Abstract
According to x-ray structure, the lactose permease (LacY) is a monomer organized into N- and C-terminal six-helix bundles that form a deep internal cavity open on the cytoplasmic side with a single sugar-binding site at the apex. The periplasmic side of the molecule is closed. During sugar/H+ symport, a cavity facing the periplasmic side is thought to open with closure of the inward-facing cytoplasmic cavity so that the sugar-binding site is alternately accessible to either face of the membrane. Double electron–electron resonance (DEER) is used here to measure interhelical distance changes induced by sugar binding to LacY. Nitroxide-labeled paired-Cys replacements were constructed at the ends of transmembrane helices on the cytoplasmic or periplasmic sides of wild-type LacY and in the conformationally restricted mutant Cys-154→Gly. Distances were then determined in the presence of galactosidic or nongalactosidic sugars. Strikingly, specific binding causes conformational rearrangement on both sides of the molecule. On the cytoplasmic side, each of six nitroxide-labeled pairs exhibits decreased interspin distances ranging from 4 to 21 Å. Conversely, on the periplasmic side, each of three spin-labeled pairs shows increased distances ranging from 4 to 14 Å. Thus, the inward-facing cytoplasmic cavity closes, and a cleft opens on the tightly packed periplasmic side. In the Cys-154→Gly mutant, sugar-induced closing is observed on the cytoplasmic face, but little or no change occurs on periplasmic side. The DEER measurements in conjunction with molecular modeling based on the x-ray structure provide strong support for the alternative access model and reveal a structure for the outward-facing conformer of LacY.
Keywords: conformational change, double electron–electron resonance, lactose permease, major facilitator superfamily, membrane transport
The lactose permease of Escherichia coli (LacY), a member of the major facilitator superfamily (MFS), utilizes free energy stored in an electrochemical H+ gradient (Δμ̃H+) to drive active transport by coupling the downhill, stoichiometric translocation of H+ with Δμ̃H+ to the uphill accumulation of galactopyranosides. In the absence of Δμ̃H+, LacY can also use free energy released from downhill translocation of galactosides in either direction to drive uphill translocation of H+ with generation of Δμ̃H+, the polarity of which depends on the direction of the sugar gradient (1, 2).
An x-ray structure of LacY (3) combined with a wealth of biochemical data (1, 2, 4, 5) has led to an alternating access model for sugar translocation across the membrane. By this means, the inward-facing cytoplasmic cavity closes with opening of an outward-facing cleft, thereby making the sugar-binding site alternatively accessible to either face of the membrane. A similar model has been proposed for the glycerol phosphate/phosphate antiporter GlpT, a related MFS protein (6). The alternating access model involves a global conformational change, which is consistent with the highly dynamic nature of LacY (2, 7–9).
Although it seems intuitively obvious that a pathway must open on the periplasmic side of LacY to allow access of sugar to the binding site, insight is foggy in this respect because all of the x-ray structures of LacY, which include the C154G mutant (3, 10) as well as the wild type (11), are in the same inward-facing conformation with an open cytoplasmic cavity and a closed periplasmic side. Nonetheless, site-directed alkylation studies (compiled in ref. 4) clearly indicate that an outward-facing conformation with a hydrophilic pathway on the periplasmic side of LacY exists during turnover.
Four-pulse double electron–electron resonance (DEER) (12, 13), combined with site-directed spin labeling (14, 15), is well suited for distance measurements in proteins in the 20- to 60-Å range, which is comparable with the size of LacY, and the method has been applied successfully to membrane proteins (16–19). The small size of the nitroxide spin-label minimizes uncertainty due to mobility of the label itself, and importantly, the signal reflects intramolecular interactions between two identical spin labels.
In this study, nine paired double-Cys mutants were labeled with a methanethiosulfonate spin label reagent (MTSL) to generate disulfide linked nitroxide side chains. The double-labeled proteins were prepared and tested for distance changes induced by sugar binding. Distances measured for apo LacY are in a good agreement with distances predicted by modeling, and the changes induced by sugar binding provide strong evidence for galactoside-induced opening of an outward-facing pathway on the periplasmic side of LacY and closing of the cytoplasmic cavity.
Results
Rationale for Construction of Mutants.
Paired double-Cys mutations were introduced into LacY at the ends of transmembrane helices in N- and C-terminal six-helix bundles on the cytoplasmic or periplasmic sides (Fig. 1). Cys replacements were made in regions of LacY predicted to undergo significant structural change as judged by (i) the proposed outward-facing conformation of LacY based on biochemical data and cross-linking experiments (3) and (ii) comparison of the LacY x-ray structure with that of EmrD (PDB ID: 2GFP), another MFS member, but in a conformation closed on both sides (20) [see supporting information (SI) Fig. 6]. In the proposed outward-facing conformation of LacY, the greatest distance changes relative to the inward-facing conformation are between the cytoplasmic ends of helices IV and V and helices X and XI, which should move toward one another upon sugar binding. As a result, the highly conserved hydrophobic residues Phe-140, Phe-334, and Tyr-350 (cytoplasmic ends of helices V, X, and XI, respectively) should facilitate closure of the inward-facing hydrophilic cavity by coming into close proximity as predicted (21). As shown in the x-ray structure, the periplasmic side of LacY in the inward-facing conformation is tightly packed and completely closed, denying access of sugar to the binding site from the periplasmic side (Fig. 1). Thus, the alternating access model predicts that in the outward-facing conformation, the periplasmic ends of the N- and C-terminal six-helix bundles should move apart to allow access to the binding site from the periplasmic side, and the cytoplasmic cavity should close.
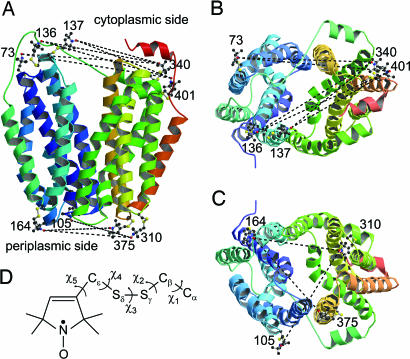
Fig. 1.
Disulfide-linked nitroxide chains are modeled on the LacY x-ray structure (PDB ID 1PV6) presented as rainbow colored ribbons from blue (helix I) to red (helix XII) with the hydrophilic cavity open to the cytoplasmic side. Nitroxides attached to the backbone of the protein are shown as balls and sticks. Interspin distances used for measurements are shown as the dashed lines. Cytoplasmic pairs R73C-S401C (helices III and XII), R73C-Q340C (helices III and X), S136C-Q340C (helices IV and X), S136C-S401C (helices IV and XII), N137C-Q340C (helices IV and X), and N137-S401C (helices IV and XII) are viewed from the side (A) and from cytoplasm (B). Periplasmic pairs V105C-T310C (helices IV and IX), I164C-T310C (helices V and IX), and I164C-S375C (helices V and XI) are viewed from side (A) and from periplasm (C). (D) Structure of the nitroxide side chain attached to the protein with indicated dihedral angles. Molscript (53) and Raster3D (54) were used in preparation of this illustration.
Molecular modeling of paired-Cys mutants modified with MTSL was carried out to predict limits for interspin distances (Fig. 1). The shortest and longest distances were estimated for the extreme positions of each nitroxide radical in sterically allowed conformations (Fig. 1D) (22–26). In each mutant, the nitroxide moieties are on the exterior face of LacY, which minimizes interactions of the attached spin labels with surrounding side chains. All distances calculated from the model are within the range favorable for DEER measurements (Table 1).
Table 1.
Properties of spin-labeled cysteine mutants of LacY
| Spin-labeled mutants | Nitroxide positions | NPGal binding* (relative to unlabeled mutant), % | Distances modeled for apo LacY† |
|||
|---|---|---|---|---|---|---|
| Ca-Ca, Å | Spin – spin distances, Å |
|||||
| Short | Long | Average | ||||
| R73C/S401C/WT | Cytoplasmic ends of helices III and XII | 69 | 41 | 44 | 52 | 48 |
| R73C/S401C/C154G | 93 | |||||
| R73C/Q340C/WT | Cytoplasmic ends of helices III and X | 63 | 36 | 38 | 48 | 43 |
| R73C/Q340C/C154G | 73 | |||||
| S136C/Q340C/WT | Cytoplasmic ends of helices IV and X | 62 | 34 | 39 | 51 | 45 |
| N137C/Q340C/WT | Cytoplasmic ends of helices IV and X | 95 | 32 | 30 | 46 | 38 |
| S136C /S401C/WT | Cytoplasmic ends of helices IV and XII | 60 | 40 | 43 | 55 | 49 |
| N137C /S401C/WT | Cytoplasmic ends of helices IV and XII | 57 | 38 | 33 | 52 | 43 |
| V105C/T310C/WT | Periplasmic ends of helices IV and IX | 80 | 34 | 30 | 50 | 40 |
| V105C/T310C/C154G | 94 | |||||
| I164C/T310C/WT | Periplasmic ends of helices V and IX | 97 | 27 | 24 | 41 | 33 |
| I164C/T310C/C154G | 88 | |||||
| I164C/S375C/WT | Periplasmic ends of helices V and XI | 94 | 33 | 30 | 45 | 38 |
*Detected by direct sugar-binding assay based on Trp-NPGal FRET as described in SI Materials and Methods.
†Estimated using LacY crystal structure (PDB ID: 1PV6) as described in Materials and Methods.
Individual Cys replacements, as well as alkylation of the single-Cys mutants with N-ethylmaleimide, have little or no effect on transport activity (27–31). Wild-type LacY was used as the background for introduction of easily accessible paired Cys replacements. With the exception of Cys-148, which is completely protected by d-galactopyranosyl-β-d-thiogalactopyranoside (TDG), the native Cys residues in wild-type LacY are not accessible under the stoichiometric conditions used for labeling (32). The conformationally constrained C154G mutant, which binds sugar as well as wild-type LacY but has very low transport activity (33–36), was used in parallel experiments for comparison. All purified mutants after modification of the Cys replacements with MTSL exhibit good sugar binding (Table 1).
Distance Changes on the Cytoplasmic Side.
Six double-Cys pairs on the cytoplasmic side of wild-type and C154G LacY were tested (Fig. 1 A and B). Results are presented in Figs. 2 and 3 where the background-corrected dipolar evolution data (time domains) and the corresponding dipolar spectra (frequency domains) are shown in columns A and B, respectively. The distance distributions shown in column C were obtained by using Tikhonov regularization (see Materials and Methods). Further analyses of distance distributions as multi-Gaussian fits with relative amounts of each distance population are also shown (black lines in panels C; also see SI Tables 2 and 3).
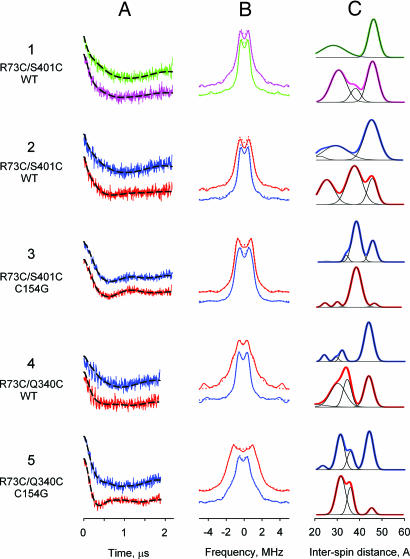
Fig. 2.
DEER characterization of sugar binding effects on interspin distances of nitroxide-labeled double-cysteine mutants located on the cytoplasmic side of LacY (helices III and XII or III and X). For each Cys pair listed on the left there are three panels: background corrected dipolar evolution data (echo amplitude recorded as a function of time) (A); dipolar spectra (Fourier transformation of the dipolar evolution data from A) (B); and distance distributions obtained by Tikhonov regularization (C). Data are shown for protein with no sugar bound (sucrose or NPGlc, green or blue, respectively) and with bound sugar (TDG or NPGal, pink or red lines, respectively). Broken lines in A and dotted lines in B are fits to the data by Tikhonov regularization. Plots in A and B are normalized by amplitude and shifted vertically for comparison. Plots in C show multi-Gaussian fits (black lines) demonstrating relative distributions of conformational populations (see also SI Table 2).
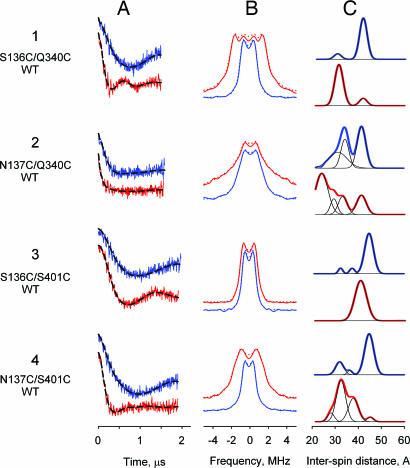
Fig. 3.
DEER characterization of sugar binding effects on interspin distances of nitroxide labeled double-cysteine mutants located on the cytoplasmic side of LacY (helices IV-X and IV-XII). Blue and red lines represent data obtained with NPGlc and NPGal, respectively. All other details are described in the legend for Fig. 2. Relative distributions of conformational populations calculated from multi-Gaussian fits are also presented in SI Table 3.
Distances between nitroxides bound at positions 73 (helix III) and 401 (helix XII) in the wild-type background (R73C/S401C/WT) were measured in the presence of four sugars: sucrose and 4-nitrophenyl-α-d-glucopyranoside (NPGlc), which do not bind to LacY; or TDG and 4-nitrophenyl-α-d-galactopyranoside (NPGal), which bind with relatively high affinity (37–40). Results with sucrose and NPGlc are practically identical. Thus, time domains, dipolar spectra and distance distributions are almost indistinguishable for both sugars [Fig. 2 1A–1C (green) and 2A–2C (blue)]. Distance distribution plots show two populations of molecules with a longer and a shorter distance between the cytoplasmic ends of helices III and XII [Fig. 2 1C (green) and 2C (blue)]. The major population corresponds to a conformer with 46 Å between nitroxide labels at positions 73 and 401, which is very close to modeled average of 48 Å (Table 1). In dramatic contrast, in the presence of either TDG or NPGal [Fig. 2 1A–1C (pink) and 2A–2C (red)], a population with a shorter interspin distance appears whereas the population exhibiting a longer distance decreases in amount [Fig. 2 1C (pink) and 2C (red) and SI Table 2]. Differences between distributions for TDG (short distance, 30 Å) and NPGal (short distances, 25 and 37 Å) are due likely to differences in binding interactions of the two sugars. Another spin-labeled Cys pair, R73C/Q340C/WT (helices III and X), shows a similar response to sugar binding (Fig. 2 4A–4C). With nonbinding NPGlc (blue), the major population of LacY exhibits an interspin distance of 44 Å. Upon NPGal binding, this population decreases significantly and a large population with a much shorter interspin distance of 30 Å predominates (red).
Shifts to shorter interspin distances in the presence of NPGal are also observed in the C154G background. Nitroxide labeled mutants R73C/S401C/C154G (helices III and XII; Fig. 2 3A–3C) or R73C/Q340C/C154G (helices III and X; Fig. 2 5A–5C) exhibit multiple populations with longest distances of 45 and 44 Å, respectively, in the presence of NPGlc. These long distance populations practically disappear upon NPGal binding with increases in populations exhibiting interspin distances of 38 and 31 Å, respectively (compare blue and red lines in Fig. 2 3C and 5C; SI Table 2).
Conformational changes on the cytoplasmic side of LacY are also observed in another direction between the ends of helix IV (position 136 or 137) and either helix X (position 340) or XII (position 401) (Fig. 1). Thus, binding of NPGal leads to decreases in interspin distances between the cytoplasmic ends of helices IV and X, as well as between helices IV and XII (Fig. 3; SI Table 3). The population of S136C/Q340C/WT molecules with an interspin distance of 41 Å shifts to a population with a distance of 30 Å in the presence of NPGal (Fig. 3 1C). Mutant N137C/Q340C/WT exhibits a much bigger shift; the major population with an interspin distance of 41 Å shifts to multiple distributions with a major population at 23 Å (Fig. 3 2C). Mutant S136C/S401C/WT in the presence of NPGlc shows an interspin distance of 44 Å that decreases to 40 Å after binding of NPGal (Fig. 3 3C). Remarkably, mutant N137C/S401C/WT with an interspin distance of 44 Å in the absence of a ligand shifts to a population with the much shorter distance of 31 Å in the presence of NPGal (Fig. 3 4C).
The observations with the cytoplasmic pairs indicate that NPGal binding leads to movement of the N- and C-terminal six-helix bundles toward each other. The large distance changes observed between helices III and X (14 Å), as well as helices III and XII (16–20 Å) should be sufficient to close the cytoplasmic cavity. Moreover, comparisons of the NPGal-induced distance changes discussed in relation to Fig. 3 indicate that neighboring positions in helix IV (136 and 137) exhibit unexpectedly different behavior, which is inconsistent with rigid body movements. The major population in the absence of ligand has the same interspin distance for positions 136 and 137 on helix IV paired with either helix X (41 Å) or XII (44 Å), but shifts differently when NPGal is bound (41 Å to 30 Å for S136C/Q340C and to 23 Å for N137C/Q340C; 44 Å to 40 Å for S136C/Q340C and to 31 Å for N137C/Q340C).
Distance Changes on the Periplasmic Side.
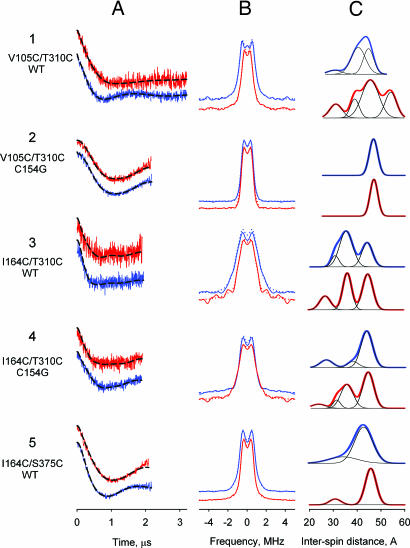
Three nitroxide-labeled Cys pairs on the periplasmic side were tested for ligand-dependent changes in interspin distance distribution (Fig. 1 A and C). The spin-labeled mutant V105C/T310C/WT (helices IV and IX) with nonbinding NPGlc displays a major population with 40 Å between the nitroxide moieties [Fig. 41A–1C (blue) and SI Table 4], which is in a good agreement with the average distance estimated from modeling (Table 1). NPGal binding causes a shift in distance distributions to 45 and 54 Å [Fig. 4 1A–1C (red) and SI Table 4]. In contrast, the same spin-labeled Cys pair in C154G background exhibits a single, narrow distance distribution of 47 Å in the presence of either NPGlc or NPGal [Fig. 4 2A–2C], although the mutant binds NPGal (Table 1). Therefore, the distance between the periplasmic ends of helices IV and IX increases upon ligand binding by 5–14 Å in wild-type LacY, and the C154G mutation completely blocks movement at these positions.
Fig. 4.
DEER characterization of sugar-binding effects on interspin distances of nitroxide-labeled double-cysteine mutants located on periplasmic side of LacY. Blue and red lines represent data obtained with NPGlc and NPGal, respectively. All other details are described in the legend for Fig. 2. Relative distributions of conformational populations calculated from multi-Gaussian fits are also presented in SI Table 4.
Nitroxide-labeled I164C/T310C/WT LacY (helices V and IX) exhibits a major population with an interspin distance of 35 Å in the presence of NPGlc (averaged modeled distance 33 Å; Table 1) and smaller population at 45 Å [Fig. 4 3A–3C (blue)]. Binding of NPGal leads to an increase in the population at 45 Å [Fig. 4 3A–3C (red) and SI Table 4] corresponding to 10 Å change in separation between the periplasmic ends of these helices. The same spin-labeled pair in the C154G background shows a predominant population at 44 Å, and no increase in distance is observed upon NPGal binding, although a population with a distance of 35 Å appears (Fig. 4 4A–4C and SI Table 4). Therefore, unlike wild-type LacY, mutant I164C/T310C/C154G does not exhibit an increase in distance between the periplasmic ends of helices V and IX in the presence of NPGal.
A third mutant examined is nitroxide-labeled I164C/S375C/WT (helices V and XI). The dipolar evolution data and corresponding dipolar spectra demonstrate a significant effect of NPGal binding with a shift in distance distribution from 42 to 46 Å (Fig. 4 5A–5C and SI Table 4). The findings are consistent with those from the other mutants in the wild-type background and with the conclusion that ligand binding induces opening on the periplasmic side of LacY.
Discussion
The observations presented here demonstrate directly that binding of ligand to wild-type LacY induces opposite movements of the cytoplasmic and periplasmic ends of transmembrane helices by 4–21 Å and 4–14 Å, respectively. As such, the DEER findings confirm and extend findings from cross-linking (41–43), site-directed alkylation (4), and single-molecule FRET (5). In addition to providing convergent support for the alternating access model, decomposition of the curves shown in Figs. 2–4, panels C, by multi-Gaussian fitting reveals three major populations of wild-type LacY conformers in many instances (Fig. 5 and SI Tables 2–4): conformer A, open on the cytoplasmic side/closed on the periplasmic side, which corresponds to the x-ray structures (3, 10, 11); conformer B, an intermediate conformer closed on both sides in a manner similar to EmrD (PDB ID: 2GFP) (20); and confomer C, open on the periplasmic side/closed on the cytoplasmic side. Distance changes between conformers A and C based on DEER measurements are very similar to the predicted differences between the inward-facing x-ray structure and the outward-facing model (3) (see SI Table 5 and SI Movie 1 for details).
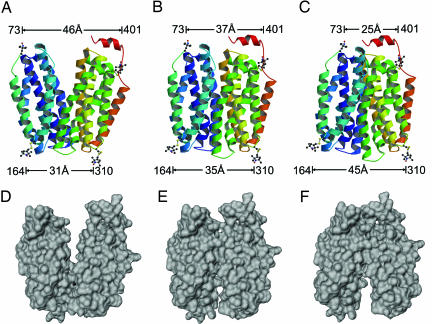
Fig. 5.
Molecular modeling of conformational changes in LacY upon sugar binding based on DEER distance measurements. Inward-facing conformation with no sugar bound (A), a transition state (B), and outward-facing conformation with bound sugar (C) correspond to three conformational populations of LacY observed in DEER measurements (see Figs. 2–4 and SI Tables 2–4). Transmembrane helices are rainbow colored from blue to red. Only two nitroxide pairs (cytoplasmic 73/401 and periplasmic 164/310) are shown as balls and sticks to demonstrate distance changes on cytoplasmic and periplasmic sides. The central loop between helices VI and VII is removed for clarity. Conformers with determined interspin distances are viewed normal to the membrane. (D–F) Space filling representations of conformers shown in A, B, and C, respectively, with helices II and VIII removed to demonstrate openings on cytoplasmic or periplasmic sides. The illustrations were prepared with Molscript (53), Raster3D (54), DINO (http://www.dino3d.org), and MSMS (55).
Conformational rearrangements between different states (Fig. 5) likely involve complex dynamics. Large changes in the lateral positions of the helices together with rotational movements including winding and unwinding of the ends of transmembrane helices may explain the dramatically different interspin distance shifts for neighboring positions 136 and 137 paired with either position 340 or 401 (Fig. 3 and SI Table 3). Such movements with tilting of helices are observed upon nucleotide binding to the β subunit of F1-ATPase (44) and upon Ca(II) binding to the transmembrane domain of Ca(II)-ATPase (45). The crystal structure of LacY also displays several transmembrane domains with kinks and bends that may provide a structural basis for flexibility of the molecule, which is apparent from the extremely fast H/D exchange observed for the peptide backbone in LacY (7, 8). With respect to the C154G mutant, both the DEER and single molecule FRET studies (5) show that, although ligand binding results in narrowing of the inward-facing cavity, no movement is observed on the periplasmic side.
Several attempts to model structural changes in LacY by using molecular dynamics approaches have been undertaken recently (46–49). DEER measurements of the type presented here should provide a powerful tool for guiding and evaluating these models.
Materials and Methods
Materials.
All galactoside derivatives were purchased from Sigma (St. Louis, MO). (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate (MTSL) was obtained from Toronto Research Chemicals (North York, Canada).
Labeling with MTSL.
Purified LacY mutants with engineered Cys pairs (40–50 μM) were incubated with MTSL (equimolar to each Cys residue introduced) in 50 mM NaPi (pH 7.2)/0.01% dodecyl-β-d-maltopyranoside (DDM) in the presence of 15 mM TDG to protect Cys-148, which is the only one of native Cys residues easily accessible for modification (32). The reaction was carried out for 15 min at room temperature. TDG and unreacted MTSL were removed by buffer exchange with 50 mM NaPi (pH 7.0)/0.01% DDM on an Amicon Ultra-15 concentrator with a 30-kDa cut-off. The samples were concentrated to 200–340 μM of protein, frozen in liquid nitrogen, and stored at −80°C before use.
Data Collection and Analysis.
Double-labeled LacY mutants dissolved in 50 mM NaPi (pH 7.4)/0.02% DDM were mixed with excess sugar (12 mM sucrose or TDG and 1 mM NPGlc or NPGal) and then with glycerol to a final concentration 20%. The samples (20 μl at a protein concentration of 140–240 μM) were loaded into quartz capillaries (1.8 mm o.d., 1.5 mm i.d.) and flash frozen in liquid nitrogen.
DEER measurements were conducted on a Bruker Elexsys 580 Pulse EPR Spectrometer equipped with a MS-2 2-mm split-ring resonator. The four-pulse DEER sequence of the Bruker assisted DEER experiment was used with pulses of 8 ns (90°) and 16 ns (180°), respectively, and evolution times of typically 1,400–3,000 ns depending on distance. The ELDOR pulse was placed at the center peak of the spectrum and the observation pulses at a 70-MHz distance away on the lowfield side. The temperature was either 50 K or 80 K, and the repetition rate adjusted to prevent saturation.
The phase-corrected dipolar evolution data were processed assuming a 3D background and Fourier transformed, and the distance distributions were obtained with Tikhonov regularization by using in-house programs developed by C.A. and also by using the DeerAnalysys2006.1 package (freely available from G. Jeschke at http://dg3.chemie.uni-konstanz.de/∼agje/G1.htm) (50). Distance distributions were analyzed by multiGaussian peak fitting by using Origin 7.5 (OriginLab Corp., Northhampton, MA).
Molecular Modeling.
Modeling of double-labeled mutants with MTSL is based on the crystal structure of unliganded protein (PDB ID: 1PV6). The side chains of amino acid residues in the protein molecule at positions of pairs 73–401, 73–340, 136–340, 137–340, 136–401, 137–401, 105–310, 164–310, and 164–375 were replaced by disulfide-linked nitroxide side chain by using XtalView (51). The dihedral angles (χ1, χ2, and χ3) for nitroxide side chain attached to the protein (Fig. 1D) were modeled according to the data presented by Guo et al. (26); χ4 and χ5 were varied to explore conformational space. The relative distances between two different nitroxide radicals were measured in XtalView (51).
To model LacY closed on the cytoplasmic side, the inward-facing conformation (PDB ID: 1PV6) was transferred into a Cartesian coordinate system so that the pivot point of the cytoplasmic and periplasmic domain movements was placed in the origin of the coordinate system. Distances corresponding to major populations of the protein and estimated for different spin-labeled pairs were used for modeling the three conformational states. LacY closed on the cytoplasmic side was produced by applying rotation matrices to both N- and C-halves of the molecule (residues 1–184 and 211–417, respectively) by using PDBSET (52). The 9- or 16-Å movements of the cytoplasmic ends of transmembrane helices toward each other and the 4- or 10-Å movements in the opposite direction at the periplasmic ends, respectively, correspond to rigid body rotation of 22° or 38° of N- and C-terminal halves of the molecule around an axis passing near Trp-151 at the apex of hydrophilic cavity in substrate binding site (Fig. 5 B and C).
All other materials, construction of mutants, protein purification, and sugar binding assay are described in SI Materials and Methods.
Acknowledgments
We thank J. Sugihara for excellent technical assistance. This work was supported in part by National Institutes of Health (NIH) Grants DK051131, DK069463, GM074929, and GM073210, as well as by National Science Foundation Grant MCB-0450970 (to H.R.K.) and by NIH Grant EY05216, the Jules Stein Professor Endowment, and the Ford Bundy and Anne Smith Bundy Foundation (to W.L.H.).
Abbreviations
- LacY
lactose/H+ symporter from Escherichia coli
- MFS
major facilitator superfamily
- NPGal
4-nitrophenyl-α-d-galactopyranoside
- NPGlc
4-nitrophenyl-α-d-glucopyranoside
- TDG
d-galactopyranosyl-β-d-thiogalactopyranoside
- DEER
double electron–electron resonance
- MTSL
methanethiosulfonate spin label (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate
- DDM
dodecyl-β-d-maltopyranoside.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708258104/DC1.
References
- 1.Kaback HR, Sahin-Toth M, Weinglass AB. Nat Rev Mol Cell Biol. 2001;2:610–620. doi: 10.1038/35085077. [DOI] [PubMed] [Google Scholar]
- 2.Guan L, Kaback HR. Annu Rev Biophys Biomol Struct. 2006;35:67–91. doi: 10.1146/annurev.biophys.35.040405.102005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 4.Kaback HR, Dunten R, Frillingos S, Venkatesan P, Kwaw I, Zhang W, Ermolova N. Proc Natl Acad Sci USA. 2007;104:491–494. doi: 10.1073/pnas.0609968104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Majumdar DS, Smirnova I, Kasho V, Nir E, Kong X, Weiss S, Kaback HR. Proc Natl Acad Sci USA. 2007;104:12640–12645. doi: 10.1073/pnas.0700969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 7.le Coutre J, Kaback HR, Patel CK, Heginbotham L, Miller C. Proc Natl Acad Sci USA. 1998;95:6114–6117. doi: 10.1073/pnas.95.11.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patzlaff JS, Moeller JA, Barry BA, Brooker RJ. Biochemistry. 1998;37:15363–15375. doi: 10.1021/bi981142x. [DOI] [PubMed] [Google Scholar]
- 9.Nie Y, Smirnova I, Kasho V, Kaback HR. J Biol Chem. 2006;281:35779–35784. doi: 10.1074/jbc.M607232200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mirza O, Guan L, Verner G, Iwata S, Kaback HR. EMBO J. 2006;25:1177–1183. doi: 10.1038/sj.emboj.7601028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan L, Mirza O, Verner G, Iwata S, Kaback HR. Proc Natl Acad Sci USA. 2007;104:15294–15298. doi: 10.1073/pnas.0707688104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. J Magn Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 13.Jeschke G. ChemPhysChem. 2002;3:927–932. doi: 10.1002/1439-7641(20021115)3:11<927::AID-CPHC927>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 14.Hubbell WL, Altenbach C. Curr Opin Struct Biol. 1994;4:566–573. [Google Scholar]
- 15.Hubbell WL, Mchaourab HA, Altenbach C, Lietzow MA. Structure (London) 1996;4:779–783. doi: 10.1016/s0969-2126(96)00085-8. [DOI] [PubMed] [Google Scholar]
- 16.Jeschke G, Wegener C, Nietschke M, Jung H, Steinhoff HJ. Biophys J. 2004;86:2551–2557. doi: 10.1016/S0006-3495(04)74310-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steigmiller S, Borsch M, Graber P, Huber M. Biochim Biophys Acta. 2005;1708:143–153. doi: 10.1016/j.bbabio.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Hilger D, Jung H, Padan E, Wegener C, Vogel KP, Steinhoff HJ, Jeschke G. Biophys J. 2005;89:1328–1338. doi: 10.1529/biophysj.105.062232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Q, Ellena JF, Kim M, Cafiso DS. Biochemistry. 2006;45:10847–10854. doi: 10.1021/bi061051x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin Y, He X, Szewczyk P, Nguyen T, Chang G. Science. 2006;312:741–744. doi: 10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasho VN, Smirnova IN, Kaback HR. J Mol Biol. 2006;358:1060–1070. doi: 10.1016/j.jmb.2006.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langen R, Oh KJ, Cascio D, Hubbell WL. Biochemistry. 2000;39:8396–8405. doi: 10.1021/bi000604f. [DOI] [PubMed] [Google Scholar]
- 23.Columbus L, Kalai T, Jeko J, Hideg K, Hubbell WL. Biochemistry. 2001;40:3828–3846. doi: 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]
- 24.Columbus L, Hubbell WL. Trends Biochem Sci. 2002;27:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 25.Lietzow MA, Hubbell WL. Biochemistry. 2004;43:3137–3151. doi: 10.1021/bi0360962. [DOI] [PubMed] [Google Scholar]
- 26.Guo Z, Cascio D, Hideg K, Kalai T, Hubbell WL. Protein Sci. 2007;16:1069–1086. doi: 10.1110/ps.062739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frillingos S, Sun J, Gonzalez A, Kaback HR. Biochemistry. 1997;36:269–273. doi: 10.1021/bi9618629. [DOI] [PubMed] [Google Scholar]
- 28.Frillingos S, Gonzalez A, Kaback HR. Biochemistry. 1997;36:14284–14290. doi: 10.1021/bi972314d. [DOI] [PubMed] [Google Scholar]
- 29.He MM, Sun J, Kaback HR. Biochemistry. 1996;35:12909–12914. doi: 10.1021/bi960876b. [DOI] [PubMed] [Google Scholar]
- 30.Frillingos S, Kaback HR. Biochemistry. 1996;35:5333–5338. doi: 10.1021/bi953068d. [DOI] [PubMed] [Google Scholar]
- 31.Sahin-Tóth M, Kaback HR. Protein Sci. 1993;2:1024–1033. doi: 10.1002/pro.5560020615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.le Coutre J, Whitelegge JP, Gross A, Turk E, Wright EM, Kaback HR, Faull KF. Biochemistry. 2000;39:4237–4242. doi: 10.1021/bi000150m. [DOI] [PubMed] [Google Scholar]
- 33.Menick DR, Sarkar HK, Poonian MS, Kaback HR. Biochem Biophys Res Commun. 1985;132:162–170. doi: 10.1016/0006-291x(85)91002-2. [DOI] [PubMed] [Google Scholar]
- 34.van Iwaarden PR, Driessen AJ, Lolkema JS, Kaback HR, Konings WN. Biochemistry. 1993;32:5419–5424. doi: 10.1021/bi00071a017. [DOI] [PubMed] [Google Scholar]
- 35.Smirnova IN, Kaback HR. Biochemistry. 2003;42:3025–3031. doi: 10.1021/bi027329c. [DOI] [PubMed] [Google Scholar]
- 36.Ermolova NV, Smirnova IN, Kasho VN, Kaback HR. Biochemistry. 2005;44:7669–7677. doi: 10.1021/bi0502801. [DOI] [PubMed] [Google Scholar]
- 37.Rudnick G, Schuldiner S, Kaback HR. Biochemistry. 1976;15:5126–5131. doi: 10.1021/bi00668a028. [DOI] [PubMed] [Google Scholar]
- 38.Wu J, Frillingos S, Voss J, Kaback HR. Protein Sci. 1994;3:2294–2301. doi: 10.1002/pro.5560031214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sahin-Tóth M, Gunawan P, Lawrence MC, Toyokuni T, Kaback HR. Biochemistry. 2002;41:13039–13045. doi: 10.1021/bi0203076. [DOI] [PubMed] [Google Scholar]
- 40.Smirnova IN, Kasho VN, Kaback HR. Biochemistry. 2006;45:15279–15287. doi: 10.1021/bi061632m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu J, Hardy D, Kaback HR. J Mol Biol. 1998;282:959–967. doi: 10.1006/jmbi.1998.2065. [DOI] [PubMed] [Google Scholar]
- 42.Wu J, Hardy D, Kaback HR. Biochemistry. 1999;38:2320–2325. doi: 10.1021/bi982288z. [DOI] [PubMed] [Google Scholar]
- 43.Kwaw I, Sun J, Kaback HR. Biochemistry. 2000;39:3134–3140. doi: 10.1021/bi992509g. [DOI] [PubMed] [Google Scholar]
- 44.Abrahams JP, Leslie AG, Lutter R, Walker JE. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 45.Toyoshima C, Nomura H. Nature. 2002;418:605–611. doi: 10.1038/nature00944. [DOI] [PubMed] [Google Scholar]
- 46.Yin Y, Jensen MO, Tajkhorshid E, Schulten K. Biophys J. 2006;91:3972–3985. doi: 10.1529/biophysj.106.085993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klauda JB, Brooks BR. J Mol Biol. 2007;367:1523–1534. doi: 10.1016/j.jmb.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bond PJ, Holyoake J, Ivetac A, Khalid S, Sansom MS. J Struct Biol. 2007;157:593–605. doi: 10.1016/j.jsb.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 49.Jensen MO, Yin Y, Tajkhorshid E, Schulten K. Biophys J. 2007;93:92–102. doi: 10.1529/biophysj.107.103994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeschke G, Chechik V, Ionita P, Godt A, Zimmermann H, Banham J, Timmel CR, Hilger D, Jung H. Appl Magn Reson. 2006;30:473–498. [Google Scholar]
- 51.McRee DE. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 52.Collaborative Computational Project No 4. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. [Google Scholar]
- 53.Kraulis PJ. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 54.Merritt EA, Bacon DJ. Methods Enzymol. 1977;277:504–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 55.Sanner MF, Olson AJ, Spehner JC. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]