Abstract
RNA interference requires efficient delivery of small double-stranded RNA molecules into the target cells and their subsequent incorporation into RNA-induced silencing complexes. Although current cationic lipids commonly used for DNA transfection have also been used for siRNA transfection, a clear need still exists for better siRNA delivery to improve the gene silencing efficiency. We synthesized a series of cationic lipids characterized by head groups bearing various aminoglycosides for specific interaction with RNA. siRNA complexation with such lipidic aminoglycoside derivatives exhibited three lipid/siRNA ratio-dependent domains of colloidal stability. Fluorescence and dynamic light-scattering experiments showed that cationic lipid/siRNA complexes were formed at lower charge ratios, exhibited a reduced zone of colloidal instability, and had smaller mean diameters compared with our previously described guanidinium-based cationic lipids. Cryo-transmission electron microscopy and x-ray-scattering experiments showed that, although the final in toto morphology of the lipid/siRNA complexes depended on the aminoglycoside type, there was a general supramolecular arrangement consisting of ordered lamellar domains with an even spacing of 67 Å. The most active cationic lipid/siRNA complexes for gene silencing were obtained with 4,5-disubstituted 2-deoxystreptamine aminoglycoside derivatives and were characterized by the siRNA being entrapped in small particles exhibiting lamellar microdomains corresponding to siRNA molecules sandwiched between the lipid bilayers. These results clearly show that lipidic aminoglycoside derivatives constitute a versatile class of siRNA nanocarriers allowing efficient gene silencing.
Keywords: gene silencing, gene transfer vectors, transfection
RNAi has become widely used for knocking down the expression of a specific target gene by a posttranscriptional silencing mechanism and thereby it allows phenotypic analysis of gene function in cells (1, 2). Therapeutic approaches involving RNAi are also actively investigated (3, 4). To achieve gene silencing, sequence-specific double-stranded small interfering RNA (siRNA) molecules have to be delivered efficiently into the cytoplasm of cells (5, 6). Various methods have already been used for siRNA delivery, in particular, cationic lipids developed for plasmid DNA transfection (see ref. 7). These cationic lipids are composed of a hydrophobic moiety linked (by a spacer) to a cationic head group bearing either a quaternary ammonium [such as the lipids DOTMA {N-[1-(2,3-dioleyloxy)propyl]-N,N,N-trimethylammonium chloride} and DOTAP (1,2-dioleoyl-3-trimethylammonium-propane)], a polycation [such as the lipid DOGS (dioctadecylamidoglycylspermine) (8) and lipopolyamine RPR120535 (9)], or guanidinium groups [such as the lipid BGTC, bis(guanidinium)-tris(2-aminoethyl)amine-cholesterol (10, 11)]. A colipid such as dioleoyl phosphatidylethanolamine (DOPE) is usually combined with the cationic lipids to form liposomes. The electrostatic interactions between the plasmid DNA and the cationic head groups lead to the formation of supramolecular assemblies whose structure has been shown to be lamellar or hexagonal, depending on the type and proportion of the colipid in the cationic liposomes (12). Nevertheless, the structural characteristics of complexes formed with siRNA are still unknown, and the relevance of the structures observed with plasmid DNA remains to be assessed in the case of siRNA. It should also be stressed that the current cationic vectors commonly used for plasmid transfection have not been optimized for the delivery of nucleic acids of low molecular weight and particular entities such as siRNA. Indeed, siRNA molecules display a highly specific structure, very different from that of plasmid DNA. A siRNA is a short (usually 21–25 nucleotides) dsRNA with a few (generally 2) nucleotides forming overhangs at the 3′ ends (13). These naturally occurring siRNAs result from the processing by Dicer, an enzyme converting long dsRNAs and hairpin RNAs into siRNAs. To bypass the dicing step, this particular structure has to be conserved in synthetic siRNAs. In contrast, plasmid DNA used for gene transfer generally contains, in addition to the prokaryotic sequences required for amplification in bacteria, a eukaryotic expression cassette with a promoter, the cDNA of interest, and a polyadenylation signal. Plasmid DNA thus forms generally large molecules, contrary to the small 21- to 25-nucleotide siRNAs. Such an important structural difference may result in a significant change in the efficiency of a particular transfection vector with one nucleic acid category or another one.
These observations prompted us to design and evaluate new classes of reagents capable of interacting with siRNA molecules in a more specific manner. Our goal was to explore the efficiency of lipidic aminoglycoside derivatives to deliver siRNA molecules and knock down gene expression in mammalian cells. Thus, we chose to synthesize a series of cationic lipids characterized by an aminoglycoside-based cationic head group linked to two hydrophobic dioleyl chains by a succinyl spacer. Indeed, we reasoned that aminoglycosides are natural compounds already widely used as antibiotics that provide a versatile polycationic framework because they are decorated with up to six amine groups and numerous hydroxyl groups. The inherent flexibility of the glycosidic linkages in aminoglycosides may also permit their remodeling for better interaction with specific RNA substructures. In addition, the aminoglycoside family allowed the synthesis of a variety of cationic head groups by using different aminoglycosides (such as kanamycin A, tobramycin, neomycin B, and paromomycin) known to interact with the major groove of duplex RNA or more generally with nucleic acids (14, 15).
In the present study, we found (i) that complexation of siRNA molecules with lipidic aminoglycoside derivatives followed the three-stage model of colloidal stability proposed for plasmid DNA (9, 11, 16–19) and (ii) that the cationic lipid/siRNA complexes were formed at lower charge ratios, exhibited a reduced zone of colloidal instability, and had smaller mean diameters compared with our guanidinium-based cationic lipids described in refs. 10 and 11. We also report the results of cryo-transmission electron microscopy (cryo-TEM) and small-angle x-ray-scattering (SAXS) experiments showing that, although the final in toto morphology of the self-assembled siRNA-condensed particles depended on the aminoglycoside type, their general supramolecular arrangement consisted of ordered lamellar domains with an even spacing of 67 Å, irrespective of the aminoglycoside moiety. Finally, we report data from in vitro gene silencing studies demonstrating that lipidic aminoglycoside derivatives displayed a remarkably high efficiency for siRNA-based gene knockdown and allowing the comparison of their efficacy with the effectiveness of cationic lipids designed for DNA transfection and of commercially available reagents for siRNA delivery.
Results
GFP Silencing by Using Current Gene Transfection Vectors.
First, we assessed the gene silencing potential of synthetic vectors that are currently used for plasmid DNA transfection by the delivery of anti-GFP siRNA molecules into human lung cancer H1299 cells engineered to express GFP (therefore termed d2GFP). Thus, anti-GFP siRNA was complexed with BGTC/DOPE or DC-Chol {3-β-[N-(N′,N′-dimethylaminoethane)carbamoyl]cholesterol}/DOPE cationic liposomes or with the cationic polymer polyethyleneimine. At day 1 after transfection, inhibition of GFP expression was monitored by measuring the GFP fluorescence of the transfected cells. The results showed that GFP fluorescence was indeed lowered and that its residual level generally decreased when the charge ratio of the vector/siRNA complexes (expressed as moles of positive charge/moles of negative charge) increased [supporting information (SI) Fig. 5]. The lowest GFP fluorescence levels obtained with the liposomes BGTC/DOPE and DC-Chol/DOPE and with the polymer polyethyleneimine were ≈32%, 52%, and 55%, respectively. These results did highlight the need to develop novel synthetic vectors that are more efficient for siRNA-mediated gene down-regulation.
Structure of the Lipidic Aminoglycoside Derivatives.
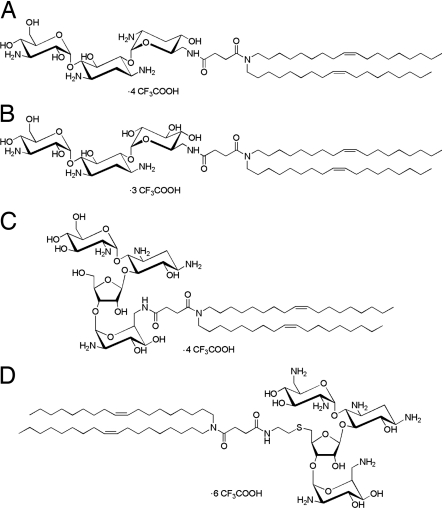
Lipidic aminoglycoside derivatives are amphiphiles with a self-aggregating hydrocarbon tail linked to an aminoglycoside head group. Fig. 1 shows the various lipidic aminoglycoside derivatives used in the present study. Schematically, their structure consisted of a tobramycin, kanamycin A, paromomycin, or neomycin B cationic head group linked to two dioleyl chains by a succinyl spacer. The four different compounds studied herein were thus the following: dioleyl succinyl kanamycin A (DOSK), dioleyl succinyl tobramycin (DOST), dioleyl succinyl paromomycin (DOSP), and dioleyl succinyl ethylthioneomycin B (DOSN). It should be stressed here that tobramycin and kanamycin A belong to the class of aminoglycosides characterized by a 4,6-disubstituted 2-deoxystreptamine (4,6-DDS) ring, whereas paromomycin and neomycin B are characterized by a 4,5-disubstituted 2-deoxystreptamine (4,5-DDS) ring. Further research on the synthesis of these compounds (20) will be reported in detail elsewhere (M.S., J.-P.V., and J.-M.L., unpublished work).
Fig. 1.
Structure of the lipidic aminoglycoside derivatives. (A) DOST. (B) DOSK. (C) DOSP. (D) DOSN.
Colloidal Stability of the Complexes Formed with Nucleic Acids.
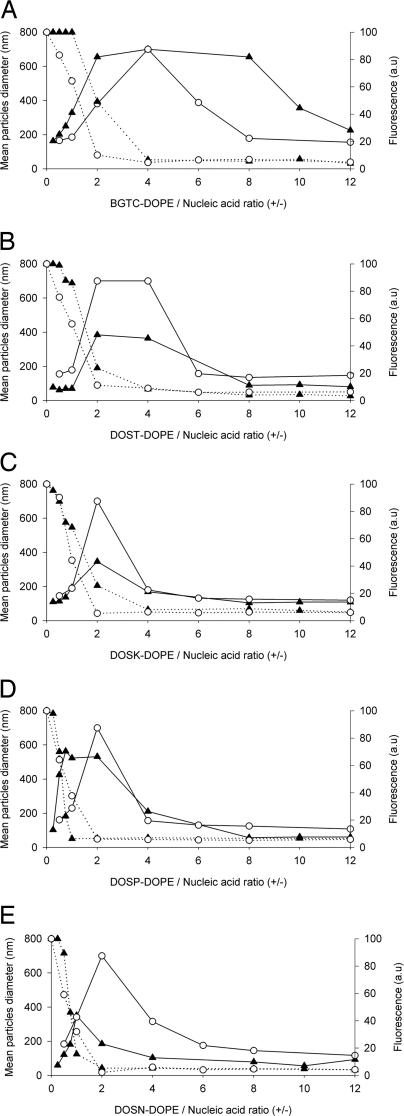
Next, we investigated the physicochemical properties of the complexes (as a function of their charge ratio) obtained by mixing plasmid DNA or siRNA molecules with lipidic aminoglycoside derivatives or BGTC-based liposomes (Fig. 2). To calculate the mean theoretical charge ratio, we assumed that 1 μg of siRNA is 3 nmol of negatively charged phosphate and that 2, 4, 3, 4, and 6 positive charges are displayed by BGTC, DOST, DOSK, DOSP, and DOSN, respectively. Dynamic light-scattering analysis of the siRNA complexes was in agreement with a three-zone model of colloidal stability. This model had actually already been described for lipopolyamine/DNA (9, 16–18) and BGTC/DNA lipoplexes (11, 19). The three different zones (named A, B, and C) were determined by the cationic lipid/nucleic acid charge ratio. In zone A at low charge ratios, negatively charged and colloidally stable complexes with partially condensed nucleic acid are formed. Zone B contains neutral, large, and colloidally unstable aggregates. In zone C, the particles are positively charged, small, and again colloidally stable. Interestingly, BGTC/siRNA complexes (Fig. 2A) were characterized by a large zone B ranging from charge ratios 2 to 8, where complexes flocculated and thus had a mean diameter >700 nm. In contrast, lipidic aminoglycoside derivatives/siRNA complexes (Fig. 2 B–E) displayed a narrower zone B and complexes were less aggregated and had a mean diameter of ≈400–500 nm. In zone C, BGTC/siRNA complexes exhibited a mean diameter of 225 nm, whereas the siRNA complexes formed by DOST, DOSK, DOSP, and DOSN liposomes had a mean diameter of 81, 105, 58, and 57 nm, respectively. Thus, DOST, DOSK, DOSP, and DOSN liposomes were able to form small particles when complexed with siRNA molecules. Note that no obvious size difference occurred between the complexes formed by BGTC and lipidic aminoglycoside derivative liposomes with plasmid DNA (Fig. 2).
Fig. 2.
Colloidal stability of cationic lipid/nucleic acid complexes as a function of their charge ratio. Complexes were obtained by mixing BGTC (A), DOST (B), DOSK (C), DOSP (D), and DOSN (E) cationic liposomes at the required concentrations with plasmid DNA (○) or siRNA (▴) at 10 μg/ml. Dynamic light-scattering analysis (solid lines) was performed to assess colloidal stability of the complexes. Ethidium bromide fluorescence measurements (dashed lines) allowed the evaluation of nucleic acid entrapment within complexes. a.u., arbitrary units. Size determination and fluorescence measurements were performed after 1 h of complexation. An arbitrary value of 700 nm was attributed to complexes that were colloidally unstable.
Next, we investigated the formation of siRNA or DNA complexes by ethidium bromide fluorescence measurements. As expected, the fluorescence intensity decreased as the cationic lipid/nucleic acid charge ratio increased. Fluorescence intensity decreased in zone A from 100% to a value close to zero and persisted at this minimum value in zones B and C. Although complexation of DNA with either BGTC or aminoglycoside derivative liposomes led to a similar decrease in fluorescence intensity, differences were observed in the siRNA case. Here, only with BGTC/siRNA complexes was there an obvious lag before the fluorescence intensity decreased. On the other hand, the slope of the decrease of the fluorescence intensity was steeper with DOSN and DOSP than with that observed with DOSK and DOST siRNA complexes. Thus, DOSN and DOSP liposomes appear to strongly complex siRNA molecules, thereby leading to rapid and total exclusion of ethidium bromide for charge ratios <2.
Structural Features of siRNA Complexes: Cryo-TEM Imaging and SAXS Experiments.
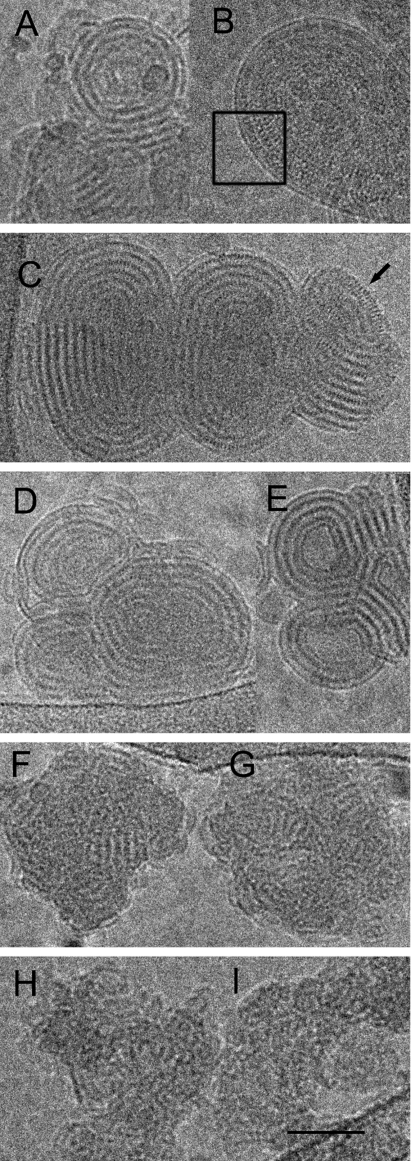
Mixing unilamellar BGTC liposomes 30–70 nm in diameter with siRNA led to the formation of discrete compact and concentric structures with a size in the 200- to 500-nm range (Fig. 3 A and B). These structures were probably made of stacks of alternating lipid bilayers and electron-dense densities corresponding to siRNA molecules. Indeed, the complexes exhibited a periodicity of ≈70 Å, a value consistent with the sum of the thickness of a lipid bilayer and the diameter of the double-stranded siRNA molecules. In addition, for well oriented structures, the micrographs also visualized fine striations with a 30-Å spacing, which might correspond to a regular arrangement of the siRNA molecules between the two lipid bilayers (box in Fig. 3B).
Fig. 3.
Cryo-TEM micrographs of cationic lipid/siRNA complexes. (A and B) Concentric “onion-like” structure of representative BGTC/siRNA complexes at high magnification. Box in B outlines the regular arrangement of the siRNA molecules between two lipid bilayers. (C) Structure of DOST/siRNA complexes at high magnification. Note the regular arrangement of the RNA molecules at the edge (black arrow). (D and E) Structure of two DOSK/siRNA complexes at high magnification. (F and G) Structure of two DOSP/siRNA complexes at high magnification. (H and I) Structure of two DOSN/siRNA complexes at high magnification. (Scale bar: 50 nm.)
When mixed with siRNA, DOST and DOSK liposomes formed concentric multilamellar (“onion-like”) structures, possibly consisting of a regular packing of lipid bilayers and siRNA molecules, which were strongly similar to those obtained with BGTC liposomes (Fig. 3 C–E). Indeed, a typical complex (Fig. 3C) again exhibited a regular spacing of ≈70 Å between two consecutive repeats, the electron-dense layer corresponding probably to the siRNA monolayer. Here again, the siRNA molecules appeared well ordered on the lipid membrane as observed on the edge of the lipoplex (black arrow in the upper-right corner of Fig. 3C) with a spacing similar to that observed with BGTC/siRNA complexes (see above).
With DOSP and DOSN liposomes, small complexes with a size of ≈60 nm were formed when adding siRNA molecules (Fig. 3 F–I), and these complexes actually exhibited a peculiar tendency to form clusters by aggregation. Most importantly, unlike the complexes formed by the three previous cationic lipid formulations (BGTC-, DOST-, and DOSK-based liposomes), the complexes obtained with DOSP and DOSN liposomes possessed a much more irregular structure. Indeed, although they were also composed of stacks of alternating lipid bilayers and siRNA molecules with a spacing of ≈70 Å, such an arrangement did not extend over a distance long enough to form fully concentric onion-like structures. Consequently, only ordered microdomains characterized by a flat fingerprint-like repetition were observed. This finding may be linked to the size of the DOSP and DOSN liposomes (diameter range, 30–50 nm). Indeed, they generally had a smaller size than those prepared with the three other cationic lipids (data not shown) and thus might not be able to induce the formation of complete onion-like structures. It is also noteworthy here that, when the liposomes prepared from the five different lipid mixtures were exposed to 300 mM NaCl, the small liposomes remained unilamellar, whereas the large ones became bilamellar (data not shown), probably because of an osmotic effect, as described in ref. 19.
SAXS scans of siRNA complexes formed by BGTC or DOSP liposomes revealed a first-order reflection peak at 67.7 and 67.5 Å, respectively (data not shown). Thus, the spacing of ≈70 Å observed by cryo-TEM imaging of DOSP and BGTC/siRNA complexes (see above) was similar to that determined by x-ray scattering. Moreover, BGTC/siRNA complexes (but not DOSP/siRNA complexes) exhibited a second-order reflection peak at 33.8 Å, which clearly indicates the presence of a lamellar structure organized over a long distance and is in good agreement with the cryo-TEM micrographs (data not shown and see above). Finally, in control experiments, BGTC/plasmid DNA lipoplexes were also studied by SAXS. A first-order reflection peak at 69.1 Å and a second-order reflection at 34.5 Å were easily detected, a finding consistent with SAXS results reported for BGTC/DNA complexes in ref. 11.
GFP Silencing Activity of the siRNA Complexes Formed by the Various Aminoglycoside Derivatives.
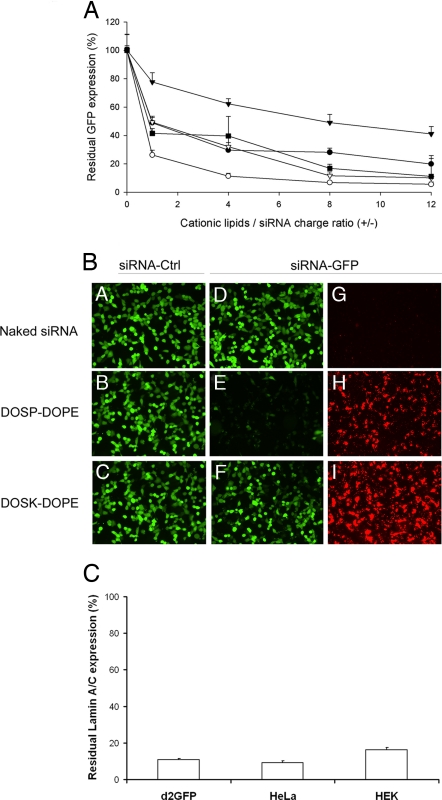
As shown in Fig. 4A, a general trend indicated that GFP fluorescence levels of d2GFP cells transfected with the various GFP-targeting siRNA complexes decreased progressively as the cationic lipid/siRNA charge ratio increased. However, cationic lipids with aminoglycoside head groups belonging to the 4,6-DDS family, i.e., DOST and DOSK, exhibited divergent behaviors because they led to either a slow (DOSK) or a fast (DOST) decrease in GFP fluorescence when the cationic lipid/siRNA charge ratio increased. In contrast, both lipidic derivatives of the 4,5-DDS class of aminoglycosides, i.e., DOSP and DOSN, led to a similar progressive decrease of GFP expression, a minimal residual GFP fluorescence level of ≈10% finally being reached. Importantly, transfections of RNA-induced silencing complex-loaded control (not GFP-targeting) siRNA by using the various cationic lipids under the same conditions allowed normalization of the residual GFP expression for each data point. Identical results were obtained with control siRNA-transfected cells and with untransfected cells that were thus used as reference for normalization. This indicates that transfection of control siRNA did not affect the endogenous GFP fluorescence level compared with nontransfected cells (data not shown), and strongly suggests that the complexes were not cytotoxic for d2GFP cells.
Fig. 4.
GFP silencing activity of the siRNA complexes formed by various aminoglycoside derivatives. (A) Residual GFP fluorescence after in vitro transfection of d2GFP cells with lipid/siRNA complexes characterized by various +/− charge ratios. GFP-expressing d2GFP cells were transfected with 400 ng of anti-GFP siRNA complexed with liposomes composed of BGTC (●), DOSK (▾), DOST (○), DOSP (■), and DOSN (▿). Residual GFP expression was expressed as the ratio (%) of GFP fluorescence in cells transfected with anti-GFP siRNA to GFP fluorescence in cells transfected with control (non-GFP targeting) siRNA. (B) Fluorescence microscopy visualization of GFP silencing and siRNA internalization. The GFP-expressing d2GFP cells were transfected with control siRNA (A–C in panel B) or 3′-rhodamine-labeled anti-GFP siRNA (D–I in panel B). The siRNA molecules (500 ng per well) were formulated in the absence (“naked” siRNA in A, D, and G in panel B) or in the presence of the cationic lipids DOSP (B, E, and H in panel B), or DOSK (C, F, and I in panel B). The transfected d2GFP cells were observed by using a FITC filter to visualize GFP fluorescence (A–F in panel B) or a rhodamine filter to visualize siRNA internalization (G–I in panel B). (C) Real-time quantitative RT-PCR analysis of human lamin A/C mRNA after transfection of various human cell lines (HEK293, HeLa, and d2GFP cells) with DOSP/siRNA lipoplexes [normalization to hypoxanthine–guanine phosphoribosyltransferase (HPRT1)]. Values are relative to cells transfected under the same experimental condition with a control siRNA.
GFP silencing efficiency of DOSP/siRNA complexes was also quantified by flow cytometry (FACS) analysis and by measuring the GFP mRNA level by real-time quantitative RT-PCR in transfected d2GFP cells. FACS results showed that 91.2% of the cells transfected with anti-GFP siRNA complexed with DOSP were undergoing the expected RNAi response (data not shown). This result was in good agreement with that obtained under the same experimental conditions by the lysis method (see above and Fig. 4A). Real-time quantitative RT-PCR analysis indicated that transfected cells had only 4.1% of GFP-mRNA compared with cells transfected with control siRNA or untransfected cells (data not shown).
Accordingly, Fig. 4B shows that delivery of anti-GFP siRNA molecules into d2GFP cells by DOSP/siRNA complexes led to a strong reduction of the number of GFP-positive cells (E), which correlated with a high cellular uptake of the siRNA molecules (as visualized in H by the red fluorescence due to the rhodamine-labeled anti-GFP siRNA). In contrast, DOSK liposomes also clearly led to efficient siRNA uptake by the cells (I), but the GFP fluorescence was only slightly reduced (F) compared with cells transfected with control siRNA (C). Naked “unreacted” siRNA was used as control of the efficiency of the cationic lipids for siRNA delivery; A, D, and G show that there was suppression of neither GFP expression nor siRNA uptake, a finding confirming the role of the vectors for siRNA delivery.
Next, we performed RNAi experiments with siRNA targeting the endogenous lamin A/C expression. Here, RT-PCR results indicated that d2GFP cells transfected with DOSP/siRNA complexes had very little residual lamin A/C mRNA compared with cells transfected with control siRNA (Fig. 4C), a finding demonstrating that DOSP/siRNA complexes can also allow the efficient knockdown of the expression of an endogenous gene in d2GFP cells. To broaden our conclusions, we also found that DOSP/anti-lamin A/C siRNA complexes were highly efficient for silencing of lamin A/C expression in other human cells lines such as HEK293 cells (human embryonic kidney cells) and HeLa cells (derived from a human epithelioid cervical cancer), a very low level of residual lamin A/C mRNA again being observed (Fig. 4C).
In additional experiments, we studied the kinetics of GFP silencing and compared the silencing efficacy of DOSP liposomes with the effectiveness of commercial reagents (see SI Text and SI Figs. 6 and 7). These studies basically showed that GFP silencing was maximal for ≈5 days and that DOSP liposomes were the most efficient among the reagents tested under our conditions. Finally, direct evaluation of the cytotoxicity of DOSP/siRNA complexes evaluated in the three different cells lines used in the present study by flow cytometry and by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay clearly showed that the viability of the transfected cells was not affected (data not shown).
Discussion
Gene silencing requires a highly efficient method for delivery of the siRNA duplexes into the cytoplasm of cells. Thus, in the present study, we have investigated the relationships between the physicochemical properties and the gene silencing efficiency of siRNA complexes formed by cationic lipids characterized by aminoglycosidic head groups. First, the colloidal stability properties of lipidic aminoglycoside derivatives/siRNA complexes were found to strongly depend on their +/− mean charge ratio, a feature allowing the identification of three different zones (termed A, B, and C) corresponding, respectively, to negative, neutral, and positive complexes. Such a colloidal stability behavior had already been observed with DNA lipoplexes formed when mixing DNA with lipopolyamine micelles (9, 16–18), BGTC (refs. 11 and 19 and this study), and quaternary ammonium cationic liposomes (21, 22). The width of the neutral zone B observed with lipidic aminoglycoside derivative/DNA complexes was in fact similar to the width of lipidic amidinium derivative/DNA complexes. However, the zone B width was clearly smaller for the siRNA complexes formed by our lipidic aminoglycoside derivatives than for those formed by lipidic amidinium derivatives. In particular, the boundary between the zones A and B was shifted toward a lower charge ratio when siRNA was complexed with 4,5-DDS aminoglycoside derivatives in comparison with 4,6-DDS derivatives and BGTC. It is also noteworthy that, in all three zones, the mean diameter of the aminoglycoside derivative/siRNA complexes was smaller than the mean diameters of lipidic amidinium derivative/siRNA complexes. In zone C, the mean diameters of the siRNA complexes obtained with DOST and DOSK were, respectively, 81 and 105 nm, and they were even smaller for the DOSP- and DOSN-based complexes (58 and 57 nm, respectively).
Cryo-TEM analysis of the siRNA complexes revealed two different types of morphologies: large, concentric lamellar structures and grape-like structures with small lamellar microdomains. Thus, most importantly, complexation with siRNA transformed the initial unilamellar liposomes into lamellar supramolecular bioassemblies consisting of either concentric multilamellar structures or ordered microdomains. A regular spacing of ≈67.7 Å, which probably corresponds to alternating lipid bilayers with intercalated siRNA molecules, was measured by SAXS and cryo-TEM in both morphological types of bioassemblies. Moreover, as shown by cryo-TEM, siRNA molecules adopted a regular arrangement with a periodicity of ≈30 Å within the monolayer they formed in the concentric multilamellar structures. It is likely that the siRNA molecules are arranged in a similar manner within the grape-like structures, although this was not observed probably because of their very small sizes. The fact that siRNA complexes with different morphologies were formed by the two classes of aminoglycoside derivatives might be explained by specific geometry features, the 4,6-DDS and 4,5-DDS derivatives adopting cylindrical and conical shapes, respectively. These features could impact the formation and size of the initial liposomes and the complexes. It has indeed been shown that a concentric multilamellar organization over a long distance was favored when using large liposomes able to disrupt and reassemble, thereby sandwiching the nucleic acid molecules. On the other hand, small liposomes may behave like spherical micelles and lead to ordered lamellar microdomains, as shown with lipopolyamine micelles in ref. 19.
As stated above, RNAi-mediated gene silencing requires the efficient delivery of siRNA molecules into the cytoplasm of the target cells. Thus, we also investigated the efficiency of our well characterized lipidic aminoglycoside derivative/siRNA complexes for gene down-regulation. Clearly, highly effective complexes for gene silencing were obtained with 4,5-DDS derivative-based zone C siRNA complexes, characterized by their small diameter and high colloidal stability. In general, it is agreed that cationic lipid/nucleic acid complexes are internalized into cells through an endocytosis process mediated by electrostatic interactions between the positively charged lipoplexes and negative residues on the cell membrane (23). Thus, regarding trafficking of the siRNA molecules into and within the cells, aminoglycoside-derived cationic lipid/siRNA complexes may actually improve both siRNA internalization and endosomal escape into the cellular cytosol because of their particular physicochemical properties and structural features. For instance, an increased endosomal escape of the siRNA may be observed as a flip-flop mechanism (24), which could be favored by the flexibility of the aminoglycoside moiety of the cationic lipid. This phenomenon may be enhanced by the existence of lamellar fingerprint-like microdomains in the siRNA complexes formed by 4,5-DDS derivatives (DOSP and DOSN) because such microdomains may have a more destabilizing effect on the endosomal membrane than the complete well ordered onion-like structures obtained with BGTC and the 4,6-DDS derivatives DOSK and DOST. Such a destabilizing process is probably critical for the release of a large amount of siRNA into the cell cytoplasm and its subsequent incorporation into functional RNA-induced silencing complexes.
In conclusion, the present study enabled us to identify a class of cationic lipids composed of synthetic derivatives of natural compounds that are highly efficient for in vitro delivery of siRNA. These cationic lipids may thus become a critical constituent of future multimodular siRNA delivery systems for in vivo cell targeting after systemic administration, such as sterically stabilized neutral siRNA complexes equipped with targeting ligands at the distal end of polyethyleneglycol (PEG) stretches.
Materials and Methods
siRNA and Preparation of Complexes.
Anti-GFP siRNA (sense sequence: GCAAGCUGACCCUGAAGUUCAU) was obtained from Eurogentec (Seraing, Belgium). 3′-Rhodamine-labeled anti-GFP siRNA and negative control siRNA (AllStars Negative Control, sense sequence: UUCUCCGAACGUGUCACGU) were provided by Qiagen (Chatsworth, CA). Human anti-Lamin A/C siRNA was from Santa Cruz Biotechnology (Santa Cruz, CA). Lipidic aminoglycoside derivatives and BGTC were synthesized as described in refs. 10, 20, 25, and 26. DC-Chol and DOPE were from Avanti Polar Lipids (Alabaster, AL). Polyethyleneimine (25 kDa) was from Sigma (St. Louis, MO). RNAiFect, X-treme-GENE, and Lipofectamine 2000 were from Qiagen, Roche Diagnostics (Mannheim, Germany), and Invitrogen (Carlsbad, CA), respectively. Lipidic aminoglycoside derivatives/DOPE (1/1, mol/mol) and BGTC/DOPE (2/3, mol/mol) cationic liposomes were prepared as described in ref. 11, with minor changes; for instance, the dispersion was sonicated for 10 min and stored at 4°C. Complexes of siRNA with lipidic aminoglycoside derivatives or BGTC-DOPE were prepared by mixing equal volumes of cationic liposomes in water with siRNA at the desired concentration in 300 mM NaCl.
Dynamic Light Scattering, Ethidium Bromide Exclusion, and Cryo-TEM.
Dynamic light scattering, ethidium bromide fluorescence measurements, and cryo-TEM experiments were performed as described in ref. 27.
GFP-Expressing Cells and siRNA Transfection.
The H1299-derived GFP-expressing human lung cancer d2GFP cells (a generous gift from A. Chauchereau, Villejuif, France), and the HEK293 and HeLa cells were cultured at 37°C in 5% CO2/humidified atmosphere, in DMEM with glucose, l-glutamine, and pyruvate, supplemented with 1% streptomycin/penicillin (GIBCO and Invitrogen Life Technologies, Carlsbad, CA) and 10% FCS (Eurobio, Courtaboeuf, France). At day −1 before transfection, the cells were transferred onto 24-well culture plates, at 65,000 cells per well, resulting in ≈70–80% confluence 24 h later. Transfection was performed by adding 50 μl of complexes in 500 μl of serum-free DMEM to each well. After 2 h, the transfection medium was replaced by 1 ml of fresh medium. Transfection experiments were performed in triplicate.
GFP Fluorescence Measurements.
At 24 h after transfection, the cells were washed twice with 500 μl of PBS and then lysed with Reporter Lysis Buffer (Promega, Madison, WI) supplemented with a protease inhibitor mixture (Roche Diagnostics), complete lysis being ensured by one freezing–thawing (−80°C/20°C) cycle. Samples were then centrifuged at 10,000 × g for 5 min. GFP fluorescence measurements were performed on a 180-μl aliquot of supernatant by using a Victor2 apparatus (PerkinElmer, Les Ulis, France). Fluorescence was normalized to the total protein concentration of the sample, determined by using a BCA assay kit (Pierce, Rockford, IL). Residual GFP expression was expressed as percentage of the fluorescence of the control sample (treated with control siRNA).
Fluorescence Microscopy of Transfected d2GFP Cells.
Fluorescence microscopy examination was carried out by using an inverted fluorescence microscope Axiovert 200 M (Carl Zeiss, Göttingen, Germany).
Real-Time Quantitative RT-PCR.
Total RNA was extracted from transfected cells by TRIzol treatment. Reverse transcription was performed with total RNA by using oligo(dT)20 primers and SuperScript III reverse transcriptase (Invitrogen). The expression of lamin A/C or GFP was quantified by real-time PCR (ABI prism 7000, Applied Biosystems, Foster City, CA). Experiments were performed by using PCR Master Mix (Applied Biosystems) with 300 nM each primer and 250 nM TaqMan MGB probes. Primers were obtained from Applied Biosystems. The cycling conditions included a hot start at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and at 60°C for 1 min. Results were normalized to the endogenous hypoxanthine–guanine phosphoribosyltransferase (HPRT1) control gene and expressed according to the 2−ΔΔCt method (28).
Supplementary Material
Acknowledgments
We thank A. Chauchereau (Villejuif, France) for providing d2GFP cells, Solenne Goubain and Clothilde Gourden (In-Cell-Art, Nantes, France) for providing excellent technical expertise in cell culture and analysis of GFP silencing, and P. Méléard (Rennes, France) for helping with the SAXS experiments. This work was supported by an Action Concertée Incitative “Jeunes Chercheurs” grant from the “Ministère Délégué à la Recherche et aux Nouvelles Technologies,” by Grant 018716 from the European Union (Strep; SyntheGeneDelivery), and by special grants from the “Association Française contre les Myopathies” (Evry, France) and “Vaincre La Mucoviscidose” (Paris, France).
Abbreviations
- BGTC
bis(guanidinium)-tris(2-aminoethyl)amine-cholesterol
- cryo-TEM
cryo-transmission electron microscope
- DC-Chol
3-β-[N-(N′,N′-dimethylaminoethane)carbamoyl]cholesterol
- DOPE
dioleoyl phosphatidylethanolamine
- 4,5-DDS
4,5-disubstituted 2-deoxystreptamine
- 4,6-DDS
4,6-disubstituted 2-deoxystreptamine
- DOSK
dioleyl succinyl kanamycin A
- DOSN
dioleyl succinyl ethylthioneomycin B
- DOSP
dioleyl succinyl paromomycin
- DOST
dioleyl succinyl tobramycin
- SAXS
small-angle x-ray scattering.
Footnotes
Conflict of interest statement: P.L., J.-M.L., and B.P. own stock in In-Cell-Art Co., which commercializes lipidic aminoglycoside derivatives.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707431104/DC1.
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Mello CC, Conte D. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 3.Hannon GJ, Rossi JJ. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 4.Dykxhoorn DM, Palliser D, Lieberman J. Gene Ther. 2006;13:541–552. doi: 10.1038/sj.gt.3302703. [DOI] [PubMed] [Google Scholar]
- 5.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;24:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 6.Li CX, Parker A, Menocal E, Xiang S, Borodyansky L, Fruehauf JH. Cell Cycle. 2006;5:2103–2109. doi: 10.4161/cc.5.18.3192. [DOI] [PubMed] [Google Scholar]
- 7.Brazas RM, Hagstrom JE. Methods Enzymol. 2005;392:112–124. doi: 10.1016/S0076-6879(04)92007-1. [DOI] [PubMed] [Google Scholar]
- 8.Behr JP, Demeneix B, Loeffler JP, Perez-Mutul J. Proc Natl Acad Sci USA. 1989;86:6982–6986. doi: 10.1073/pnas.86.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pitard B, Aguerre O, Airiau M, Lachagès AM, Boukhnikachvili T, Byk G, Dubertret C, Herviou C, Scherman D, Mayaux J-F, Crouzet J. Proc Natl Acad Sci USA. 1997;94:14412–14417. doi: 10.1073/pnas.94.26.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vigneron J-P, Oudrhiri N, Fauquet M, Vergely L, Bradley J-C, Basseville M, Lehn P, Lehn J-M. Proc Natl Acad Sci USA. 1996;93:9682–9686. doi: 10.1073/pnas.93.18.9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pitard B, Oudhriri N, Vigneron JP, Hauchecorne M, Aguerre O, Toury R, Airiau M, Ramasawmy R, Scherman D, Crouzet J, et al. Proc Natl Acad Sci USA. 1999;96:2621–2626. doi: 10.1073/pnas.96.6.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pitard B. Somat Cell Mol Genet. 2002;27:5–15. doi: 10.1023/a:1022923721335. [DOI] [PubMed] [Google Scholar]
- 13.Grünweller A, Hartmann RK. Curr Med Chem. 2005;12:3143–3161. doi: 10.2174/092986705774933489. [DOI] [PubMed] [Google Scholar]
- 14.Moazed D, Noller HF. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- 15.Ogle JM, Brodersen DE, Clemons WM, Tarry MJ, Carter AP, Ramakrishnan V. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 16.Kreiss P, Cameron B, Rangara R, Mailhe P, Aguerre-Charriol O, Airiau M, Scherman D, Crouzet J, Pitard B. Nucleic Acids Res. 1999;27:3792–3798. doi: 10.1093/nar/27.19.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turek J, Dubertret C, Jaslin G, Antonakis K, Scherman D, Pitard B. J Gene Med. 2000;2:32–40. doi: 10.1002/(SICI)1521-2254(200001/02)2:1<32::AID-JGM78>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 18.Wetzer B, Byk G, Frederic M, Airiau M, Blanche F, Pitard B, Scherman D. Biochem J. 2001;15:747–756. doi: 10.1042/0264-6021:3560747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pitard B, Oudrhiri N, Lambert O, Vivien E, Masson C, Wetzer B, Hauchecorne M, Scherman D, Rigaud JL, Vigneron JP, et al. J Gene Med. 2001;3:478–487. doi: 10.1002/jgm.211. [DOI] [PubMed] [Google Scholar]
- 20.Sainlos M. Paris: Univ Pierre et Marie Curie; 2004. PhD thesis. [Google Scholar]
- 21.Lasic D, Strey H, Stuart M, Podgornik R, Frederik PM. J Am Chem Soc. 1997;119:832–833. [Google Scholar]
- 22.Radler JO, Koltover I, Salditt T, Safinya CR. Science. 1997;275:810–814. doi: 10.1126/science.275.5301.810. [DOI] [PubMed] [Google Scholar]
- 23.Mukherjee S, Ghosh RN, Maxfield FR. Physiol Rev. 1997;77:759–803. doi: 10.1152/physrev.1997.77.3.759. [DOI] [PubMed] [Google Scholar]
- 24.Xu Y, Szoka FC., Jr Biochemistry. 1996;7:5616–5623. doi: 10.1021/bi9602019. [DOI] [PubMed] [Google Scholar]
- 25.Sainlos M, Hauchecorne M, Oudrhiri N, Zertal-Zidani S, Aissaoui A, Vigneron J-P, Lehn J-M, Lehn P. ChemBioChem. 2005;6:1023–1033. doi: 10.1002/cbic.200400344. [DOI] [PubMed] [Google Scholar]
- 26.Belmont P, Aissaoui A, Hauchecorne M, Oudrhiri N, Petit L, Vigneron JP, Lehn J-M, Lehn P. J Gene Med. 2002;4:517–526. doi: 10.1002/jgm.297. [DOI] [PubMed] [Google Scholar]
- 27.Pitard B, Bello-Roufai M, Lambert O, Richard R, Desigaux L, Fernandes S, Lanctin C, Pollard H, Zeghal M, Rescan PY, Escande D. Nucleic Acids Res. 2004;32:e159. doi: 10.1093/nar/gnh153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.