Abstract
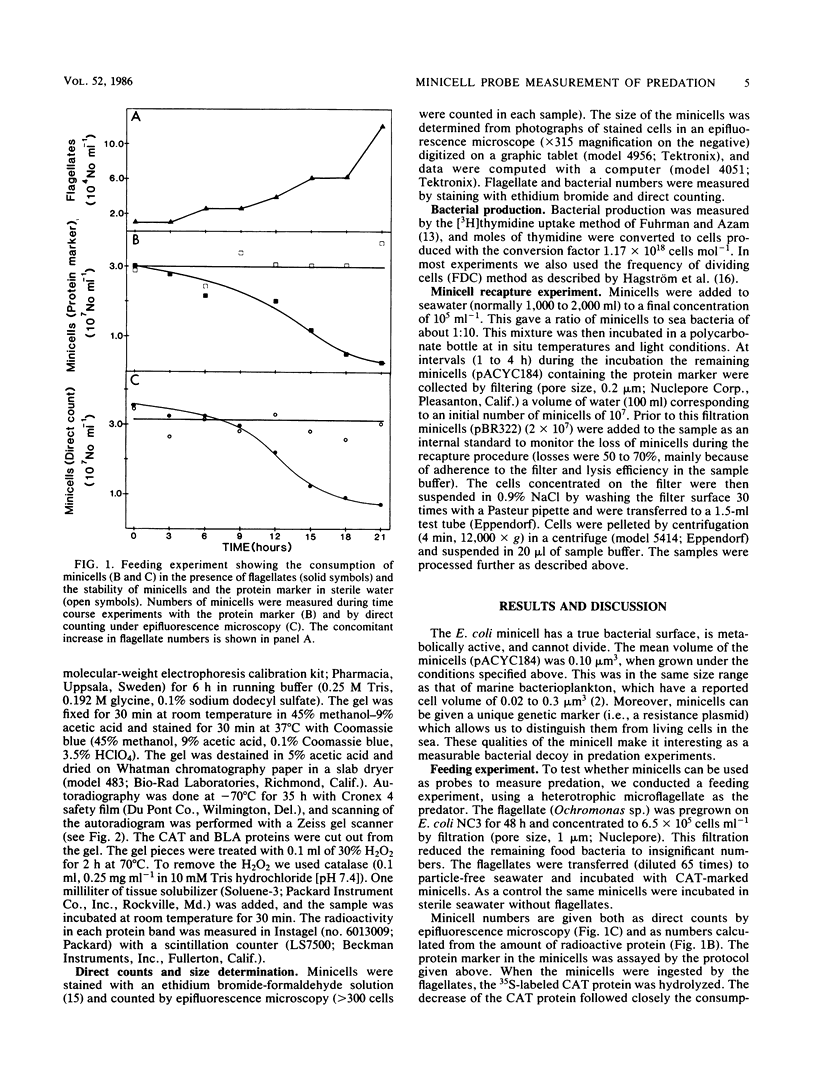
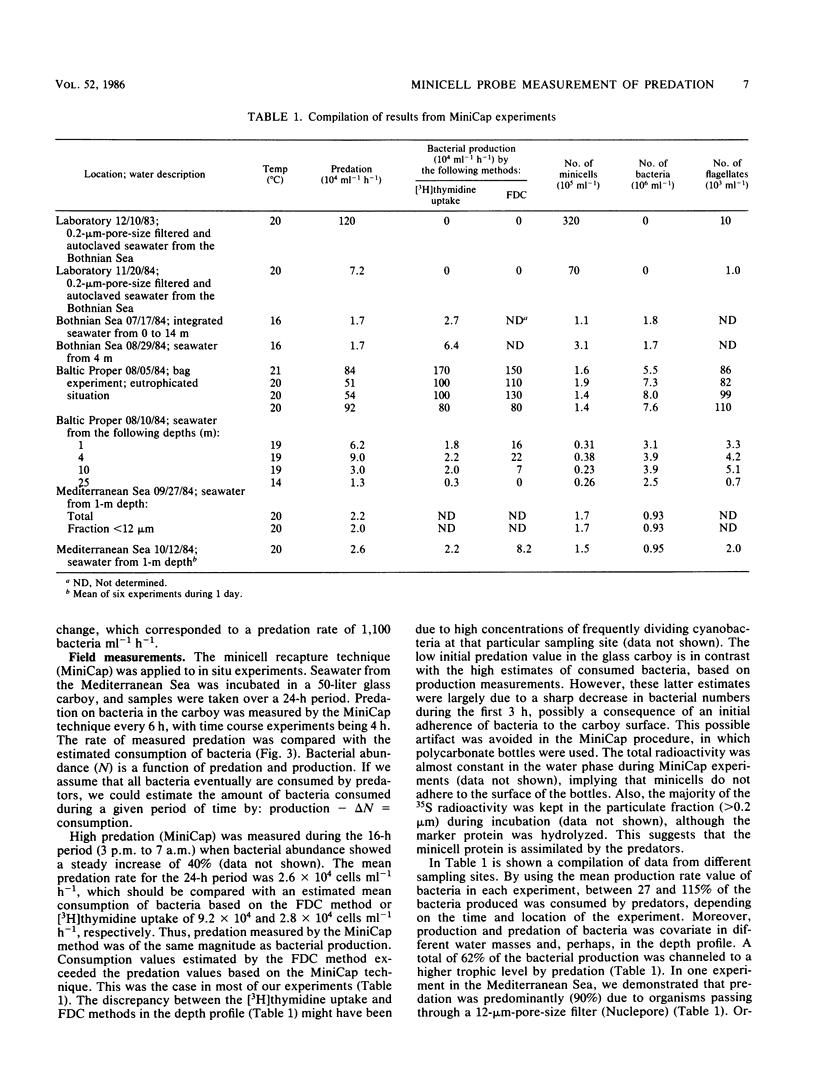
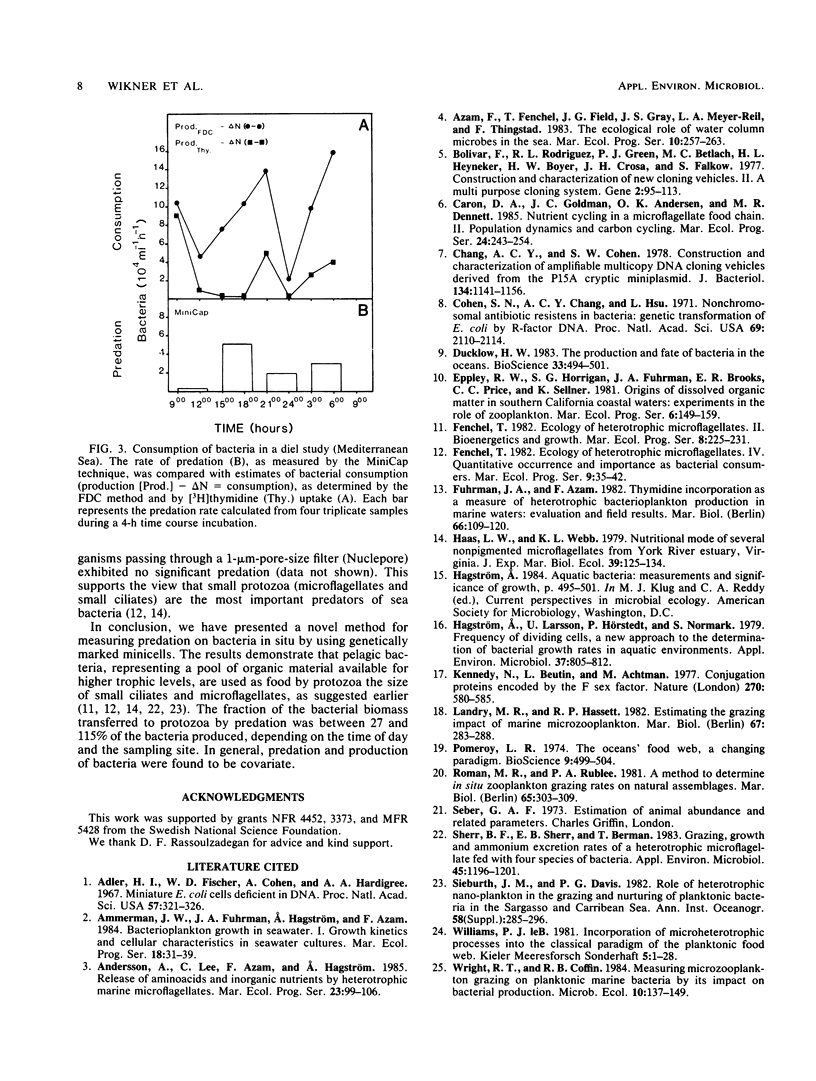
Minicells produced by Escherichia coli M2141 were used as probes to measure predation on pelagic bacteria in situ. The minicells, labeled with [35S]methionine in one specific protein, were shown to disappear in the presence of a microflagellate (Ochromonas sp.), as seen by a decrease in the amount of labeled marker protein with time. Incubation in filtered (pore size, 0.2 μm) and autoclaved seawater did not affect the amount of labeled marker protein in the minicell. The generation time of flagellates feeding on minicells was determined to be similar to that found for flagellates grown on seawater bacteria or living E. coli NC3. Data indicate that minicells are seen as true food particles by the flagellates. The minicell probe was used in recapture experiments, in which predation in situ on pelagic bacteria was demonstrated. The rate of bacterial production showed a clear covariation with the rate of predation, both in different sea areas and in depth profiles. The obtained results (11 field experiments) showed that the rate of predation, on average, accounts for the consumption of 62% of the bacteria produced.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler H. I., Fisher W. D., Cohen A., Hardigree A. A. MINIATURE escherichia coli CELLS DEFICIENT IN DNA. Proc Natl Acad Sci U S A. 1967 Feb;57(2):321–326. doi: 10.1073/pnas.57.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagström A., Larsson U., Hörstedt P., Normark S. Frequency of dividing cells, a new approach to the determination of bacterial growth rates in aquatic environments. Appl Environ Microbiol. 1979 May;37(5):805–812. doi: 10.1128/aem.37.5.805-812.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy N., Beutin L., Achtman M., Skurray R., Rahmsdorf U., Herrlich P. Conjugation proteins encoded by the F sex factor. Nature. 1977 Dec 15;270(5638):580–585. doi: 10.1038/270580a0. [DOI] [PubMed] [Google Scholar]
- Sherr B. F., Sherr E. B., Berman T. Grazing, growth, and ammonium excretion rates of a heterotrophic microflagellate fed with four species of bacteria. Appl Environ Microbiol. 1983 Apr;45(4):1196–1201. doi: 10.1128/aem.45.4.1196-1201.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]




