130 | MATa elp3Δ::LEU2. | Wittschieben et al. (1999) |
| JSY131 | MATα elp3Δ::LEU2. | Wittschieben et al. (1999) |
| JSY141 | MATa gcn5Δ::HIS3 | this study |
| JSY142 | MATα gcn5Δ::HIS3 | this study |
| JSY143 | MATa elp3Δ::LEU2 gcn5Δ::HIS3 | this study |
| JSY144 | MATα elp3Δ::LEU2 gcn5Δ::HIS3 | this study |
| JSY411 | MATα elp1Δ::LEU2 gcn5Δ::HIS3 | this study |
| JSY150 | MATa elp2Δ::LEU2 gcn5Δ::HIS3 | this study |
| JSY151 | MATα elp2Δ::LEU2 gcn5Δ::HIS3 | this study |
| JSY330 | MATα elp3Δ::LEU2 spt3Δ::HIS3 | this study |
| JSY331 | MATa elp3Δ::LEU2 spt3Δ::HIS3 | this study |
| JSY332 | MATa spt3Δ::HIS3 | this study |
| JSY456 | MATa ahc1Δ::HIS3 | this study |
| JSY457 | MATα ahc1Δ::HIS3 | this study |
| JSY458 | MATa elp3Δ::LEU2 ahc1Δ::HIS3 | this study |
| JSY459 | MATα elp3Δ::LEU2 ahc1Δ::HIS3 | this study |
| JSY407 | MATa spt20Δ::HIS3 | this study |
| JSY408 | MATα spt20Δ::HIS3 | this study |
| JSY409 | MATa elp3Δ::LEU2 spt20Δ::HIS3 | this study |
| JSY410 | MATα elp3Δ::LEU2 spt20Δ::HIS3 | this study |
| DY5939 | MATa elp3::LEU2 gcn5::HIS3 hda1::URA3 | this study |
| DY5937 | MATa elp3::LEU2 gcn5::HIS3 hos2::TRP1 | this study |
| DY5935 | MATa elp3::LEU2 gcn5::HIS3 hos2::TRP1 hda1::URA3 | this study |
| DY5993 | MATα elp3::LEU2 gcn5::HIS3 rpd3::KanMX | this study |
| DY5733 | MATa ade2 can1 his3 leu2 lys2 trp1 ura3 hht1-hhf1Δ::LEU2 hht2-hhf2Δ::kanMX3 + YCp50:HHT2-HHF2 (URA3) | this study |
| DY5734 | MATα ade2 can1 his3 leu2 lys2 trp1 ura3 hht1-hhf1Δ::LEU2 hht2-hhf2Δ::kanMX3 + YCp50:HHT2-HHF2 (URA3) | this study |
| JSY316 | MATα ade2 can1 his3 leu2 lys2 trp1 ura3 elp3::ADE2 hht1-hhf1Δ::LEU2 hht2-hhf2Δ::kanMX3 + YCp50:HHT2-HHF2 (URA3) | this study |
ELP3 coding sequence with upstream regulatory sequences as well as downstream terminator sequences was copied from yeast genomic DNA using Pfu polymerase (Stratagene) and primers containing BamHI and KpnI sites, respectively. Products were cloned into the BamHI and KpnI sites of pRS316 (Sikorski and Hieter, 1989) to create the plasmid pBOP60-13 (pWT). Single point mutations in sequences encoding the HAT domain of Elp3 were created by PCR mutagenesis using primers encoding the mutation as well as BamHI or SalI sites. Mutated versions of the HAT catalytic domain were then exchanged with wild-type fragments, yielding pBOP60-14 (pY540A) and pBOP60-15 (pY541A). Mutations that had been introduced were confirmed by sequencing. TRP1-marked plasmids concomitantly expressing Gcn5 and Elp3 alleles from their cognate promoters were constructed by cloning PCR products generated from pBOP60-13 and pBOP60-14 with EagI ends into the EagI site of pRS314 (Sikorski and Hieter, 1989), pMK125 and pMK125 F221A (Kuo et al., 1998). These plasmids were named pBOP60-26 (wild-type Elp3), pBOP60-31 (Elp3 Y540A), pBOP60-32 (wild-type Elp3; wild-type Gcn5) and pBOP60-35 (Elp3 Y540A; Gcn5 F221A). The experiments described were performed using plasmids where the ELP3 gene pointed towards the GCN5 gene, but similar results were obtained with either orientation. Details of cloning procedures are available on request.
Plasmid shuffle experiments
Plasmid shuffle of histone alleles carried on TRP1-marked plasmids into ELP3 (DY5734) or elp3Δ (JSY316) cells carrying wild-type histones on a URA3-marked plasmid was performed as described (Zhang et al., 1998). Concomitant experiments with similar gcn5Δ cells confirmed the results previously reported using another parental strain (Zhang et al., 1998). S.Roth (University of Texas) kindly provided all the histone plasmids and the parental histone shuffle strain used to construct a W303 counterpart. After growth in URA+ media, cells that had lost the URA3 plasmid were selected on 5-fluoroorotic acid (5-FOA)-containing media. Survivors were checked for the loss of ability to grow on ura– plates. For temperature sensitivity assays, cells that had lost the wild-type URA3-marked histone alleles on 5-FOA were plated on yeast extract/peptone/dextrose (YPD) and incubated at 37°C for 3–4 days.
We repeatedly observed that the gcn5Δelp3Δ strain very rapidly picked up suppressor mutations. This was further enhanced when experiments with point-mutated versions of the genes were attempted, and we therefore adopted the plasmid shuffle technique to circumvent the problem. gcn5Δelp3Δ cells carrying wild-type ELP3 on a URA3-marked plasmid were transformed with TRP1-marked plasmids concomitantly expressing ELP3 and GCN5 HAT alleles. These cells were thus phenotypically ELP3 gcn5Δ. Cells that allowed the loss of the URA3 plasmid were selected on 5-FOA-containing media. This approach allowed a reproducible comparison of the severity of deleting GCN5 and ELP3 with merely mutating the respective HAT domains.
In-gel HAT assay
HAT activity assays of Elp3 expressed in insect cells were performed as described (Wittschieben et al., 1999) using either bovine serum albumin (BSA) or core histones (Sigma). Gels were typically exposed to autoradiograms for 3–6 days to reveal Elp3-positive HAT signals. Relative amounts of wild-type and mutant Elp3 to be used for the activity comparisons were determined by western blotting of dilution series from the respective HAT fractions. Quantification of acetylation levels was performed by scanning autoradiographs with a Umax Powerlook 2000 scanner, using NIH Image 1.61 software for data processing. The relatively modest decreases in the activity of Elp3 HAT mutants compared with those obtained by a similar mutation in Gcn5 (Kuo et al., 1998) might at least partly be a reflection of the different assays used (in-gel assay at high protein concentration versus liquid assay).
Acknowledgments
Acknowledgements
We would like to thank the Imperial Cancer Research Fund (ICRF) service facilities, particularly the fermentation and photography services, for their help. We also thank Sharon Roth for providing plasmids and strains and Alain Verreault, Peter Verrijzer, Brad Cairns, Rick Wood and John Diffley for carefully reading and commenting on various versions of the manuscript. The project was supported by grants from the ICRF and the Human Frontier Science Project (RG0193/97) to J.Q.S., an EU fellowship to B.Ø.W and a National Institutes of Health grant (GM39067) to D.J.S.
References
- Angus-Hill M.L., Dutnall,R.N., Tafrov,S.T., Sternglanz,R. and Ramakrishnan,V. (1999) Crystal structure of the histone acetyltransferase Hpa2: a tetrameric member of the Gcn5-related N-acetyltransferase superfamily. J. Mol. Biol., 294, 1311–1325. [DOI] [PubMed] [Google Scholar]
- Ayer D.E. (1999) Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol., 9, 193–198. [DOI] [PubMed] [Google Scholar]
- Biggar S.R. and Crabtree,G.R. (1999) Continuous and widespread roles for the Swi–Snf complex in transcription. EMBO J., 18, 2254–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeden L. and Nasmyth,K. (1987) Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell, 48, 389–397. [DOI] [PubMed] [Google Scholar]
- Brown C.E., Lechner,T., Howe,L. and Workman,J.L. (2000) The many HATs of transcription coactivators. Trends Biochem. Sci., 25, 15–19. [DOI] [PubMed] [Google Scholar]
- Brownell J.E. and Allis,C.D. (1995) An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc. Natl Acad. Sci. USA, 92, 6364–6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell J.E., Zhou,J., Ranalli,T., Kobayashi,R., Edmondson,D.G., Roth,S.Y. and Allis,C.D. (1996) Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell, 84, 843–851. [DOI] [PubMed] [Google Scholar]
- Cairns B.R. (1998) Chromatin remodelling machines: similar motors, ulterior motives. Trends Biochem. Sci., 23, 20–25. [DOI] [PubMed] [Google Scholar]
- Cosma M.P., Tanaka,T. and Nasmyth,K. (1999) Ordered recruitment of transcription and chromatin remodelling factors to a cell cycle- and developmentally-regulated promoter. Cell, 97, 299–311. [DOI] [PubMed] [Google Scholar]
- Cote J., Quinn,J., Workman,J.L. and Peterson,C.L. (1994) Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science, 265, 53–60. [DOI] [PubMed] [Google Scholar]
- Dutnall R.N., Tafrov,S.T., Sternglanz,R. and Ramakrishnan,V. (1998) Structure of the histone acetyltransferase Hat1: a paradigm for the GCN5-related N-acetyltransferase superfamily. Cell, 94, 427–438. [DOI] [PubMed] [Google Scholar]
- Eberharter A., Sterner,D.E., Schieltz,D., Hassan,A., Yates,J.R.,III, Berger,S.L. and Workman,J.L. (1999) The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 6621–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellows J., Erdjument-Bromage,H., Tempst,P. and Svejstrup,J. (2000) The Elp2 subunit of Elongator and elongating RNA polymerase II holoenzyme is a WD40 protein. J. Biol. Chem., 275, 12896–12899. [DOI] [PubMed] [Google Scholar]
- Georgakopoulos T. and Thireos,G. (1992) Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J., 11, 4145–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P.A. et al. (1997) Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev., 11, 1640–1650. [DOI] [PubMed] [Google Scholar]
- Gregory P.D., Schmid,A., Zavari,M., Lui,L., Berger,S.L. and Horz,W. (1998) Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Mol. Cell, 1, 495–505. [DOI] [PubMed] [Google Scholar]
- Gregory P.D., Schmid,A., Zavari,M., Munsterkotter,M. and Horz,W. (1999) Chromatin remodelling at the PHO8 promoter requires SWI–SNF and SAGA at a step subsequent to activator binding. EMBO J., 18, 6407–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn J.N., Brown,S.A., Clark,C.D. and Winston,F. (1992) Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev., 6, 2288–2298. [DOI] [PubMed] [Google Scholar]
- Imbalzano A.N., Kwon,H., Green,M.R. and Kingston,R.E. (1994) Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature, 370, 481–485. [DOI] [PubMed] [Google Scholar]
- Kadosh D. and Struhl,K. (1997) Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell, 89, 365–371. [DOI] [PubMed] [Google Scholar]
- Kleff S., Andrulis,E.D., Anderson,C.W. and Sternglanz,R. (1995) Identification of a gene encoding a yeast histone H4 acetyltransferase. J. Biol. Chem., 270, 24674–24677. [DOI] [PubMed] [Google Scholar]
- Krebs J.E., Kuo,M.H., Allis,C.D. and Peterson,C.L. (1999) Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev., 13, 1412–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M.H., Zhou,J., Jambeck,P., Churchill,M.E. and Allis,C.D. (1998) Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev., 12, 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H., Imbalzano,A.N., Khavari,P.A., Kingston,R.E. and Green,M.R. (1994) Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature, 370, 477–481. [DOI] [PubMed] [Google Scholar]
- Lorenz M.C., Muir,R.S., Lim,E., McElver,J., Weber,S.C. and Heitman,J. (1995) Gene disruption with PCR products in Saccharomyces cerevisiae. Gene, 158, 113–117. [DOI] [PubMed] [Google Scholar]
- Lucchini G., Hinnebusch,A.G., Chen,C. and Fink,G.R. (1984) Positive regulatory interactions of the HIS4 gene of Saccharomyces cerevisiae. Mol. Cell. Biol., 4, 1326–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzen C.A. and Allis,C.D. (1998) Linking histone acetylation to transcriptional regulation. Cell. Mol. Life Sci., 54, 6–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald A.F. and Landsman,D. (1997) GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci., 22, 154–155. [DOI] [PubMed] [Google Scholar]
- Otero G., Fellows,J., Li,Y., de Bizemont,T., Dirac,A.M.G., Gustafsson,C.M., Erdjument-Bromage,H., Tempst,P. and Svejstrup, J.Q. (1999) Elongator, a multi-subunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell, 3, 109–118. [DOI] [PubMed] [Google Scholar]
- Parthun M.R., Widom,J. and Gottschling,D.E. (1996) The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell, 87, 85–94. [DOI] [PubMed] [Google Scholar]
- Perez-Martin J. and Johnson,A.D. (1998) Mutations in chromatin components suppress a defect of Gcn5 protein in Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 1049–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard K.J. and Peterson,C.L. (1997) Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol. Cell. Biol., 17, 6212–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard K.J. and Peterson,C.L. (1998) Chromatin remodelling: a marriage between two families? BioEssays, 20, 771–780. [DOI] [PubMed] [Google Scholar]
- Reifsnyder C., Lowell,J., Clarke,A. and Pillus,L. (1996) Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nature Genet., 14, 42–49. [DOI] [PubMed] [Google Scholar]
- Roberts S.M. and Winston,F. (1997) Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics, 147, 451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas J.R., Trievel,R.C., Zhou,J., Mo,Y., Li,X., Berger,S.L., Allis,C.D. and Marmorstein,R. (1999) Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature, 401, 93–98. [DOI] [PubMed] [Google Scholar]
- Ruiz-Garcia A.B., Sendra,R., Galiana,M., Pamblanco,M., Perez-Ortin, J.E. and Tordera,V. (1998) HAT1 and HAT2 proteins are components of a yeast nuclear histone acetyltransferase enzyme specific for free histone H4. J. Biol. Chem., 273, 12599–12605. [DOI] [PubMed] [Google Scholar]
- Rundlett S.E., Carmen,A.A., Kobayashi,R., Bavykin,S., Turner,B.M. and Grunstein,M. (1996) HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl Acad. Sci. USA, 93, 14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.R., Eisen,A., Gu,W., Sattah,M., Pannuti,A., Zhou,J., Cook,R.G., Lucchesi,J.C. and Allis,C.D. (1998) ESA1 is a histone acetyl transferase that is essential for growth in yeast. Proc. Natl Acad. Sci. USA, 95, 3561–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg P.W., Stern,M.J., Clark,I. and Herskowitz,I. (1987) Activation of the yeast HO gene by release from multiple negative controls. Cell, 48, 567–577. [DOI] [PubMed] [Google Scholar]
- Sterner D.E., Grant,P.A., Roberts,S.M., Duggan,L.J., Belotserkovskaya, R., Pacella,L.A., Winston,F., Workman,J.L. and Berger,S.L. (1999) Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol., 19, 86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- Sudarsanam P., Cao,Y., Wu,L., Laurent,B.C. and Winston,F. (1999) The nucleosome remodelling complex, Snf/Swi, is required for the maintenance of transcription in vivo and is partially redundant with the histone acetyltransferase, Gcn5. EMBO J., 18, 3101–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B.J. and Rothstein,R. (1989) Elevated recombination rates in transcriptionally active DNA. Cell, 56, 619–630. [DOI] [PubMed] [Google Scholar]
- Travers A. (1999) Chromatin modification by DNA tracking. Proc. Natl Acad. Sci. USA, 96, 13634–13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utley R.T., Ikeda,K., Grant,P.A., Cote,J., Steger,D.J., Eberharter,A., John,S. and Workman,J.L. (1998) Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature, 394, 498–502. [DOI] [PubMed] [Google Scholar]
- Wade P.A., Pruss,D. and Wolffe,A.P. (1997) Histone acetylation: chromatin in action. Trends Biochem. Sci., 22, 128–132. [DOI] [PubMed] [Google Scholar]
- Wang L., Liu,L. and Berger,S.L. (1998) Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev., 12, 640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben B.O. et al. (1999) A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell, 4, 123–128. [DOI] [PubMed] [Google Scholar]
- Workman J.L. and Kingston,R.E. (1998) Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem., 67, 545–579. [DOI] [PubMed] [Google Scholar]
- Zhang W., Bone,J.R., Edmondson,D.G., Turner,B.M. and Roth,S.Y. (1998) Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J., 17, 3155–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]