Abstract
Amyloid β-protein (Aβ) oligomers may be the proximate neurotoxins in Alzheimer's disease (AD). Recently, to elucidate the oligomerization pathway, we studied Aβ monomer folding and identified a decapeptide segment of Aβ, 21Ala–22Glu–23Asp–24Val–25Gly–26Ser–27Asn–28Lys–29Gly–30Ala, within which turn formation appears to nucleate monomer folding. The turn is stabilized by hydrophobic interactions between Val-24 and Lys-28 and by long-range electrostatic interactions between Lys-28 and either Glu-22 or Asp-23. We hypothesized that turn destabilization might explain the effects of amino acid substitutions at Glu-22 and Asp-23 that cause familial forms of AD and cerebral amyloid angiopathy. To test this hypothesis, limited proteolysis, mass spectrometry, and solution-state NMR spectroscopy were used here to determine and compare the structure and stability of the Aβ(21–30) turn within wild-type Aβ and seven clinically relevant homologues. In addition, we determined the relative differences in folding free energies (ΔΔGf) among the mutant peptides. We observed that all of the disease-associated amino acid substitutions at Glu-22 or Asp-23 destabilized the turn and that the magnitude of the destabilization correlated with oligomerization propensity. The Ala21Gly (Flemish) substitution, outside the turn proper (Glu-22–Lys-28), displayed a stability similar to that of the wild-type peptide. The implications of these findings for understanding Aβ monomer folding and disease causation are discussed.
Abundant evidence links the amyloid β-protein (Aβ) with the neuropathogenesis of Alzheimer's disease (AD) (for recent reviews, see refs. 1 and 2). Aβ is a normal metabolite of the Aβ precursor (AβPP), from which Aβ is produced by endoproteolysis (3). Two predominant forms of Aβ exist in vivo, Aβ40 and Aβ42, which are 40- and 42-aa in length, respectively (2, 4, 5). Recent experimental and clinical evidence suggests that the primary neurotoxins in AD are Aβ oligomers or protofibrils (1, 6–10). Understanding the folding and oligomerization of nascent Aβ monomers thus has become an especially important aspect of current strategies for understanding AD etiology and developing therapeutic agents.
We have applied a multidisciplinary approach to the Aβ assembly problem. Initial studies used limited proteolysis coupled with mass spectrometry to determine whether monomeric Aβ possessed any stable or quasistable structure that could protect the peptide from proteolysis. Surprisingly, a 10-residue segment within both Aβ40 and Aβ42, Ala-21–Ala-30, was identified (11). The homologous decapeptide, Aβ(21–30), displayed protease resistance identical to that of full-length Aβ, suggesting that this region could organize monomer folding and thus be a folding nucleus. This suggestion was consistent with the observation that many folding nuclei studied in isolation are structurally stable (12–16). In fact, NMR studies of the Aβ(21–30) peptide revealed a turn in the Val-24–Lys-28 region that was stabilized by hydrophobic interactions between the isopropyl and n-butyl side chains of Val-24 and Lys-28, respectively, and by long-range electrostatic interactions between the Nϵ cation of Lys-28 and the side-chain carboxylate anions of Glu-22 or Asp-23 (11).
To study the structural dynamics of turn formation within the Aβ(21–30) region at high resolution, Borreguero et al. (17) used discrete MD and a “united-atom” protein model in in silico studies. Representative structures were remarkably similar to those determined by NMR. All-atom MD (18) or replica-exchange MD (19) simulations in explicit water have produced similar results. The conformational dynamics of Aβ(21–30) also have been simulated by using activation–relaxation techniques coupled with a coarse-grained force field (OPEP) (20). These studies revealed three families of structures, two similar to those observed by NMR (11) and a third family displaying a more open structure.
The aforementioned biochemical, NMR, and computational studies emphasize the potential importance of Glu-22 and Asp-23 in controlling the nucleation of Aβ monomer folding. Nature itself has provided evidence for the biological importance of residues in the Aβ monomer folding nucleus, because mutations within the Aβ coding region of AβPP that produce single amino acid substitutions in the turn region cause familial forms of AD (FAD) and cerebral amyloid angiopathy (CAA) (21). These diseases are referred to by the ethnicities of the kindreds in which they were discovered and include the Flemish (A21G) (22), Arctic (E22G) (23, 24), Dutch (E22Q) (25, 26), Italian (E22K) (27), and Iowa (D23N) (28) disease forms [supporting information (SI) Table 2]. We have hypothesized that the biophysical basis for the diseases occurring in patients expressing FAD- and CAA-linked amino acid substitutions at Glu-22 and Asp-23 is alteration of the stability of the monomer folding nucleus (11). We report here results of experimental studies testing this hypothesis.
Results
Probing Peptide Folding by Limited Proteolysis.
To examine the structural stability of the folding nuclei of wild-type Aβ and seven mutant** Aβ peptides (SI Table 2), we applied the technique of limited proteolysis coupled with mass spectrometry. Prior enzymological (11), spectroscopic (11), and computational (17–19) studies consistently showed the involvement of Lys-28 in stabilization of the monomer folding nucleus. Therefore, we used the Lys-specific protease trypsin to probe peptide structure.
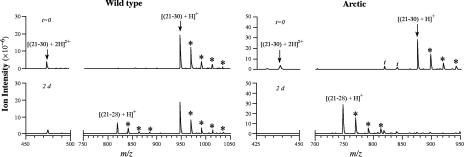
Representative mass spectra from wild-type and Arctic Aβ(21–30) are shown in Fig. 1. In the absence of trypsin, only protonated (m/z = 947.5 and 474.3) or sodiated (m/z = 969.5, 991.4, 1,013.3, and 1,035.3) molecular ion peaks were observed in the wild-type sample (Fig. 1). After 2 h of digestion, the intensities of the molecular ion peaks decreased and the Aβ(21–28) fragment ion appeared at m/z = 819.4 atomic mass units (data not shown). Smaller peaks at 841.4, 863.4, and 885.3 atomic mass units, corresponding to singly charged sodium adducts of Aβ(21–28), also were observed. The intensities of the Aβ(21–28) ions did not increase significantly in samples analyzed during prolonged digestion periods (up to 3 d; data not shown). Based on analysis of ion intensities, ≈27% of wild-type Aβ(21–30) was digested, on average. Arctic Aβ(21–30), in contrast, was digested quite efficiently (Fig. 1). Greater than half the peptide was digested by 6 h of incubation and the digestion was essentially complete by 1 d.
Fig. 1.
LC/MS of tryptic digests of Aβ(21–30) and Arctic Aβ(21–30). Samples were analyzed at the indicated times (t = 0 is peptide alone, no trypsin). Identities of ions are noted above arrows. Asterisks denote peaks of singly charged, multiply substituted sodium adducts separated by 22 atomic mass units.
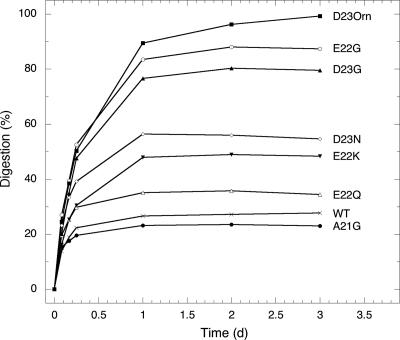
Analogous experiments were performed by using Flemish, Dutch, Italian, and Iowa Aβ(21–30) peptides. In addition, we substituted Gly or ornithine (Orn; 2,5-diaminopropanoic acid) for Asp-23 to produce Arctic- or Italian-type homologues, respectively, at this position (see SI Fig. 5). Fig. 2 presents progress curves for the digestion of each of the eight peptides. The representative mass spectra used to produce these curves are shown in Figs. 1 and SI Fig. 6. As with wild-type and Arctic Aβ(21–30), prominent molecular ions (singly or doubly protonated or singly or multiply sodiated) of each peptide were observed in the absence of trypsin. Rapid increases in the intensities of ions corresponding to each of the respective Aβ(21–28) tryptic fragments were observed within the first 2 h of digestion. Significant increases continued during the subsequent 24 h. Levels of the Flemish, Dutch, Italian, and Iowa tryptic fragments did not increase significantly upon extended incubation (up to 3 d). The intensities of the Arctic and Italian Asp-23 analogues (Asp23Gly and Asp23Orn, respectively) increased very modestly between 1 and 3 d.
Fig. 2.
Tryptic digestion progress curves. Percent digestion was calculated according to the formula (I0 − It/I0), where It is ion intensity at time t and I0 is intensity at t = 0. Each intensity was obtained by summing the intensities of all Aβ(21–28) ions (protonated and sodiated). The resulting percentage is plotted against incubation time.
In total, ≈35% of the Dutch decapeptides and ≈23% of the Flemish decapeptides were cleaved, relative to ≈25% for wild-type Aβ(21–30). The Iowa and Italian peptides were more protease sensitive. Approximately 50–55% digestion was observed for each. The Italian mutation creates an additional cleavage site, after Lys-22. Consistent with this fact, after 2 h, in addition to a prominent Aβ(21–28) fragment ion, smaller amounts of Aβ(23–28) and Aβ(23–30) were observed. The ion intensities of both the Aβ(21–28) and Aβ(23–28) fragments increased significantly during the first 24 h of digestion. The Arctic- and Italian-type substitutions at Asp-23 produced two of the three most proteolytically sensitive peptides (the third being the natural Arctic mutation) (Fig. 2). In total, ≈87% of the Asp23Gly and ≈99% Asp23Orn peptides were digested. In summary, the rank order of protease sensitivity, as measured by percent digestion at day 3, was Asp23Orn > Arctic > Asp23Gly ≫ Iowa ≈ Italian > Dutch > wild type ≥ Flemish (Table 1). The magnitudes of the inequalities specified above reflect the clustering of the sensitivities into three broad groups from most to least sensitive: (group 1) Asp23Orn, Glu22Gly, and Asp23Gly; (group 2) Asp23Asn and Glu22Lys; and (group 3) Glu22Gln, wild type, and Ala21Gly.
Table 1.
Aβ turn dynamics and oligomerization propensity
| Peptide | Digestion, % | IT | IR | ΔΔGf | Oligomerization propensity |
|
|---|---|---|---|---|---|---|
| Aβ40 | Aβ42 | |||||
| Flemish (A21G) | 23 | + | + | −0.3 | 6 | 6 |
| Wild type | 27 | +++ | + | 0 | 5 | 5 |
| Dutch (E22Q) | 35 | ++ | + | −0.1 | 4 | 1 |
| Italian (E22K) | 48 | ± | − | 1.2 | 2–3 | 2–3 |
| Iowa (D23N) | 55 | + | − | 0.9 | 2–3 | 2–3 |
| “Arctic 23” (D23G) | 80 | ± | − | 1.9 | ND | ND |
| Arctic (E22G) | 87 | + | − | 2.0 | 1 | 4 |
| “Italian 23” (D23Orn) | 99 | ± | − | 2.6 | ND | ND |
Shown are the final (plateau) trypsin digestion levels, as specified in Fig. 1; the Lys-28HζN TOCSY peak intensity (IT) (±, very weak; +, weak; ++, medium; +++, strong); the Glu-22αH–Gly-29NH, Glu-22αH–Ala-30NH ROESY cross-peak intensities (IR) (+, present; −, absent); the free energies of folding relative to wild type Aβ(21–30) in units of kBT (uncertainty in ΔΔGf values is ≈0.2 kBT); and the oligomerization propensity values for Aβ40 and Aβ42, determined by photoinduced cross-linking of unmodified proteins (36), with the highest propensity being 1.
Probing Peptide Folding by 2D Homonuclear NMR.
To examine the structure of the folding nucleus at higher resolution, we used solution-phase NMR. The backbone amide (NH) resonances of Aβ(21–30) were resolved in 1D 1H NMR spectra (data not shown) collected on a 1 mM peptide sample at 10°C, a temperature at which the NH proton chemical shift dispersion was optimal. The narrow line widths of the backbone NH proton resonances of Asp-22–Ala-30 remained unchanged across various temperatures, suggesting that Aβ(21–30) adopts a single global fold in solution (data not shown). All of the proton resonance assignments for the mutant Aβ(21–30) peptides were completed by using 2D 1H total correlation spectroscopy (TOCSY) and rotating-frame Overhauser enhancement (ROE) spectroscopy (ROESY) spectra and previously determined resonance assignments for wild-type Aβ(21–30) (11).
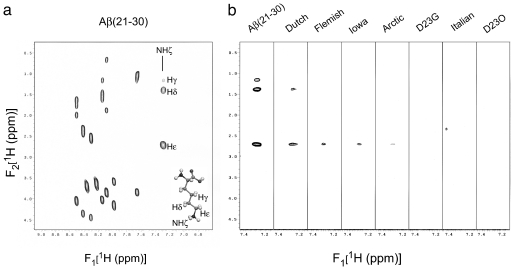
A representative TOCSY spectrum of wild-type Aβ(21–30) collected by using a 45-msec mixing time shows a single set of resolved resonances for each residue (Fig. 3a). In addition, the Hε-H;ζN, Hδ-HζN and Hγ-HζN side-chain resonances of Lys-28 were observed in the 7.2- to 7.4-ppm (F1) region of the spectrum, suggesting that this extended side-chain butyl group is immobilized on the NMR time scale at 10°C. Structure calculations of Aβ(21–30) revealed that attractive side-chain contacts between Glu-22 and Lys-28 and between Asp-23 and Lys-28, along with hydrophobic contacts between the isopropyl and n-butyl side chains of Val-24 and Lys-28, respectively, stabilize the turn structure within the Val-24–Lys-28 region of Aβ(21–30).
Fig. 3.
Two-dimensional 1H TOCSY NMR spectra. (a) The NH-αH region (6.6–9.1 ppm) of the spectrum of wild-type Aβ(21–30) in 25 mM d4-acetate, pH 6.0/H2O:2H2O at 9:1, collected at 10°C with a 45-msec mixing time. The amino acid Lys is rendered at the bottom right with side-chain protons labeled. Note that the individual HζN protons have degenerate chemical shifts. (b) Two-dimensional TOCSY spectra (7.2–7.4 ppm; NHζ cross-peak region) of wild-type and mutant Aβ(21–30) peptides collected under identical sample and experimental conditions.
TOCSY spectra then were collected for the Dutch, Iowa, Arctic, Flemish, Italian, Asp23Gly, and Asp23Orn peptides. Analysis of the spectral region from 7.2 to 7.4 ppm and comparisons of the intensity of Lys-28 HζN side-chain resonances to those in the wild-type peptide revealed the relative loss of immobilization (increased movement) of the Lys-28 side chain in each of the mutant peptides (Fig. 3b). Modest protection of the Lys-28 side-chain HζN correlation cross-peak intensity from exchange broadening was observed only in the Dutch peptide TOCSY data. Weak protection of the Lys-28 side-chain butyl group cross-peak intensity was observed in the Flemish and Iowa peptide TOCSY data. For the Arctic, Asp23Gly, Italian, and Asp23Orn peptides, the Lys-28 butyl group correlation cross-peaks were almost or completely absent, suggesting that protection of the Lys butyl group from exchange broadening is highly reduced in each of these homologues relative to the wild-type decapeptide (Fig. 3b). To various degrees, all of the amino acid substitutions studied thus increased Lys-28 side-chain mobility (Table 1).
Spatial Proton Connectivities in 2D ROESY Data.
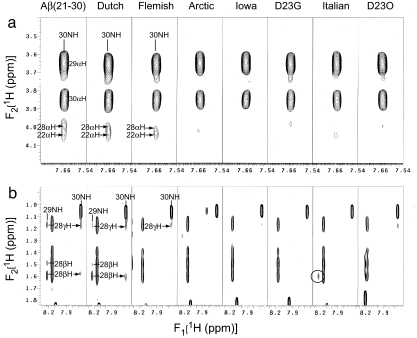
To examine further the effects of amino acid substitutions on folding nucleus stability, we examined sequential, mid- and long-range ROE cross-peaks in 2D ROESY spectra. Sequential Lys-28αH–Ala-30NH(i, i + 2) and long-range Glu-22αH–Ala-30NH connectivity ROE cross-peaks in expanded NH-αH proton regions of the ROESY spectra are shown in Fig. 4a. The single long-range, weak ROE between the αH proton of residue 22 and the amide proton of Ala-30 previously provided evidence of a folded backbone turn in wild-type Aβ(21–30) (11). These sequential Lys-28αH–Ala-30NH(i, i+ 2) and long-range Glu-22αH–Ala-30NH connectivities are partially preserved in the Dutch, significantly reduced in the Flemish, and not observed in the Arctic, Iowa, Asp23Gly, Italian, or Asp23Orn peptides. These data are consistent with the respective magnitudes of loss of Lys-28 side-chain butyl group cross-peaks in the TOCSY data (Fig. 3 and Table 1).
Fig. 4.
Two-dimensional 1H ROESY NMR spectra. (a) Expanded regions from each of the 2D ROESY spectra of wild-type and mutant Aβ(21–30) peptides, as indicated, in 25 mM acetate, pH 6.0/H2O:2H2O at 9:1, collected at 10°C with a mixing time of 300 msec. Connectivities are illustrated for sequential Lys-28αH–Ala-30NH(i, i + 2) and long-range Glu-22αH–Ala-30NH resonances with labeled arrows. (b) Connectivities for sequential Lys-28βH–Gly-29NH(i, i + 1) and Lys-28γH–Gly-29NH(i, i + 1), and Lys-28βH–Ala-30NH(i, i + 1) and Lys-28γH–Ala-30NH(i, i + 1) resonances. The circle in the Italian spectral segment highlights the Lys-28βH–Gly-29NH(i, i + 1) cross-peak that is missing in all other mutants.
Fig. 4b shows an expanded NH–βH/γH proton region of the ROESY spectrum for each peptide. Weak ROEs between Lys-28 side-chain protons and the backbone amide proton of Gly-29 or Ala-30 suggest stabilization of the Lys-28 side-chain in wild-type Aβ(21–30). These ROEs are preserved in the Dutch peptide, significantly reduced in the Flemish homologue, and absent in the Arctic, Iowa, Asp23Gly, Italian, and Asp23Orn homologues. Our data suggest that interresidue interactions in Aβ(21–30) between Lys-28 and Gly-29 and between Lys-28 and Ala-30 depend on Glu-22/Asp-23–Lys-28 electrostatic interactions.
The correlation between the turn conformation of Aβ(21–30) and stability of the Glu-22/Asp-23–Lys-28 electrostatic interactions is further supported by additional observations in the ROESY data for the mutant peptides compared with wild-type peptide. For example, as shown in SI Fig. 7, the adjacent ROE cross-peaks correlating Ser-26βH–Asn-27NH and Asn-27βH–Lys-28NH are weakened in all mutant peptides in which protection of the side-chain n-butyl group of Lys-28 from exchange broadening (i.e., side-chain immobilization) in TOCSY studies was completely or almost completely lost (namely, the Iowa, Asp23Gly, Italian, and Asp23Orn peptides) (Fig. 3b).
Discussion
The impetus for the experiments reported here was the identification and characterization of a monomer folding nucleus, common to Aβ40 and Aβ42, between Ala-21 and Ala-30 (11). This finding was significant because five of seven FAD/CAA-linked AβPP mutations cluster within the gene region encoding this decapeptide (22–25, 27, 28). We postulated that the mechanistic effect of the amino acid substitutions resulting from these clustered AβPP mutations was alteration in the stability of the folding nucleus (11). To test this hypothesis, here we systematically altered the primary structure of the Aβ folding nucleus, Aβ(21–30), and then probed its conformation and stability. Consistent with our postulation, we found that amino acid substitutions at Glu-22 and Asp-23 destabilized the folding nucleus. In addition, engineered changes in Asp-23 side-chain structure, analogous to those produced at Glu-22 by the Italian and Arctic mutations, resulted in profound destabilization of the folding nucleus. In contrast, the Flemish substitution, which affects an amino acid not found to be involved in turn stabilization (11), had little effect.
Structural Factors Controlling Folding Nucleus Stability.
The rank order of turn stability among the peptides studied (Table 1) emphasizes the importance of residues within the turn proper (Glu-22 and Asp-23) that stabilize its structure through side-chain–side-chain interactions. Substitutions for these residues probed predominately two classes of effects, electrostatic and entropic. All six substitutions eliminated the electrostatic interaction between Lys-28 and the side chains of amino acids at positions 22 or 23. Three of the four most destabilizing substitutions were at Asp-23. The “Italian-type” substitution at Asp-23, Asp23Orn, had the greatest effect, as evidenced by the complete digestion of the Asp23Orn decapeptide. The actual Italian Glu22Lys substitution produced half that level of digestion. Similarly, the Iowa (Asp23Asn) carboxylate→carboxyamide substitution caused approximately twice the level of destabilization as did the structurally analogous Dutch (Glu22Gln) mutation. The lesser destabilization by the carboxyamide side-chain substitutions relative to the Gly and Orn substitutions may result from the ability of the carboxyamides to hydrogen bond with the Nζ atom of Lys-28.
Our NMR data emphasize the importance of electrostatic stabilization of the Lys-28 side chain and suggest that this stabilization primarily, but not exclusively, depends on the electrostatic character of Asp-23. We observed very strong destabilization of the HζN Lys-28 side-chain n-butyl group when the anionic carboxylate side chain of Asp-23 was eliminated, e.g., as in the Iowa (Asp23Asn), Asp23Gly, and Asp23Orn peptides. These data are consistent with results of simulations of Aβ folding and assembly that reveal that Asp-23 plays a central role in organizing the Aβ(21–30) turn motif (19, 29). Destabilization of the Lys-28 n-butyl group also was observed in mutants that preserve the anionic carboxylate side chain of Asp-23 (Dutch, Flemish, and Arctic). The Asp-23–Lys-28 electrostatic interaction is partially destabilized by neutral charge substitutions at Glu-22 (Dutch and Arctic) or Gly substitution at Ala-21 (Flemish) and completely eliminated by the Italian Glu22Lys substitution. For the Dutch substitution, results of all-atom MD simulations of Aβ(10–35)-amide conformational dynamics, a model for full-length Aβ monomer folding (30), also emphasize fold destabilization as a key mechanistic explanation for the pathogenetic effects of the substitution (31). The Italian data are interesting because some (≈50%) protease resistance is observed in the absence of stabilizing electrostatic interactions. A formal, experimentally determined explanation for this phenomenon does not exist, but reasonable speculation can be offered. Specifically, our sequential ROE data show that some structure is retained in the Ser-26–Gly-29 segment of the Italian peptide. For example, a weak Lys-28βH–Gly29NH ROESY cross-peak was observed (Fig. 4b, circled intensity). It also is possible that new or increased stabilizing hydrophobic interactions may occur among the Lys-22 n-butyl side chain and residues such as Ala-30 and Val-24 or between Lys-28 and Val-24. Such protease resistance requires some peptide structure, but this structure may differ from that of the wild-type Aβ(21–30) peptide.
Gly substitutions, as in the Arctic E22G and “Arctic-type” D23G peptides, were highly destabilizing (≈80–90% trypsin digestion). Elimination of the carboxylate groups not only prevents electrostatic interactions but also significantly expands the conformational space accessible to the peptide. This creates an entropic “penalty” for turn formation. Gly is a well known disruptor of regular secondary structure in proteins, e.g., of α-helices (32). In our experiments, only the Asp23Orn substitution was more destabilizing than the Gly substitutions but by only ≈10–20%. The conformational contributions of residues 21 and 22 to the electrostatic stabilization of the Asp-23–Lys-28 side-chain interaction in Aβ(21–30) are revealed by the NMR data for the Flemish and Arctic peptides. Insertion of Gly at either of these positions increases the local conformational flexibility, resulting in the destabilization of the electrostatic Asp-23–Lys-28 interaction. In addition, the finding that the Asp-23–Lys-28 interaction in the Arctic peptide is destabilized more than in the Dutch peptide suggests that the charge neutralization of Glu-22 per se and the increased conformational flexibility provided by Gly substitution act additively to decrease stability. We also note that side-chain elimination precludes hydrophobic interactions involving the ethylene or methyl portions of the Ala-21 and Glu-22 residues, respectively.
In conclusion, we infer from our data that electrostatic interactions between Asp-23 and Lys-28 are particularly important, relative to Glu-22, in organizing the decapeptide fold and that entropic and hydrophobic factors can significantly alter folding.
Thermodynamic Analysis of Nucleus Stability.
The trypsin proteolysis progress curves (Fig. 2) reveal effects of mutations on the endoproteinase sensitivity of the Aβ folding nucleus. The cause of these effects is the relative difference in the free energies of folding (ΔΔGf) (Table 1) among wild-type and mutant peptides. Here, we formulate a kinetic scheme consistent with experimental observations and summarize key mathematical aspects of the analysis that yields ΔΔGf values. (A full discussion is provided in SI Text.)
In the presence of active trypsin, the Lys-28–Gly-29 peptide bond eventually should be cleaved completely in all peptides. Furthermore, one would expect similar progress curves for all eight peptides because bond scission involves the same sequence of residues and thus the binding and proteolysis processes should be equivalent. Our data (Fig. 2) do not show these expected behaviors. The most obvious explanation for incomplete proteolysis is trypsin autodigestion (33, 34). The large differences among the progress curves must be ascribed to peptide-specific differences in folding and the effect of this folding on enzyme:substrate (E:S) complex formation. We thus incorporate terms for enzyme inactivation and peptide unfolding and complexation with enzyme in our formulation (see SI Figs. 8 and 9). Eqs. 1 and 2 below define the kinetic evolution of our system, where A is concentration of Aβ, Z is concentration of trypsin, σ is the rate constant for peptide proteolysis within the E:S complex, K is the equilibrium constant for E:S complex formation, and γ and α are rates for first- and second-order enzyme inactivation processes.
It is important to note that K is equal to the product of two equilibrium constants, Kf, that for the two-state transition between the unfolded peptide monomer and the turn structure comprising the folding nucleus, and Kb, that for the binding of unfolded substrate to enzyme. Eq. 1 (Eq. 9 in SI Text) stipulates that the rate of substrate consumption is proportional to the concentration of E:S complex, AZ/(1 + KA). Eq. 2 (Eq. 11 in SI Text) describes the temporal change in active enzyme concentration. The inactivation rate is proportional to the concentration of free enzyme, Z/(1 + KA), and may be first- or second-order. The final digestion level A∞/A0 is given by Eq. 3, in which Z0 is initial enzyme concentration and κ = (αZ0/γ):
We determined γ and αZ0 experimentally (see SI Text) and thus were able to determine κ. That reduces the description of Aβ proteolysis to the two variables K and σ. Using the known initial E:S ratio (Z0/A0) of 1/100, we fit the peptide digestion progress curves by numerically solving Eqs. 1 and 2 and optimizing the two parameters KA0 and σA0/γ that define the solutions to these equations. The resulting fits to the peptide digestion progress curves were excellent for all peptides (see SI Fig. 10).
As discussed above, K = Kf × Kb, and Kb should be approximately the same for all unfolded peptides. If the latter assumption and our consideration of the structural similarity of the peptides around the scissile peptide bond Lys-28–Gly-29 are true, we would expect similar rates of proteolysis of bound substrate. In fact, when we interpret our formulation in the context of Michaelis–Menten kinetics by determining the substrate turnover rate λ = σK, we find that the magnitude of λ is similar among all peptides (SI Table 3). Therefore, because ΔG = −RT ln(Kf/Kb), we can extract ΔΔGf values for each mutant peptide by calculating the quotient e(Ki/KWT) (see Table 1 and SI Table 3). The ΔΔGf values clustered in three groups analogous to the clustering observed in proteolysis, namely, D23Orn, Arctic, and D23G (1.6–2.6 kBT), Iowa and Italian (0.7–1.2 kBT), and Dutch and Flemish (−0.2–0.2 kBT). It is useful conceptually to consider these energy differences in terms of probabilities that the peptides will be in an unfolded state (digestible). The differences thus provide a quantitative measure of the stability of the folding nucleus. Relative to the wild-type peptide, the most readily digested peptides are ≈7- to 12-fold (e1.9–e2.6) less stable, the second cluster peptides are ≈3-fold less stable (e0.9–e1.2), and the remaining peptides are of approximately equivalent stability (e−0.3–e−0.1).
Nucleus Stability and Aβ Assembly.
To monitor the initial oligomerization of Aβ40 and Aβ42, Bitan et al. (35–37) used photochemical cross-linking [photoinduced cross-linking of unmodified proteins (35)] to “freeze” the highly dynamic monomer–oligomer equilibria (36). Aβ40 oligomer order was restricted primarily to n < 5, with similar amounts of each oligomer formed, whereas the Aβ42 distribution was characterized by nodes at n ≈ 6, 12, and 18 (37). Examination of the same FAD/CAA mutations studied here revealed that the propensity of Aβ40 to form large oligomers (n > 4) followed the rank order Arctic > Italian ≈ Iowa > Dutch > wild type > Flemish (36), with the Arctic having the highest propensity. This order correlated perfectly with nucleus instability (Table 1 and SI Table 3). A similar correlation was observed between nucleus instability and the propensity of Aβ42 to form dodecamers, octadecamers, and higher-order assemblies. The exception was an exchange in rank order between the Dutch and Arctic peptides, which reflects isoform-specific differences in assembly. In addition to studies of lower-order oligomerization, the effects of substitutions at position 22 on protofibril formation have been examined. Päiviö et al. (38) showed that the rank order of protofibril propensity was Arctic > Dutch > wild type, identical to the Aβ40 rank order of oligomerization.
We now consider how the existence of a common nuclear fold may be integrated into isoform-specific folding and assembly and how this integration may explain the biophysical effects of mutations. Our original discovery of a turn-type monomer folding nucleus common to Aβ40 and Aβ42 was consistent with the identity of primary structure in the N-terminal 40 aa of these peptides (11). Experimental data show that formation of the turn is a kinetically favorable folding event that facilitates the interaction between the central hydrophobic cluster (CHC; 17Leu–18Val–19Phe–20Phe–21Ala) and C terminus. This interaction likely explains the isoform-specific oligomerization differences discussed above, because the C terminus defines the two isoforms. In fact, computational studies have yielded contact maps showing extensive interactions between the Aβ42 C terminus and the CHC that are not observed in Aβ40 (39). Subsequent simulations also have shown that Aβ42 forms a unique hairpin structure stabilized by C-terminal–CHC interactions (M. F. Yang and D.B.T., unpublished data). Importantly, recent scanning tyrosine intrinsic fluorescence studies of Aβ40 and Aβ42 fibril assembly revealed significant differences in the environment at position 20, directly adjacent to the folding nucleus (40). In particular, substantial structural rearrangement of the Aβ40 monomer occurs. The necessity for such rearrangement during Aβ40 fibril formation is consistent with the especially strong correlation noted above between nucleus instability and oligomerization order.
Our data suggest that rearrangement of the monomer folding nucleus is necessary for higher-order peptide assembly. Recent NMR studies of Aβ fibrils provide evidence supporting this postulation (41, 42). Petkova et al. (41) used solid-state NMR to probe the structure of the Aβ40 fibril. Strong electrostatic interactions were observed between D23 and K28 that were modeled best by intermolecular interactions between the cognate chains of peptide monomers i and i + 2 [“STAG(+2)”]. Luhrs et al. (42) used hydrogen/deuterium exchange NMR to study the structure of Aβ42 fibrils. They also observed an intermolecular Lys-28–Asp-23 salt bridge. The necessity for pre facto turn destabilization in monomer conformational transitions also has been suggested in in silico studies of the formation of compact collapsed coil and α-helix monomer structures from turn-containing precursors (43). Recent simulations of wild-type and A21G isoforms of Aβ40 and Aβ42 have revealed both length-dependent (Aβ40 versus Aβ42) and sequence-dependent (Ala-21 versus Gly-21) differences in the frequency distribution of intra- and intermolecular salt bridges involving Glu-22, Asp-23, and Lys-28 (44). Taken together, these results provide an explanation for the ostensibly counterintuitive observation that mutations destabilizing the initial Aβ fold lead to disease: They drive the intermolecular peptide interactions necessary for higher-order peptide assembly.
Finally, and importantly, our data also may be of relevance to understanding AβPP processing. In studies of γ-secretase, the enzyme complex that releases Aβ from AβPP through cleavage at the Aβ C terminus (45), Zhang et al. (46) have shown that the region of AβPP immediately preceding the transmembrane domain [equivalent to Aβ(10–28)] affects the intramembranous proteolytic processing that produces Aβ40, Aβ42, and related Aβ isoforms. In particular, Ser-26 and Lys-28 have been found to be key residues controlling this process (47). It has been hypothesized that γ-secretase is a “two-site” enzyme that contains a binding site for the AβPP substrate, from which the substrate is translated into a second site, the active site, in which peptide bond cleavage occurs (45). One or both of these interactions may be affected by the folding of the Aβ(21–30) region of AβPP, which would alter the interactions of amino acids in this juxtamembranous portion of AβPP with contact residues in γ-secretase.
Materials and Methods
Peptides.
The primary structure of Aβ(21–30) is 21Ala–22Glu–23Asp–24Val–25Gly–26Ser–27Asn–28Lys–29Gly–30Ala (SI Table 2 provides a list of all peptides). Each peptide was synthesized chemically as previously described (11).
Limited Proteolysis.
Peptides were dissolved in 25 mM ammonium acetate, pH 7.1, at a concentration of ≈0.5 mg/ml. Enzymatic digestions were done at 25°C using trypsin (Roche Applied Science, Indianapolis, IN) at an E:S ratio of 1:100 (wt/wt) as described previously (11). Four independent experiments were performed using each peptide, with the exception of the Dutch peptide, with which five experiments were performed.
Liquid Chromatography/Mass Spectrometry.
Liquid chromotography/mass spectrometry was performed essentially as described previously (11). Before analysis, aliquots of peptide digests were acidified with 1% (vol/vol) trifluoroacetic acid in water.
NMR Spectroscopy.
One-dimensional 1H and 2D 1H–1H (homonuclear) NMR data were recorded as described previously (11).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants NS038328, and AG027818 and by a Zenith Grant from the Alzheimer's Association.
Abbreviations
- AD
Alzheimer's disease
- Aβ
amyloid β-protein
- AβPP
Aβ precursor
- CAA
cerebral amyloid angiopathy
- E:S
enzyme:substrate
- FAD
familial AD
- MD
molecular dynamics
- Orn
ornithine
- ROE
rotating-frame Overhauser enhancement
- ROESY
ROE spectroscopy
- TOCSY
total correlation spectroscopy.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705197104/DC1.
The term “mutant,” as used here in the context of peptides, distinguishes those peptides containing disease-linked amino acid substitutions from wild-type Aβ peptides.
References
- 1.Walsh DM, Selkoe DJ. Neuron. 2004;44:181–193. doi: 10.1016/j.neuron.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 2.Lazo ND, Maji SK, Fradinger EA, Bitan G, Teplow DB. In: Amyloid Proteins-The Beta Sheet Conformation and Disease. Sipe JC, editor. Weinheim, Germany: Wiley–VCH; 2005. pp. 385–492. [Google Scholar]
- 3.Wilquet V, De Strooper B. Curr Opin Neurobiol. 2004;14:582–588. doi: 10.1016/j.conb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Mori H, Takio K, Ogawara M, Selkoe DJ. J Biol Chem. 1992;267:17082–17086. [PubMed] [Google Scholar]
- 5.Roher AE, Lowenson JD, Clarke S, Woods AS, Cotter RJ, Gowing E, Ball MJ. Proc Natl Acad Sci USA. 1993;90:10836–10840. doi: 10.1073/pnas.90.22.10836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirkitadze MD, Bitan G, Teplow DB. J Neurosci Res. 2002;69:567–577. doi: 10.1002/jnr.10328. [DOI] [PubMed] [Google Scholar]
- 7.Klein WL, Stine WB, Jr, Teplow DB. Neurobiol Aging. 2004;25:569–580. doi: 10.1016/j.neurobiolaging.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 8.Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, Morgan TE, Rozovsky I, Trommer B, Viola KL, et al. Proc Natl Acad Sci USA. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pitschke M, Prior R, Haupt M, Riesner D. Nat Med. 1998;4:832–834. doi: 10.1038/nm0798-832. [DOI] [PubMed] [Google Scholar]
- 10.Haass C, Selkoe DJ. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 11.Lazo ND, Grant MA, Condron MC, Rigby AC, Teplow DB. Protein Sci. 2005;14:1581–1596. doi: 10.1110/ps.041292205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neira JL, Fersht AR. Fold Des. 1996;1:231–241. doi: 10.1016/s1359-0278(96)00034-x. [DOI] [PubMed] [Google Scholar]
- 13.Honda S, Kobayashi N, Munekata E. J Mol Biol. 2000;295:269–278. doi: 10.1006/jmbi.1999.3346. [DOI] [PubMed] [Google Scholar]
- 14.Kammerer RA, Schulthess T, Landwehr R, Lustig A, Engel J, Aebi U, Steinmetz MO. Proc Natl Acad Sci USA. 1998;95:13419–13424. doi: 10.1073/pnas.95.23.13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuroda Y. Biochemistry. 1993;32:1219–1224. doi: 10.1021/bi00056a004. [DOI] [PubMed] [Google Scholar]
- 16.Ramirez-Alvarado M, Serrano L, Blanco FJ. Protein Sci. 1997;6:162–174. doi: 10.1002/pro.5560060119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borreguero JM, Urbanc B, Lazo ND, Buldyrev SV, Teplow DB, Stanley HE. Proc Natl Acad Sci USA. 2005;102:6015–6020. doi: 10.1073/pnas.0502006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cruz L, Urbanc B, Borreguero JM, Lazo ND, Teplow DB, Stanley HE. Proc Natl Acad Sci USA. 2005;102:18258–18263. doi: 10.1073/pnas.0509276102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baumketner A, Bernstein SL, Wyttenbach T, Lazo ND, Teplow DB, Bowers MT, Shea JE. Protein Sci. 2006;15:1239–1247. doi: 10.1110/ps.062076806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen W, Mousseau N, Derreumaux P. J Chem Phys. 2006;125 doi: 10.1063/1.2337628. 08491101–08491108. [DOI] [PubMed] [Google Scholar]
- 21.Selkoe DJ, Podlisny MB. Annu Rev Genomics Hum Genet. 2002;3:67–99. doi: 10.1146/annurev.genom.3.022502.103022. [DOI] [PubMed] [Google Scholar]
- 22.Hendriks L, van Duijn CM, Cras P, Cruts M, Van Hul W, van Harskamp F, Warren A, McInnis MG, Antonarakis SE, Martin JJ, et al. Nat Genet. 1992;1:218–221. doi: 10.1038/ng0692-218. [DOI] [PubMed] [Google Scholar]
- 23.Kamino K, Orr HT, Payami H, Wijsman EM, Alonso E, Pulst SM, Anderson L, O'dahl S, Nemens E, White JA, et al. Am J Hum Genet. 1992;51:998–1014. [PMC free article] [PubMed] [Google Scholar]
- 24.Nilsberth C, Westlind-Danielsson A, Eckman CB, Condron MM, Axelman K, Forsell C, Stenh C, Luthman J, Teplow DB, Younkin SG, et al. Nat Neurosci. 2001;4:887–893. doi: 10.1038/nn0901-887. [DOI] [PubMed] [Google Scholar]
- 25.Levy E, Carman MD, Fernandez-Madrid IJ, Power MD, Lieberburg I, van Duinen SG, Bots GTAM, Luyendijk W, Frangione B. Science. 1990;248:1124–1126. doi: 10.1126/science.2111584. [DOI] [PubMed] [Google Scholar]
- 26.van Broeckhoven C, Haan J, Bakker E, Hardy JA, Hul WV, Vegter-Van Der Vlis M, Roos RAC. Science. 1990;248:1120–1122. doi: 10.1126/science.1971458. [DOI] [PubMed] [Google Scholar]
- 27.Tagliavini F, Rossi G, Padovani A, Magoni M, Andora G, Sgarzi M, Bizzi A, Savioardo M, Carella F, Morbin M, et al. Alzheimer's Rep. 1999;2(Suppl):S28. [Google Scholar]
- 28.Grabowski TJ, Cho HS, Vonsattel JPG, Rebeck GW, Greenberg SM. Ann Neurol. 2001;49:697–705. doi: 10.1002/ana.1009. [DOI] [PubMed] [Google Scholar]
- 29.Yun SJ, Urbane B, Cruz L, Bitan G, Teplow D, Stanley HE. Biophys J. 2007;92:1–14. doi: 10.1529/biophysj.106.097766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang S, Iwata K, Lachenmann MJ, Peng JW, Li S, Stimson ER, Lu Y, Felix AM, Maggio JE, Lee JP. J Struct Biol. 2000;130:130–141. doi: 10.1006/jsbi.2000.4288. [DOI] [PubMed] [Google Scholar]
- 31.Massi F, Straub JE. Biophys J. 2001;81:697–709. doi: 10.1016/S0006-3495(01)75734-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serrano L, Neira JL, Sancho J, Fersht AR. Nature. 1992;356:453–455. doi: 10.1038/356453a0. [DOI] [PubMed] [Google Scholar]
- 33.Anson ML, Mirsky AE. J Gen Physiol. 1934;17:393–398. doi: 10.1085/jgp.17.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fraser D, Powell RE. J Biol Chem. 1950;187:803–820. [PubMed] [Google Scholar]
- 35.Bitan G, Teplow DB. Acc Chem Res. 2004;37:357–364. doi: 10.1021/ar000214l. [DOI] [PubMed] [Google Scholar]
- 36.Bitan G, Vollers SS, Teplow DB. J Biol Chem. 2003;278:34882–34889. doi: 10.1074/jbc.M300825200. [DOI] [PubMed] [Google Scholar]
- 37.Bitan G, Kirkitadze MD, Lomakin A, Vollers SS, Benedek GB, Teplow DB. Proc Natl Acad Sci USA. 2003;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Päiviö A, Jarvet J, Gräslund A, Lannfelt L, Westlind-Danielsson A. J Mol Biol. 2004;339:145–159. doi: 10.1016/j.jmb.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 39.Urbanc B, Cruz L, Yun S, Buldyrev SV, Bitan G, Teplow DB, Stanley HE. Proc Natl Acad Sci USA. 2004;101:17345–17350. doi: 10.1073/pnas.0408153101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maji SK, Amsden JJ, Rothschild KJ, Condron MM, Teplow DB. Biochemistry. 2005;44:13365–13376. doi: 10.1021/bi0508284. [DOI] [PubMed] [Google Scholar]
- 41.Petkova AT, Yau WM, Tycko R. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luhrs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Doeli H, Schubert D, Riek R. Proc Natl Acad Sci USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Straub JE, Guevara J, Huo SH, Lee JP. Acc Chem Res. 2002;35:473–481. doi: 10.1021/ar010031e. [DOI] [PubMed] [Google Scholar]
- 44.Huet A, Derreumaux P. Biophys J. 2006;91:3829–3840. doi: 10.1529/biophysj.106.090993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolfe MS. Biochemistry. 2006;45:7931–7939. doi: 10.1021/bi060799c. [DOI] [PubMed] [Google Scholar]
- 46.Zhang JM, Ye WJ, Wang R, Wolfe MS, Greenberg BD, Selkoe DJ. J Biol Chem. 2002;277:15069–15075. doi: 10.1074/jbc.M105375200. [DOI] [PubMed] [Google Scholar]
- 47.Ren Z, Schenk D, Basi GS, Shapiro IP. J Biol Chem. 2007 doi: 10.1074/jbc.M702739200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.