Abstract
Interleukin-1 (IL-1) is a potent regulator of cell proliferation, inflammation, and contraction of cardiovascular cells. It has been proposed that the IL-1/IL-1ra (IL-1 receptor antagonist) ratio influences these functions. Other members of the IL-1 family and the related caspase-1 also contribute to regulation of IL-1–mediated functions. We determined the mRNA expression of caspase-1, caspase-3, IL-1α , IL-1β , IL-18, IL-1 receptor type I (IL-1-RI), and IL-1ra in left ventricle tissue of hearts from patients with ischemic or dilated cardiomyopathy (ICM or DCM) and in control tissues from unused donor transplant hearts in RT-PCR experiments. We show that the expression of caspase-1, caspase-3, IL-1β , and IL-1-RI mRNA was not different between patients and control tissues. Furthermore, we did not find detectable amounts of IL-1α mRNA in any of these adult myocardial tissues. On the other hand, expression of IL-18 RNA was lower in myocardium of both patient groups compared with control hearts. Furthermore, IL-1ra mRNA expression was significantly lower in tissues of DCM patients compared with ICM patients and controls. This was in line with a trend towards lower IL-1ra protein levels in myocardial tissues of DCM patients. In contrast with the adult tissues discussed above, which did not express IL-1α mRNA, commercially available human fetal tissue expressed IL-1α mRNA. On the other hand IL-1β mRNA was present in fetal and in adult human heart tissue. Our data provide evidence for an altered ratio of IL-1/IL-1ra in DCM patients. This dysregulation may contribute to pathogenesis and/or progression of heart disease by modulating the otherwise balanced IL-1–mediated functions in cardiovascular cells.
INTRODUCTION
Proinflammatory cytokines are potent modulators of cardiovascular diseases such as acute myocardial infarction, atherosclerosis, and chronic heart failure (1–6). Accordingly, altered levels of cytokines have been observed in the plasma of patients with various heart diseases (7–10). The role of plasma cytokines in the pathogenesis of heart diseases still remains to be elucidated in more detail. In addition to plasma cytokine measurements, detection of cytokine mRNA in heart tissues may also add information regarding the significance of these mediators in various heart diseases. It has been reported that, besides many other cytokines, tumor necrosis factor-α (TNF-α ) (11,12) and interleukin-1 (IL-1) (13,14) are expressed in the heart. Both mediators are potently involved in innate immune responses and may contribute to regulation of cardiovascular diseases. IL-1β mRNA and protein expression has been shown in myocardium of patients with dilated cardiomyopathy (DCM) (14,15), viral myocarditis (16,17) and in coronary arteries or within the myocardium of patients with ischemic heart disease (14,18). Furthermore, in animal models, IL-1β was detected in hearts of mice with myocarditis following inoculation with coxsackie B3 virus (19) and in rat hearts following myocardial infarction (20).
IL-1 is thought to contribute to regulation of proliferation, inflammation, and contractility of cardiovascular cells (21,22). It has been shown that IL-1 stimulates proliferation of vascular smooth muscle cells (23). It potently induces cytokine production of vessel wall or heart cells (24–26), as well as expression of chemokines and adhesion molecules (27). Furthermore, IL-1 also exerts negative inotropic effects on whole hearts (28), isolated muscle preparations (29,30), and cultured cardiac myocytes (31,32). Besides the above functions, IL-1 can also regulate levels of intracellular Ca2+ of myocytic cells (33) and induce apoptosis in cardiac myocytes (34,35).
A variety of proteins belong to the IL-1 family, such as IL-1α and IL-1β , IL-18, IL-33, IL-1 receptors and related proteins, IL-1 receptor antagonist (IL-1ra), as well as caspase-1. IL-1 is a central mediator in the cytokine network (36), important for the regulation of innate and/or inflammatory processes. These processes contribute to cardiovascular diseases including atherosclerosis (37–39). Both IL-1 isoforms (IL-1α and IL-1β ) are produced as precursor proteins. IL-1α is active in the precursor and the mature form, whereas IL-1β is only active in the mature, caspase-1–cleaved form. IL-1α preferentially remains cell or cell-surface-associated, whereas mature IL-1β is released. Another agonist of the IL-1 family is IL-18, a protein related to IL-1 in terms of its structure, receptor class, and the fact that it is also activated by the same enzyme as IL-1β , the caspase-1 [IL-1β –converting enzyme (ICE)]. Another recently described caspase-1 substrate belonging to the IL-1 family is IL-33, which binds to the IL-1 family receptor ST2 (40). Caspase-1, the enzyme cleaving IL-1β , IL-18, and IL-33, was the 1st caspase reported, and a number of other caspases have been described subsequently. Some of them are preferentially involved in inflammatory processes (caspases 1, 4, and 5); some, such as caspase-3, are preferentially involved in regulation of apoptosis. These enzymes are activated by enzymatic processes in multiprotein complexes, the “inflammasomes” or “apoptosomes.” Like its substrates IL-1β , IL-18, and IL-33, caspase-1 is cleaved after aspartate.
Besides the agonists mentioned above, a unique antagonist, the IL-1 receptor antagonist (IL-1ra), belongs to the IL-1 family. This antagonist exists in a released form and in additional intracellular forms resulting from differential splicing. IL-1ra binds to the same receptor as the agonists IL-1α and IL-1β , thereby blocking the binding of both IL-1 isoforms. Binding of IL-1ra to the receptor does not result in activation of signal transduction. Thus, the IL-1 receptor antagonist is an important regulator of IL-1–induced functions. The important role of this cytokine in the maintenance of normal cellular functions was emphasized in experiments with IL-1ra knockout animals. Both, complete removal of IL-1ra in IL-1ra–/– mice, as well as the diminished IL-1ra content in IL-1ra+/– heterozygotes resulted in the onset of spontaneous inflammatory and autoimmune processes in the animals (41,42). A variety of diseases have been associated with an IL-1/IL-1ra imbalance. Thus, the decreased mucosal ratio of IL-1ra to IL-1 in patients with inflammatory bowel disease, resulting from enhanced IL-1β expression, correlated with the severity of the disease (43). A role of the IL-1/IL-1ra ratio in myocarditis was derived from transfection experiments with infected rats, where transfection with an IL-1ra–Ig chimera resulted in reduced myocarditis area, reduced ANP expression, and improved cardiac function (44). In leukocytes of patients with chronic myelogenous leukemia, IL-1β production in monocytes of patients was enhanced, compared with healthy controls, and was associated with disease progression. The IL-1ra levels were not altered in these patients, whereas in the accelerated phase of the disease IL-1ra was lower, thereby contributing to the advancement of the disease (45). Furthermore, an altered IL-1/IL-1ra ratio was present in supernatants of pieces of cultured synovium derived from patients with rheumatoid arthritis (46). The imbalance resulted from enhanced IL-1ra and reduced IL-1β release in response to IL-4 and IL-10, the latter reducing IL-1β but not IL-1ra production.
It is obvious that the IL-1 function is regulated at various levels. Its function in tissues is modulated not only by its own concentration or by synergism with other cytokines, but also by 1) the activation of IL-1β through caspase-1, 2) the presence of functional receptors, and 3) the expression of its inhibitor, IL-1ra. Thus, a synergistic action of IL-1 and IL-18 on myocardial function has been proposed recently (47), and a role of IL-1ra in the maintenance of normal cardiac function has been suggested (48). Although some researchers have investigated the expression of single members of the IL-1 and caspase families, their expression has not been compared in patients suffering from ischemic or dilated cardiomyopathy (ICM or DCM).
In the present study we analyze the mRNA expression of the IL-1 family members IL-1α , IL-1β , IL-1ra, IL-1-R1, and IL-18, as well as caspase-1 and caspase-3 in non-failing human donor heart tissue and in myocardium of patients with ischemic or dilated cardiomyopathy. We demonstrate reduced IL-1ra mRNA expression in left ventricular tissue of patients with dilated cardiomyopathy, as compared to tissues of patients with ischemic cardiomyopathy and tissues of donor hearts. IL-18 mRNA expression was lower in both patient groups. Expression of IL-1β , IL-1-RI, caspase-1 and caspase-3 was not significantly different in the groups studied. IL-1α mRNA was detectable only in human fetal myocardium, but not in the adult tissues. The results indicate that in patients suffering from DCM the IL-1/IL-1ra ratio may be altered by the reduced expression of IL-1ra. The lower IL-1ra expression, accompanied by the unaltered IL-1β expression, may result in enhanced inflammation, apoptosis and disturbance of contractility in the diseased heart.
MATERIALS AND METHODS
Patients and Myocardial Tissue Samples
For the present investigations, we used biopsies of left ventricular tissues from explanted hearts of patients with terminal stages of ICM or DCM. They were compared with tissues of control hearts of organ donors scheduled for transplantation, which were not used for technical reasons. The use of these tissues was approved by the local ethics committee. Six male patients and 2 female patients (53.9 ± 6.6 years) had ICM (Table 1), 9 male patients and 1 female patient (50.1 ± 9.2 years) suffered from DCM, and 9 organ donors (6 male, 3 female; 42.6 ± 14.5 years) served as nonfailing controls. The “Human Cardiovascular Multiple Tissue cDNA (MTCTM) Panel,” obtained from Clontech (cat. no. K1427-1; Clontech Laboratories Inc; Palo Alto, CA, USA), served as a source of fetal human tissue.
Table 1.
Patient data
| Control | ICM | DCM | |
|---|---|---|---|
| n | 9 | 8 | 10 |
| Sex, M/F | 6/3 | 6/2 | 9/1 |
| Age, years | 42.6 ± 14.5 | 53.9 ± 6.6 | 50.1 ± 9.2 |
| Hemodynamics | |||
| EF, % | 25.0 ± 1.0 | 26.0 ± 8.0 | |
| NYHA grade | 3.5 ± 0.2 | 3.7 ± 0.2 | |
| Heart rate, bpm | 83.0 ± 7.0 | 80.0 ± 8.0 | |
| PCW, mmHg | 16.0 ± 3.0 | 20.0 ± 3.0 | |
| MAP, mmHg | 75.0 ± 2.0 | 79.0 ± 2.0 | |
| Cardiac index | 2.5 ± 0.5 | 2.0 ± 0.2 | |
| Comorbidity, n | |||
| Atrial fibrillation | 1 | 7 | |
| Hypertension | 4a | 1 | |
| Diabetes | 4 | 1 | |
| Hyperlipidemia | 5 | 2 | |
| Pre-explantation treatments, n | |||
| Beta-blockers | 5 | 7 | |
| Glycoside | 4 | 8 | |
| Other antiarrhythmics | 1 | 3 | |
| Anticoagulation | 5 | 10 | |
| ACE inhibitor/AT1 blocker | 8 | 9 | |
| Nitrate | 7 | 6 | |
| Lipid-lowering drug | 5 | 2 | |
Hypertension was not relevant for cardiac failure according to the judgments of the treating physician and the explanting surgeon. ACE, angiotensin converting enzyme; AT1, angiotensin II type 1 receptor; EF, ejection fraction; MAP, mean arterial pressure; NYHA, New York Heart Association score; PCW, pulmonary capillary wedge pressure.
Reverse-Transcription Polymerase Chain Reaction
Total RNA was isolated from the specimen by mechanical pulverization after storage in liquid nitrogen, following the protocol of Chirgwin et al. (49). RNA yield and purity were determined by UV-spectrophotometry, and total RNA (1 μg) was reverse-transcribed in a 20-μL reaction using SuperScript Plus reverse transcriptase (Gibco BRL). The complete RT-sample was diluted 1:5 with water, and 5 μL of this sample was used as template for PCR, using Taq DNA Polymerase (Gibco BRL) in a volume of 50 μL. RT and PCR reactions were performed according to the manufacturer’s recommendations. Primer sequences, expected sizes of PCR products, type of PCR protocols used, annealing temperatures, and number of the respective cycles are given in Table 2. RNA that had not been reverse-transcribed was used as a control. It did not yield PCR products using 18S rRNA primers, indicating that the samples did not contain contaminating genomic DNA. Each experiment also included a negative control (buffer alone), which was always negative and a positive control consisting of cDNA derived from SMC mRNA. The PCR conditions were not within the plateau phase of amplification, as determined in preliminary experiments (data not shown). The PCR products were separated in 1.6% agarose gels containing ethidium bromide and visualized by UV-transillumination.
Table 2.
Primers used for RT-PCR
| Primer sequence (5′-3′ ) | PCR product size, bp | Protocola | Primer annealing, °C | PCR cycles | |
|---|---|---|---|---|---|
| IL-1α | ATG-GCC-AAA-GTT-CCA-GAC-ATG
TAC-GCC-TGG-TTT-TCC-AGT-ATC-TG |
817 | I | 56 | 30 |
| IL-1β | GAG-GAT-GAC-TTG-TTC-TTT-GAA-G
GTT-GCT-CAT-CAG-AAT-GTG-GGA-G |
1096 | I | 50 | 29 |
| IL-1ra | ATG-GCT-TTA-GAG-ACG-ATC-TGC
AGT-AGA-ATT-TGG-TGA-CCA-TGA-C |
465 | III | 56 | 23 |
| IL-18 | ATA-TGG-ATC-CAT-GGC-TGC-TGA-ACC-AGT-AG
ATA-TCT-GCA-GCT-AGT-CTT-CGT-TTT-GAA-CAG |
604 | II | 60 | 30 |
| IL-1-RI | ATG-AGA-CAA-TGG-AAG-TAG-AC
TAG-ATG-AAA-ACA-GAA-CAC-AC |
372 | III | 56 | 21 |
| Caspase-1 | ATA-TGG-ATC-CGA-CAA-CCC-AGC-TAT-GCC-C
ATA-TCT-GCA-GAT-GTC-CTG-GGA-AGA-GGT-A |
880 | III | 56 | 22 |
| Caspase-3 | ATA-AAG-GGA-TCC-ATG-GAG-AAC-ACT-GAA-AAC-TCA-G
AAC-CTG-CAG-TTA-GTG-ATA-AAA-ATA-GAG-TTC-TTT-TGT-GAG-C |
857 | II | 64 | 30 |
| 18S rRNA | GTT-GGT-GGA-GCG-ATT-TGT-CTG-G
AGG-GCA-GGG-ACT-TAA-TCA-ACG-C |
348 | IV | 60 | 21 |
| β-actin | GTC-AGA-AGG-ATT-CCT-ATG-TGG
GGA-GCA-ATG-ATC-TTG-ATC-TTC |
855 | I | 50 | 28 |
After the initial denaturation step (94°C; 2.5 min) 1 of the 4 following RT-PCR protocols was performed (D, denaturation; A, annealing; E, elongation). I: A (30 s, 94°C; 30 s at primer-specific annealing temperature); E (90 s, 72°C). II: same as I except annealing time 90 s. III (touch down PCR): after the denaturation step (30 s, 94°C) in the first cycle, A and E were 60 s at 66°C; A and E were reduced 1°C each following cycle until they reached 57°C; followed by cycles: D (30 s, 94°C); A (30 s, 56°C); E (90 s, 72°C). IV: D (30 s, 94°C); A (30 s, 60°C); E (30 s, 72°C).
Normalization and Quantification of the PCR Results
Equal aliquots of the positive control (mRNA of unstimulated vascular smooth muscle cells), applied to each gel, served as an internal standard. After measuring the optical density of the PCR products with the imaging system (AIDA 1D Densitometry; Raytest Isotopenmessgeräte GmbH, Berlin, Germany), the mean of two independent PCR reactions of each sample with the same pair of primers was calculated. Subsequently, the ratio of the density of the analyzed PCR-product versus the PCR-product of the 18S rRNA was determined for each sample (normalization procedure). Finally, mean and SEM of all normalized values were calculated for each group (ICM, DCM, or control). The data were analyzed with the independent t-test using Microsoft Office Excel 2003. A value of P < 0.05 was considered to be statistically significant.
Determination of IL-1ra Protein
Frozen tissues of independent samples were rapidly homogenized in a homogenization buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% NP-40, and protease inhibitors (protease inhibitor cocktail; Sigma, Taufkirchen, Germany). The samples were sonicated, incubated on ice for 15 min, and centrifuged (15 min; 16,000 x g). The supernatants were collected and the protein concentration was quantified using the BCA Protein Assay (Pierce, Bonn, Germany). Samples containing 100 μg protein were prepared in dilution buffer, and the IL-1ra was measured in IL-1ra ELISA (R&D Systems, Wiesbaden, Germany). Mean and SEM of the samples were calculated and data expressed as pg IL-1ra per 100 μg tissue. Statistical analysis was performed using 1-way ANOVA.
RESULTS
Analysis of Caspase-1, Caspase-3, IL-1α , IL-1β , IL-18, IL-1-RI, and IL-1ra mRNA Expression in Cardiac Tissues of Patients with ICM or DCM
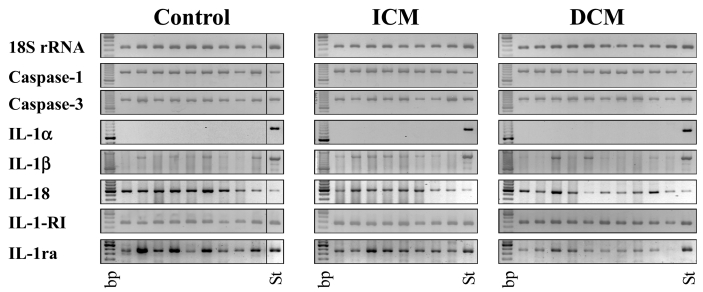
We quantified the mRNA expression of caspase-1, caspase-3, IL-1α , IL-1β , IL-18, IL-1-RI, and IL-1ra by RT-PCR (Figure 1). Because the number of the tested samples is low and hampers the quality of the statistical analysis, we included this presentation of all samples, along with the presentation of the statistical analysis expressed in arbitrary units in Figure 2. 18S rRNA, used as the control, was present in all samples in comparable amounts.
Figure 1.
Expression of 18S rRNA, caspase-1, caspase-3, IL-1α , IL-1β , IL-18, IL-1-RI, and IL-1ra mRNA in ICM and DCM patients. RT-PCR analysis was preformed as described in Material and Methods. Expected sizes of the PCR products are given in Table 2. Two PCR experiments each were performed with equal results. bp, 100-bp DNA ladder, the brightest band at 600 bp; St, standardization sample: an aliquot of PCR with human smooth muscle cell cDNA, used as positive control, was applied to the gel and used for quantification of PCR products.
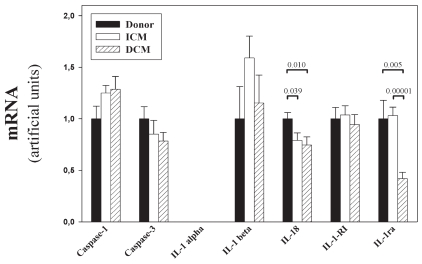
Figure 2.
Quantification of RT-PCR products. PCR products were quantified by densitometric scanning of ethidium bromide–stained gels. Relative values are shown for each gene after adjusting to the standardization sample (St in Figure 1), normalization to 18S rRNA, and calculation of mean ± SEM for each studied group (control, ICM, DCM). The P values are included in the figure if the differences were significant (<0.05).
mRNA expression of the IL-1 processing enzyme caspase-1 and the apoptosis-related caspase-3 was similar in the two patient groups (ICM, DCM) and the control. Quantification of the bands revealed no significant differences (P > 0.05) in the expression of these genes (Figure 2) in the tested groups. For IL-1α , we detected no mRNA expression in any of the tested samples, which were derived from adult donors. Enhancement of the PCR cycle-number did not change this result, but led to appearance of unspecific bands (data not shown). On the other hand, the IL-1β gene was expressed in most of the adult samples. The percentage of positive samples was similar in all studied groups. Although 2 DCM patients expressed potently increased IL-1β mRNA, a situation similar to the data reported by Ukimura et al. (15), statistical analysis revealed no evidence for a significant difference. On the other hand mRNA expression of the IL-18 gene was detectable in all samples and was lower in tissues derived from ICM and DCM patients, compared with control tissues. Quantification and statistical analysis showed that the reduction was significant (ICM vs. control, P = 0.039; DCM vs. control, P = 0.01) and that the IL-18 expression in the 2 patient groups was comparable (ICM vs. DCM, P = 0.47). The IL-1 isoforms IL-1α and IL-1β will bind to IL-1RI in order to induce signaling. The PCR measurements showed that IL-1-RI mRNA expression was similar in the tested groups.
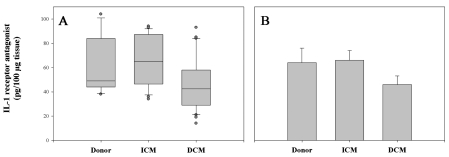
Finally, we investigated IL-1ra mRNA expression in the tissues. The IL-1ra–specific PCR bands were expressed equally in the myocardium of controls and ICM patients; however, the expression was much lower in the left ventricle of patients with DCM. Statistical analysis confirmed these results—differences of the IL-1ra mRNA level between DCM patients and the two other groups were statistically significant (DCM vs. ICM, P = 0.00001; DCM vs. control, P = 0.005), whereas the IL-1ra levels of ICM patients and control were similar (ICM vs. control, P = 0.884). These data were in line with the protein measurements performed in ELISA (Figure 3) and the determination of the IL-1/IL-1ra protein ratio (data not shown), although these results did not reach statistical significance.
Figure 3.
IL-1 receptor antagonist protein expression. Frozen tissue of independent samples was homogenized in a homogenization buffer, centrifuged, and 100 μg protein was analyzed in IL-1ra ELISA. Data of 6 control tissues, 8 ICM tissues, and 10 DCM tissues were analyzed by 1-way ANOVA and presented as box blot (A) and bar chart representing the mean ± SEM (B).
Agonist and Antagonist Expression in Adult and Fetal/Neonatal Tissue
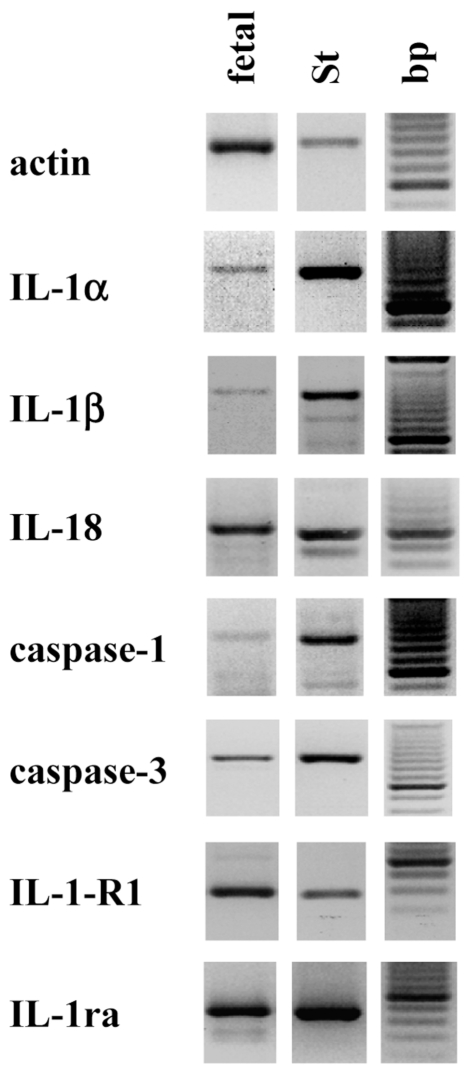
The reason for the lack of IL-1α expression in the tested adult samples was unclear. Previous reports of our group showed expression of functional IL-1α activity in neonatal rat cardiac cells, and tissue samples of adult rat heart also expressed IL-1α mRNA (50). Thus, we investigated the expression of IL-1α and IL-1β in fetal human tissues. For the PCR experiments, we used a commercially available “human cardiovascular MTC panel” that was the only source of human fetal material available. We show that the mRNA of IL-1α and IL-1β , as well as β-actin, IL-18, caspase-1, caspase-3, IL-1-RI, IL-1ra, and both IL-1 isoforms, was present in fetal human heart tissue (Figure 4).
Figure 4.
IL-1α mRNA is expressed in fetal human heart tissue. RT-PCR analysis was performed with cDNA of fetal heart tissue [fetal; human cardiovascular multiple tissue cDNA (MTC) panel]. PCR experiments with two MTC panels were performed with equal results. bp, 100-bp DNA ladder, the brightest band at 600 bp; St, the cDNA of SMC was used as the positive control.
DISCUSSION
IL-1 is a multifunctional cytokine (36). It has the capacity to modulate various cardiovascular functions (21,22). IL-1–mediated functions are regulated at different levels, e.g. by activation of the IL-1 proform by caspase-1; production and release of IL-1; interaction of this mediator with other cytokines, such as IL-18; receptor expression (e.g., IL-1-RI); and antagonistic pathways (e.g., IL-1ra). Thus, we studied the expression of genes of the IL-1 and caspase families known to participate in or interfere with IL-1 functions in nonfailing donor tissues or failing human heart. We describe significantly lowered IL-18 mRNA expression in tissues of ICM and DCM hearts, as well as significantly lowered IL-1ra mRNA expression and a trend to lower IL-1ra protein levels in the myocardium of DCM patients, compared with control tissues derived from hearts scheduled for transplantation. The lower IL-1ra level may contribute to enhanced inflammatory status, resulting in dysregulation of heart cell function due to enhanced IL-1 activity.
mRNA expression of caspase-1 (the enzyme necessary for IL-1β maturation) and caspase-3 (used as an example of an important apoptotic caspase) was not statistically different between control and patient tissues. Narula et al. (51) investigated caspase-3 activity in cardiomyopathic hearts. Their data show caspase-3 activity in patient tissues, but not in control tissues. The data reported by us are still in accordance with their Western blot results, because they showed that the caspase-3 proform is expressed in patients and controls. In our experiments, owing to the small amount of material, we were not able to perform analysis of caspase protein expression. More recently, experiments by the same group in a sheep heart failure model showed increased caspase-3 activity only at the onset of myocardial dysfunction (52).
Caspase-1 activates the proform of IL-1β . IL-1 is a potent activator of heart and vessel wall cells (24–26). It has been shown that cardiac overexpression of IL-1 results in left ventricular hypertrophy (53). Here we investigated the mRNA expression of both IL-1 isoforms. We found IL-1β mRNA expression in the myocardium of ICM and DCM patients, as well as in control tissues. IL-1β expression has been shown previously in coronary arteries of ICM patients (14,18), heart tissues of DCM patients (14,15), infarcted rat heart tissue (20), and the healing zones of postischemic tissues (54). Although Francis et al. (14) reported a difference between ICM and DCM patients, we did not find a significant difference between these groups. However, our results are in line with previous findings in biopsies of transplanted hearts, showing only low or even no IL-1β expression in the tissues (55). Furthermore, other authors reported that myocarditis patients, but not DCM patients, expressed IL-1β mRNA (16). In T3-induced rat heart hypertrophy, neither IL-1α nor IL-1β were expressed in control rats, but only after incubation with 3,3,5-triiodothyronine (56). Our findings are in line with the variable and low expression of the IL-1β mRNA that others (15) described.
We also investigated IL-1α expression in the tissues. We did not find IL-1α mRNA expression in any of the samples derived from tissues of adult patients or control hearts. Han et al. (17), who studied endomyocardial biopsies from patients with dilated cardiomyopathy, inflammatory myocarditis, and nonfailing human hearts, also did not find IL-1α mRNA. IL-1α mRNA was also not present in NMRI mice before virus infection (57). We have previously observed IL-1 activity and IL-1α protein expression in neonatal rat heart cells (50) and detected IL-1α mRNA in fetal and adult rat heart tissue. Thus, we investigated IL-1 mRNA expression in fetal human heart by using commercially available cDNAs derived from human undiseased heart tissues. Again, none of the adult heart tissues expressed IL-1α mRNA. However, we found IL-1α gene expression in fetal human heart cDNA. We did not find other reports showing IL-1α expression in fetal human heart; only one other report showed IL-1α expression in the developing heart, in experiments with rat tissue (58). Others showed expression of IL-1α and IL-1β protein in young and old mice, with higher expression of IL-1β in the hearts, compared with IL-1α (59). Taken together, these data indicate a differential expression of IL-1α mRNA in murine (fetal and adult) and human (fetal, not adult) heart tissues.
IL-18, like IL-1β , is activated by caspase-1. After ischemia/reperfusion, both IL-1β and IL-18 are elevated and are thought to contribute to impaired contractile force (47). This was suggested by inhibition studies with caspase-1 inhibitors. Application of IL-1ra or IL-18 binding protein also resulted in improved contractile force in isolated trabeculae. We show here that all human myocardial samples studied expressed IL-18 mRNA. However, the IL-18 mRNA expression in ICM and DCM patients was significantly lower compared with controls. Others reported enhanced IL-18 plasma levels in ICM and DCM patients (60); however, the IL-18 mRNA levels reported by them were only enhanced in ICM patients. In light of the report by Pomerantz et al. (47), who showed enhancement of IL-18 protein and mRNA in human trabeculae after ischemia-reperfusion treatment, it appears possible that the control tissues investigated by us may contain enhanced IL-18 mRNA levels due to ischemic activation during the transplantation procedure, rather than that the patient tissues contain reduced levels. However, this hypothesis is probably not relevant for this study, because it has been reported that IL-1β mRNA is enhanced in tissues following ischemia-reperfusion (54). However, IL-1β mRNA levels were not different in the samples tested by us, indicating that no profound (ischemic) activation was measurable. Further studies are needed to understand the situation.
In line with our results, it has been shown previously that IL-1-RI was not altered in tissues from cardiomyopathic patients compared with nonfailing hearts, but was reduced in myocarditis (17). In healthy mouse hearts, IL-1-RI was detected in low expression (61).
Finally, we investigated the expression of IL-1ra mRNA in these tissue samples. IL-1ra is a competitor of IL-1 and has been postulated to be an important counterbalance for IL-1 in tissues (36,48). The important role of IL-1ra in tissue inflammation has been reported previously in investigations using mice lacking the IL-1ra gene. These animals suffered considerably from arterial inflammation (42). Likewise, the local administration of IL-1ra in a vector expression system reduced mortality and inflammation in mouse hearts suffering from experimental coxsackievirus myocarditis (62), and administration of IL-1ra by osmotic pumps reduced injury in a myocarditis model (63). Also, transfection with IL-1ra reduced heart inflammation and apoptosis in rats and provided cardioprotection (64). The alteration of the IL-1/IL-1ra balance has been proposed to be one of the mechanisms promoting progression of cardiovascular diseases. It has been described that coronary arteries of ICM patients expressed more IL-1ra mRNA and immunoreactive IL-1ra protein than DCM arteries (65). In heart tissues, higher expression of IL-1β mRNA was detected in DCM compared with ICM patients, and it may be assumed that the IL-1/IL-1ra balance could be disturbed by this enhanced IL-1 production (14). Here we show that IL-1ra mRNA and protein expression in heart tissues of DCM patients is lower compared with ICM or control tissues. Thus, we conclude that IL-1ra could participate in pathogenesis of DCM by shifting the balance of IL-1ra and IL-1 in cardiac tissues, thereby enhancing inflammatory potential in the tissue, resulting in dysregulation of cell growth, contraction, and subsequent worsening of heart failure.
A genetic predisposition of DCM patients could be responsible for the observed reduction of IL-1ra gene expression. The IL-1ra gene is polymorph and contains a variable number of an 86-bp repeat sequence in intron 2. Allele 1 (IL-1RN*1; frequency 0.74) contains 4 repeats, and allele 2 (IL-1RN*2; frequency 0.21) contains 2 repeats (66). Although the data concerning association of IL-1RN*2 with IL-1ra production are controversial, and upregulation or reduction of IL-1ra content seems to be dependent on the cell type producing the protein (67), some reports show association of allele 2 with decreased IL-1ra synthesis. For example, lowered total IL-1ra protein production was reported in PBMCs from healthy individuals and ulcerative colitis (UC) patients carrying the IL-1RN*2 allele compared with those carrying the IL-1RN*1 allele (68). In columnar epithelial cells, IL-1RN*2 has been shown to be associated with reduced protein content (69). In Dewberry et al. (65), the association of IL-1RN*2 with decreased icIL-1ra protein production in HUVECs was also documented. Association of IL-1RN*2 with DCM could be one possible explanation of reduced IL-1ra gene expression in DCM myocardium. In contrast, no clear-cut association with ICM was reported (70), whereas an association with coronary artery disease (SVD, but not MVD) was described (71). Thus, the suggestion that IL-1ra expression is dependent on the polymorphism remains to be proven by further investigations.
Taken together, we studied the mRNA expression of various genes possibly involved in regulation of IL-1 function in myocardial tissues of heart-failure patients and non-failing organ donors. We show a lower IL-18 mRNA expression in ICM and DCM patients and lower IL-1ra gene expression in DCM patients, as compared to controls. The data support the hypothesis that an imbalance of IL-1 and its antagonist may contribute to progression of heart disease.
ACKNOWLEDGMENTS
We thank Claudia Pilowski and Beate Heinze for their expert technical assistance. This work was supported by a grant (project 06; Forschungsverbund—Myocard Hypertrophie) of the BMBF to K.W. and H.L. and grant Lo385/4-1 of the DFG to H.L.
Footnotes
Online address: http://www.molmed.org
REFERENCES
- 1.Hansson GK, Libby P. The immune response in atherosclerosis: a double-edged sword. Nat Rev Immunol. 2006;6:508–19. doi: 10.1038/nri1882. [DOI] [PubMed] [Google Scholar]
- 2.Libby P. Atherosclerosis: the new view. Sci Am. 2002;286:46–55. doi: 10.1038/scientificamerican0502-46. [DOI] [PubMed] [Google Scholar]
- 3.Seta Y, Shan K, Bozkurt B, Oral H, Mann DL. Basic mechanisms in heart failure: the cytokine hypothesis. J Card Fail. 1996;2:243–9. doi: 10.1016/s1071-9164(96)80047-9. [DOI] [PubMed] [Google Scholar]
- 4.Dinarello CA, Pomerantz BJ. Proinflammatory cytokines in heart disease. Blood Purif. 2001;19:314–21. doi: 10.1159/000046960. [DOI] [PubMed] [Google Scholar]
- 5.Sasayama S, Matsumori A, Kihara Y. New insights into the pathophysiological role for cytokines in heart failure. Cardiovasc Res. 1999;42:557–64. doi: 10.1016/s0008-6363(99)00050-4. [DOI] [PubMed] [Google Scholar]
- 6.Werdan K. The activated immune system in congestive heart failure: from dropsy to the cytokine paradigm. J Intern Med. 1998;243:87–92. doi: 10.1046/j.1365-2796.1998.00265.x. [DOI] [PubMed] [Google Scholar]
- 7.Biasucci LM, Vitelli A, Liuzzo G, et al. Elevated levels of interleukin-6 in unstable angina. Circulation. 1996;94:874–7. doi: 10.1161/01.cir.94.5.874. [DOI] [PubMed] [Google Scholar]
- 8.Levine B, Kalman J, Mayer L, Fillit HM, Packer M. Elevated circulating levels of tumor necrosis factor in severe chronic heart failure. N Engl J Med. 1990;323:236–41. doi: 10.1056/NEJM199007263230405. [DOI] [PubMed] [Google Scholar]
- 9.Nozaki N, Yamaguchi S, Shirakabe M, Nakamura H, Tomoike H. Soluble tumor necrosis factor receptors are elevated in relation to severity of congestive heart failure. Jpn Circ J. 1997;61:657–64. doi: 10.1253/jcj.61.657. [DOI] [PubMed] [Google Scholar]
- 10.Torre-Amione G, Kapadia S, Benedict C, Oral H, Young JB, Mann DL. Proinflammatory cytokine levels in patients with depressed left ventricular ejection fraction: a report from the studies of left ventricular dysfunction (SOLVD) J Am Coll Cardiol. 1996;27:1201–6. doi: 10.1016/0735-1097(95)00589-7. [DOI] [PubMed] [Google Scholar]
- 11.Birks EJ, Owen VJ, Burton PB, et al. TNF-α is expressed in donor heart and predicts right ventricular failure after human heart transplantation. Circulation. 2000;102:326–31. doi: 10.1161/01.cir.102.3.326. [DOI] [PubMed] [Google Scholar]
- 12.Doyama K, Fujiwara H, Fukumoto M, et al. Tumour necrosis factor is expressed in cardiac tissues of patients with heart failure. Int J Cardiol. 1996;54:217–25. doi: 10.1016/0167-5273(96)02607-1. [DOI] [PubMed] [Google Scholar]
- 13.Shioi T, Matsumori A, Kihara Y, et al. Increased expression of interleukin-1β and monocyte chemotactic and activating factor/monocyte chemoattractant protein-1 in the hypertrophied and failing heart with pressure overload. Circ Res. 1997;81:664–71. doi: 10.1161/01.res.81.5.664. [DOI] [PubMed] [Google Scholar]
- 14.Francis SE, Holden H, Holt CM, Duff GW. Interleukin-1 in myocardium and coronary arteries of patients with dilated cardiomyopathy. J Mol Cell Cardiol. 1998;30:215–23. doi: 10.1006/jmcc.1997.0592. [DOI] [PubMed] [Google Scholar]
- 15.Ukimura A, Terasaki F, Fujioka S, Deguchi H, Kitaura Y, Isomura T, Suma H. Quantitative analysis of cytokine mRNA expression in hearts from patients with nonischemic dilated cardiomyopathy (DCM) J Card Surg. 2003;18(Suppl 2):S101–8. doi: 10.1046/j.1540-8191.18.s2.8.x. [DOI] [PubMed] [Google Scholar]
- 16.Satoh M, Tamura G, Segawa I, Tashiro A, Hiramori K, Satodate R. Expression of cytokine genes and presence of enteroviral genomic RNA in endomyocardial biopsy tissues of myocarditis and dilated cardiomyopathy. Virchows Arch. 1996;427:503–9. doi: 10.1007/BF00199511. [DOI] [PubMed] [Google Scholar]
- 17.Han RO, Ray PE, Baughman KL, Feldman AM. Detection of interleukin and interleukin-receptor mRNA in human heart by polymerase chain reaction. Biochem Biophys Res Comm. 1991;181:520–3. doi: 10.1016/0006-291x(91)91219-3. [DOI] [PubMed] [Google Scholar]
- 18.Galea J, Armstrong J, Gadsdon P, Holden H, Francis SE, Holt CM. Interleukin-1β in coronary arteries of patients with ischemic heart disease. Arterioscler Thromb Vasc Biol. 1996;16:1000–6. doi: 10.1161/01.atv.16.8.1000. [DOI] [PubMed] [Google Scholar]
- 19.Shioi T, Matsumori A, Sasayama S. Persistent expression of cytokine in the chronic stage of viral myocarditis in mice. Circulation. 1996;94:2930–7. doi: 10.1161/01.cir.94.11.2930. [DOI] [PubMed] [Google Scholar]
- 20.Ono K, Matsumori A, Shioi T, Furukawa Y, Sasayama S. Cytokine gene expression after myocardial infarction in rat hearts: possible implication in left ventricular remodeling. Circulation. 1998;98:149–56. doi: 10.1161/01.cir.98.2.149. [DOI] [PubMed] [Google Scholar]
- 21.Loppnow H, Werdan K, Reuter G, Flad H-D. The interleukin-1 and interleukin-1-converting enzyme families in the cardiovascular system. Eur Cytokine Netw. 1998;9:675–80. [PubMed] [Google Scholar]
- 22.Long CS. The role of interleukin-1 in the failing heart. Heart Fail Rev. 2001;6:81–94. doi: 10.1023/a:1011428824771. [DOI] [PubMed] [Google Scholar]
- 23.Libby P, Warner SJC, Friedman GB. Interleukin-1: a mitogen for human vascular smooth muscle cells that induces the release of growth-inhibitory prostanoids. J Clin Invest. 1988;81:487–98. doi: 10.1172/JCI113346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loppnow H, Libby P. Proliferating or interleukin-1-activated human vascular smooth muscle cells secrete copious interleukin-6. J Clin Invest. 1990;85:731–8. doi: 10.1172/JCI114498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loppnow H, Libby P. Adult human vascular endothelial cells express the IL-6 gene differentially in response to LPS or IL-1. Cell Immunol. 1989;122:493–503. doi: 10.1016/0008-8749(89)90095-6. [DOI] [PubMed] [Google Scholar]
- 26.Müller-Werdan U, Schumann H, Loppnow H, et al. Endotoxin and tumor necrosis factor-α exert a similar proinflammatory effect in neonatal rat cardiomyocytes, but have different cardiodepressant profiles. J Mol Cell Cardiol. 1998;30:1027–36. doi: 10.1006/jmcc.1998.0667. [DOI] [PubMed] [Google Scholar]
- 27.Kacimi R, Karliner JS, Koudssi F, Long CS. Expression and regulation of adhesion molecules in cardiac cells by cytokines: response to acute hypoxia. Circ Res. 1998;82:576–86. doi: 10.1161/01.res.82.5.576. [DOI] [PubMed] [Google Scholar]
- 28.Hosenpud JD, Campbell SM, Mendelson DJ. Interleukin-1-induced myocardial depression in an isolated beating heart preparation. J Heart Transplant. 1989;8:460–4. [PubMed] [Google Scholar]
- 29.Evans HG, Lewis MJ, Shah AM. Interleukin-1β modulates myocardial contraction via dexamethasone sensitive production of nitric oxide. Cardiovasc Res. 1993;27:1486–90. doi: 10.1093/cvr/27.8.1486. [DOI] [PubMed] [Google Scholar]
- 30.Cain BS, Meldrum DR, Dinarello CA, Meng XZ, Joo KS, Banerjee A, Harken AH. TNF and IL-1 synergistically depress human myocardial function. Crit Care Med. 1999;27:1309–18. doi: 10.1097/00003246-199907000-00018. [DOI] [PubMed] [Google Scholar]
- 31.Stein B, Frank P, Schmitz W, Scholz H, Thoenes M. Endotoxin and cytokines induce direct cardiodepressive effects in mammalian cardiomyocytes via induction of nitric oxide synthase. J Mol Cell Cardiol. 1996;28:1631–9. doi: 10.1006/jmcc.1996.0153. [DOI] [PubMed] [Google Scholar]
- 32.Kumar A, Thota V, Dee L, Olson J, Uretz E, Parrillo JE. Tumor necrosis factor-α and interleukin-1β are responsible for in vitro myocardial cell depression induced by human septic shock serum. J Exp Med. 1996;183:949–58. doi: 10.1084/jem.183.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bick RJ, Liao JP, King TW, LeMaistre A, McMillin JB, Buja LM. Temporal effects of cytokines on neonatal cardiac myocyte Ca2+ transients and adenylate cyclase activity. Am J Physiol. 1997;41:H1937–44. doi: 10.1152/ajpheart.1997.272.4.H1937. [DOI] [PubMed] [Google Scholar]
- 34.Arstall MA, Sawyer DB, Fukazawa R, Kelly RA. Cytokine-mediated apoptosis in cardiac myocytes: The role of inducible NO synthase induction and peroxynitrite generation. Circ Res. 1999;85:829–40. doi: 10.1161/01.res.85.9.829. [DOI] [PubMed] [Google Scholar]
- 35.Ing DJ, Zang J, Dzau VJ, Webster KA, Bishopric NH. Modulation of cytokine-induced cardiac myocyte apoptosis by nitric oxide, Bak, and Bcl-x. Circ Res. 1999;84:21–33. doi: 10.1161/01.res.84.1.21. [DOI] [PubMed] [Google Scholar]
- 36.Dinarello CA. Biologic basis for interleukin-1 in disease. Blood. 1996;87:2095–147. [PubMed] [Google Scholar]
- 37.Libby P. Inflammation in atherosclerosis. Nature. 2002;420:868–74. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 38.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352:1685–95. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 39.Ross R. Atherosclerosis: an inflammatory disease. N Engl J Med. 1999;340:115–26. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 40.Schmitz J, Owyang A, Oldham E, et al. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23:479–90. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 41.Horai R, Saijo S, Tanioka H, et al. Development of chronic inflammatory arthropathy resembling rheumatoid arthritis in interleukin-1 receptor antagonist-deficient mice. J Exp Med. 2000;191:313–20. doi: 10.1084/jem.191.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicklin MJH, Hughes DE, Barton JL, Ure JM, Duff GW. Arterial inflammation in mice lacking the interleukin-1 receptor antagonist gene. J Exp Med. 2000;191:303–11. doi: 10.1084/jem.191.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Casini-Raggi V, Kam L, Chong YJ, Fiocchi C, Pizarro TT, Cominelli F. Mucosal imbalance of IL-1 and IL-1 receptor antagonist in inflammatory bowel disease: a novel mechanism of chronic intestinal inflammation. J Immunol. 1995;154:2434–40. [PubMed] [Google Scholar]
- 44.Liu H, Hanawa H, Yoshida T, et al. Effect of hydrodynamics-based gene delivery of plasmid DNA encoding interleukin-1 receptor antagonist-Ig for treatment of rat autoimmune myocarditis: possible mechanism for lymphocytes and noncardiac cells. Circulation. 2005;111:1593–1600. doi: 10.1161/01.CIR.0000160348.75918.CA. [DOI] [PubMed] [Google Scholar]
- 45.Wetzler M, Kurzrock R, Estrov Z, Kantarjian H, Gisslinger H, Underbrink MP, Talpaz M. Altered levels of interleukin-1β and interleukin-1 receptor antagonist in chronic myelogenous leukemia: clinical and prognostic correlates. Blood. 1994;84:3142–7. [PubMed] [Google Scholar]
- 46.Chomarat P, Vannier E, Dechanet J, Rissoan MC, Banchereau J, Dinarello CA, Miossec P. Balance of IL-1 receptor antagonist/IL-1β in rheumatoid synovium and its regulation by IL-4 and IL-10. J Immunol. 1995;154:1432–9. [PubMed] [Google Scholar]
- 47.Pomerantz BJ, Reznikov LL, Harken AH, Dinarello CA. Inhibition of caspase-1 reduces human myocardial ischemic dysfunction via inhibition of IL-18 and IL-1ß. Proc Natl Acad Sci U S A. 2001;98:2871–6. doi: 10.1073/pnas.041611398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsukawa A, Fukumoto T, Maeda T, Ohkawara S, Yoshinaga M. Detection and characterization of IL-1 receptor antagonist in tissues from healthy rabbits: IL-1 receptor antagonist is probably involved in health. Cytokine. 1997;9:307–15. doi: 10.1006/cyto.1996.0170. [DOI] [PubMed] [Google Scholar]
- 49.Chirgwin JM, Pryzbyla AE, Macdonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–9. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 50.Westphal E, Chen L, Pilowski C, et al. Endotoxin-activated cultured neonatal rat cardiomyocytes express functional surface-associated interleukin 1. J Endotoxin Res. 2007;13:25–34. doi: 10.1177/0968051907078609. [DOI] [PubMed] [Google Scholar]
- 51.Narula J, Pandy P, Arbustini E, et al. Apoptosis in heart failure: release of cytochrome c from mitochondria and activation of caspase-3 in human cardiomyopathy. Proc Natl Acad Sci U S A. 1999;96:8144–9. doi: 10.1073/pnas.96.14.8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moorjani N, Ahmad M, Catarino P, et al. Activation of apoptotic caspase cascade during the transition to pressure overload-induced heart failure. J Am Coll Cardiol. 2006;48:1451–8. doi: 10.1016/j.jacc.2006.05.065. [DOI] [PubMed] [Google Scholar]
- 53.Nishikawa K, Yoshida M, Kusuhara M, Ishigami N, Isoda K, Miyazaki K, Ohsuzu F. Left ventricular hypertrophy in mice with a cardiac-specific overexpression of interleukin-1. Am J Physiol. 2006;291:H176–83. doi: 10.1152/ajpheart.00269.2005. [DOI] [PubMed] [Google Scholar]
- 54.Herskowitz A, Choi S, Ansari AA, Wesselingh S. Cytokine mRNA expression in postischemic/reperfused myocardium. Am J Pathol. 1995;146:419–28. [PMC free article] [PubMed] [Google Scholar]
- 55.Van Hoffen E, Van Wichen D, Stuij I, et al. In situ expression of cytokines in human heart allografts. Am J Pathol. 1996;149:1991–2003. [PMC free article] [PubMed] [Google Scholar]
- 56.Ziegelhoffer-Mihalovicova B, Briest W, Baba HA, Rassler B, Zimmer HG. The expression of mRNA of cytokines and of extracellular matrix proteins in triiodothyronine-treated rat hearts. Mol Cell Biochem. 2003;247:61–8. doi: 10.1023/a:1024153003249. [DOI] [PubMed] [Google Scholar]
- 57.Schmidtke M, Glück B, Merkle I, Hofmann P, Stelzner A, Gemsa D. Cytokine profiles in heart, spleen, and thymus during the acute stage of experimental coxsackievirus B3-induced chronic myocarditis. J Med Virol. 2000;61:518–26. [PubMed] [Google Scholar]
- 58.Nakagawa M, Terracio L, Carver W, Birkedal-Hansen H, Borg TK. Expression of collagenase and IL-1α in developing rat hearts. Dev Dyn. 1992;195:87–99. doi: 10.1002/aja.1001950203. [DOI] [PubMed] [Google Scholar]
- 59.Hacham M, Argov S, White RM, Segal S, Apte RN. Different patterns of interleukin-1alpha and interleukin-1beta expression in organs of normal young and old mice. Eur Cytokine Netw. 2002;13:55–65. [PubMed] [Google Scholar]
- 60.Mallat Z, Heymes C, Corbaz A, et al. Evidence for altered interleukin 18 (IL)-18 pathway in human heart failure. FASEB J. 2004;18:1752–4. doi: 10.1096/fj.04-2426fje. [DOI] [PubMed] [Google Scholar]
- 61.Deyerle KL, Sims JE, Dower SK, Bothwell MA. Pattern of IL-1 receptor gene expression suggests role in noninflammatory processes. J Immunol. 1992;149:1657–65. [PubMed] [Google Scholar]
- 62.Lim BK, Choe SC, Shin JO, et al. Local expression of IL-1ra by plasmid DNA improves mortality and decreases myocardial inflammation in experimental coxsackieviral myocarditis. Circulation. 2002;105:1278–81. [PubMed] [Google Scholar]
- 63.Neumann DA, Lane JR, Allen GS, Herskowitz A, Rose NR. Viral myocarditis leading to cardiomyopathy: do cytokines contribute to pathogenesis? Clin Immunol Immunopathol. 1993;68:181–90. doi: 10.1006/clin.1993.1116. [DOI] [PubMed] [Google Scholar]
- 64.Suzuki K, Murtuza B, Smolenski RT, Sammut IA, Suzuki N, Kaneda Y, Yacoub MH. Overexpression of interleukin-1 receptor antagonist provides cardioprotection against ischemia-reperfusion injury associated with reduction in apoptosis. Circulation. 2001;104(12 Suppl 1):I-308–13. doi: 10.1161/hc37t1.094871. [DOI] [PubMed] [Google Scholar]
- 65.Dewberry RM, Holden H, Crossman DC, Francis SE. Interleukin-1 receptor antagonist expression in human endothelial cells and atherosclerosis. Arterioscler Thromb Vasc Biol. 2000;20:2394–400. doi: 10.1161/01.atv.20.11.2394. [DOI] [PubMed] [Google Scholar]
- 66.Tarlow JK, Blakemore AI, Lennard A, Solari R, Hughes HN, Steinkasserer A, Duff GW. Polymorphism in human IL-1 receptor antagonist gene intron 2 is caused by variable numbers of an 86-bp tandem repeat. Hum Genet. 1993;91:403–4. doi: 10.1007/BF00217368. [DOI] [PubMed] [Google Scholar]
- 67.Arend WP, Guthridge CJ. Biological role of IL-1 receptor antagonist isoforms. Ann Rheum Dis. 2000;59 (Suppl 1):i60–4. doi: 10.1136/ard.59.suppl_1.i60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tountas NA, Casini-Raggi V, Yang H, et al. Functional and ethnic association of allele 2 of the interleukin-1 receptor antagonist gene in ulcerative colitis. Gastroenterology. 1999;117:806–13. doi: 10.1016/s0016-5085(99)70338-0. [DOI] [PubMed] [Google Scholar]
- 69.Carter MJ, di Giovine FS, Jones S, Mee J, Camp NJ, Lobo AJ, Duff GW. Association of the interleukin-1 receptor antagonist gene with ulcerative colitis in northern European Caucasians. Gut. 2001;48:461–7. doi: 10.1136/gut.48.4.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Manzoli A, Andreotti F, Varlotta C, et al. Allelic polymorphism of the interleukin-1 receptor antagonist gene in patients with acute or stable presentation of ischemic heart disease. Cardiologia. 1999;44:825–30. [PubMed] [Google Scholar]
- 71.Francis SE, Camp NJ, Dewberry RM, et al. Interleukin-1 receptor antagonist gene polymorphism and coronary artery disease. Circulation. 1999;99:861–6. doi: 10.1161/01.cir.99.7.861. [DOI] [PubMed] [Google Scholar]