Abstract
Lipodystrophies are syndromes of adipose tissue degeneration associated with severe defects in lipid and glucose homeostasis. We report here the generation and analysis of Ppargldi, a targeted allele that confers conditional dominant lipodystrophy in mice. The Ppargldi allele was generated by insertion of the Tet activator (tTA) and a tTA-regulated Flag-Pparg1 transgene into the Pparg gene. Unexpectedly, tTA elicits mild lipodystrophy, insulin resistance, and dyslipidemia, and the Flag-PPARγ1 transgene surprisingly exacerbates these traits. Doxycycline can both completely prevent and reverse these phenotypes, providing a mouse model of inducible lipodystrophy. Embryonic fibroblasts from either Ppargldi/+ or the phenotypically similar aP2-nSrebp1c (Sr) transgenic mice undergo robust adipogenesis, suggesting that neither strain develops lipodystrophy because of defective adipocyte differentiation. In addition, Ppargldi/+ adipose tissue shares extensive gene expression aberrations with that of Sr mice, authenticating the phenotype at the molecular level and revealing a common expression signature of lipodystrophic fat. Thus, the Ppargldi/+ mouse provides a conditional animal model for studying lipodystrophy and its associated physiology and gene expression.
Keywords: doxycycline, adipocyte, insulin resistance
Generalized and partial lipodystrophies are rare syndromes of heritable or acquired adipose tissue degeneration and misdistribution, often accompanied by severe defects in lipid and glucose homeostasis (reviewed in refs. 1 and 2). Genetic causes have been identified for several familial forms of the disease in humans and include mutations in the AGPAT2, BSCL2 (Seipin), LMNA (Lamin A), ZMPSTE24 (a lamin-processing protease), PPARG, or AKT2 genes. Acquired lipodystrophies are either idiopathic or arise because of immunological disorders, such as autoimmunity or complement activation. An increasingly prevalent type of acquired lipodystrophy occurs in HIV-infected patients subject to prolonged protease inhibitor therapy (3). This lipodystrophic syndrome, whose molecular etiology is unclear, is typified by loss of s.c., peripheral, facial, and gluteal fat and accumulation of excess fat around the neck and trunk, along with insulin resistance, type II diabetes, and dyslipidemia.
Several mouse models have been developed and characterized to date for the study of lipodystrophy and its consequences. Mice with deliberate subversion of adipocyte differentiation or viability include: aP2-DTA mice, in which lipoatrophy was engineered by adipocyte-specific expression of diphtheria toxin (4); A-ZIP/F mice, which were rendered lipoatrophic by artificial interference with the adipogenic C/EBP transcription factors (5); and FAT-ATTAC mice, which undergo transient lipoatrophy after temporal induction of adipocyte apoptosis (6). These models are useful for exploring the outcomes of lipodystrophy and general treatment modalities. Models in which lipodystrophy arose expectedly or serendipitously because of mutation or misexpression of endogenous genes include the following: aP2-nSrebp1c mice (Sr), which express a truncated, constitutively active Srebp-1c transgene in adipocytes, and whose lipodystrophic phenotype is neither expected nor mechanistically understood (7); fld (“fatty liver dystrophy”) mice, which suffer from severe lipoatrophy because of spontaneous mutations in the Lpin1 gene (8); and several knockout mouse strains with varying degrees of lipodystrophy, including mice deficient for Lmna (9), Zmpste24 (10), ribosomal S6 kinase (Rsk2) (11), Irs1/Irs3 double-knockout mice (12), and adipocyte-specific Pparg-null mice (13–15). These mouse models are useful for studying both the etiology and consequences of lipodystrophy.
Here, we report a mouse model of lipodystrophy, which is the serendipitous outcome of a genetically engineered allele of peroxisome proliferator-activated receptor γ (PPARγ), termed Ppargldi. The Ppargldi allele confers dominantly heritable lipodystrophy, which shares extensive anatomic, metabolic, and gene expression properties with Sr transgenic mice. The pathology is caused by unexpected combined effects of a knocked-in Tet-activator protein, tTA (16), and a Tet-regulated Flag-Pparg1 transgene and is completely inhibited by doxycycline, making the Ppargldi/+ mouse a unique model of conditional lipodystrophy. We further show that neither Ppargldi/+ nor Sr fibroblasts exhibit major adipogenic defects, suggesting that both arise subsequent to normal adipocyte differentiation. The Ppargldi/+ mouse offers an ideal inducible platform for studying both systemic and adipose tissue-specific attributes of lipodystrophy.
Results
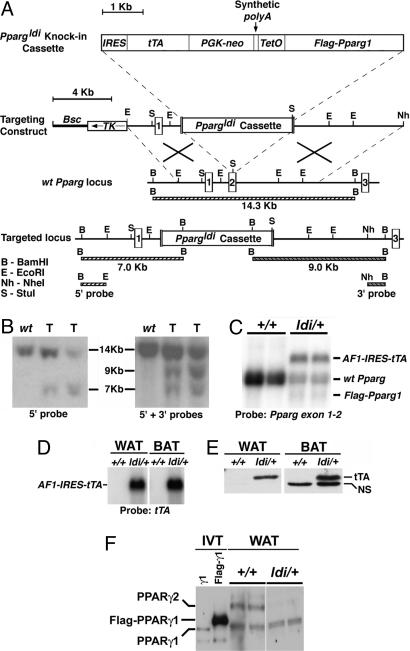
The Ppargldi Allele.
The Ppargldi allele was originally designed for substituting the endogenous Pparg gene with recombinant, inducible PPARγ. It was generated by homologous disruption of the second coding exon of Pparg with a cassette encoding the tetracycline-regulated transactivator (tTA; ref. 16), Pgk-Neo, and Tet operator-controlled Flag-Pparg1 cDNA (Fig. 1 A and B). The predicted fusion transcript between the 5′ portion of the endogenous Pparg gene and the knocked-in tTA (AF1-IRES-tTA) was abundantly expressed in adipose tissue of Ppargldi/+ mice, as was the tTA protein (Fig. 1 C–E). An ≈1.5-kb transcript, corresponding to Tet operator-driven Flag-Pparg1 cDNA, was modestly expressed in Ppargldi/+ adipose tissue (Fig. 1C). However, expression of the Flag-PPARγ1 protein in adipose tissue was below the detection threshold (Fig. 1F). Both tTA and Flag-Pparg1 were poorly expressed in other Pparg-expressing tissues of Ppargldi/+ mice, such as placenta and liver (data not shown). Interestingly, endogenous PPARγ2, but not the PPARγ1 isoform, was drastically reduced in the heterozygous fat pads, despite integrity of the companion WT Pparg allele (Fig. 1F).
Fig. 1.
The Ppargldi allele. (A) The Ppargldi targeting cassette contains, from 5′ to 3′, the following: an internal ribosome entry site (IRES), the Tet activator gene, tTA (15), Pgk-neo, a synthetic polyA module, and a Tet operator (TetO)-driven Flag-Pparg1 cDNA. The cassette was targeted into the second coding exon of the mouse Pparg gene via homologous recombination, as shown. (B) Southern blots of BamHI-digested genomic DNA were hybridized to 5′ and 3′ probes external to the genomic arms of the targeting construct (see A). Correctly targeted Ppargldi/+ ES cell clones (T) exhibit the predicted restriction fragment lengths. (C) A Northern blot of two WT (+/+) and two Ppargldi/+ epididymal WAT pads was hybridized to the 5′ portion of Pparg cDNA. Ppargldi/+ WAT expresses both WT Pparg and an abundant fusion transcript of the 5′ part of Pparg, IRES, and tTA (AF1-IRES-tTA), as well as modest levels of an ≈1.5-kb transcript, corresponding to the transgenic Flag-Pparg1; expression and identity of the latter transcript were confirmed by RNase protection analysis (data not shown). (D) A tTA probe corroborates the identity of the AF1-IRES-tTA transcript in Ppargldi/+ WAT and BAT. (E) Anti-VP16 antibodies detect ample tTA in Ppargldi/+ WAT and BAT protein extracts (tTA is a Tet repressor–VP16 fusion protein). BAT exhibits an additional, nonspecific reactive band (NS). (F) PPARγ isoforms in WT and Ppargldi/+ WAT. A SDS/7.5% PAGE Western blot was reacted with a custom-made anti-PPARγ antibody. Identities of PPARγ1, Flag-PPARγ1, and PPARγ2 proteins were determined based on migration patterns of in vitro-translated counterparts (IVT) (PPARγ2 data not shown). Flag-PPARγ1 protein could not be detected in Ppargldi/+ WAT extracts, whereas PPARγ2 was suppressed.
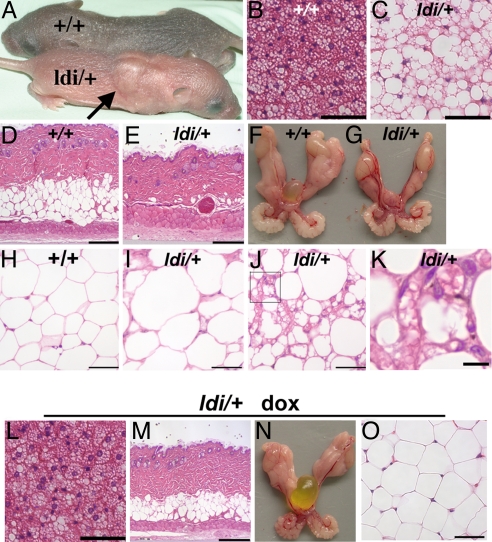
Ppargldi/+ Mice Exhibit Severe, Doxycycline-Treatable Lipodystrophy.
Unexpectedly, heterozygous Ppargldi/+ mice derived from three independent ES cell clones exhibited overt, dominantly inherited lipodystrophy. Anatomical and histological aspects of this phenotype included: (i) “buffalo humps” (Fig. 2A), comprising swollen interscapular fat pads due to hypertrophy and unilocular lipid deposition in mutant brown adipocytes (compare Fig. 2C with Fig. 2B); (ii) complete depletion of s.c. adipocytes (compare Fig. 2E with Fig. 2D); (iii) severely reduced size of gonadal white adipose tissue (WAT) (Fig. 2G vs. Fig. 2F); (iv) irregular shapes and sizes of residual adipocytes, ranging from overtly hypertrophic (compare Fig. 2I with Fig. 2H) to minuscule cells (Fig. 2J); and (v) fibrotic WAT stroma filled with fragmented lipid droplets and leukocytes (Fig. 2K). Importantly, all of these defects were completely prevented by continuous administration of doxycycline (dox) in the drinking water from midgestation onward (Fig. 2 L–O). Likewise, a 6-wk dox treatment of pubertal Ppargldi/+ mice fully reversed all of their defects, with the exception of WAT, whose histology was entirely corrected without regaining normal size (data not shown).
Fig. 2.
The histological Ppargldi/+ phenotype. (A) A characteristic buffalo hump in a 5 day-old Ppargldi/+ pup (arrow). (B and C) BAT of WT (+/+) and Ppargldi/+ mice. Ppargldi/+ BAT exhibits markedly enlarged lipid droplets with unilocular vs. multilocular deposition (C vs. B). (D and E) WT and Ppargldi/+ skin. Subdermal fat is eliminated in Ppargldi/+ mice. (F and G) Epididymal fat pads (EFP) of WT and Ppargldi/+ mice. White adipose mass is substantially decimated in Ppargldi/+ mice. (H–K) Histology of WT and Ppargldi/+ WAT. (I) A domain of marked adipocyte hypertrophy in the Ppargldi/+ tissue. (J) Another domain in the same EFP with minuscule cells, fragmented lipid droplets, and marked leukocyte infiltration. (K) A higher power micrograph of J Inset, with clear leukocytes and fragmented lipid droplets. (L–O) Complete phenotypic reversal in Ppargldi/+ mice treated with dox from gestation onwards. Shown are BAT (L), skin (M), EFP (N), and WAT (O); compare with untreated mutants in C, E, G, I–J, respectively. All tissue specimens, except in A, are from 10-wk-old male cohorts. (Scale bars: B, C, H–J, L, and, O, 50 μm; D, E, and M, 250 μm; K, 10 μm.)
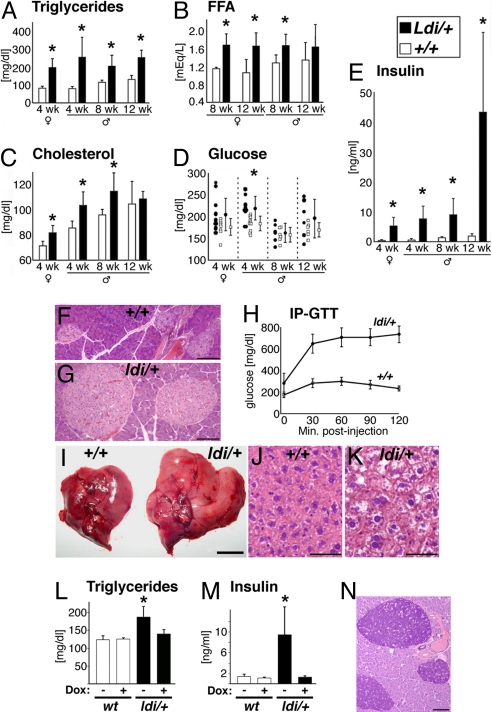
Ppargldi/+ mice displayed typical metabolic sequelae of lipodystrophy. These included the following: (i) early onset increases in the levels of circulating triglycerides, cholesterol, and free fatty acids (Fig. 3 A–C); (ii) hyperglycemia in prepubertal Ppargldi/+ males (P < 0.05), which normalized later in life, with sporadic, transient hyperglycemia in individual mutant mice (Fig. 3D); (iii) hyperinsulinemia (Fig. 3E), pancreatic islet hyperplasia (compare Fig. 3G with Fig. 3F), and severe glucose intolerance (Fig. 3H); and (iv) a 60–150% increase in liver size (Fig. 3I) accompanied by hepatocyte hypertrophy and vacuolization, but not the prototypic lipid droplet accumulation observed in classical steatosis (Fig. 3 J and K). Like the histological defects, all metabolic anomalies of Ppargldi/+ mice were completely reversed by dox treatment (Fig. 3 L and M and data not shown). Thus, the Tet-regulated components of the Ppargldi allele, and not structural interference with endogenous Pparg, are responsible for the unexpected lipodystrophic phenotype in its entirety.
Fig. 3.
The metabolic Ppargldi/+ phenotype. (A–E) Levels of plasma triglycerides (A), free fatty acids (B), cholesterol (C), glucose (D), and insulin (E) in matched, 6-h-fasted WT (white bars; n = 6–10) and Ppargldi/+ cohorts (black bars; n = 6–10). Values are presented for 4-wk-old females and a longitudinal analysis of males aged 4–12 wk. Asterisks designate statistically significant differences (P < 0.05) between Ppargldi/+ and WT mice. (F and G) Pancreatic islet hyperplasia in Ppargldi/+ mice. (H) Severe glucose intolerance in Ppargldi/+ (n = 5) as compared with WT mice (n = 5). (I) Hepatomegaly in Ppargldi/+ mice. (J and K) Hepatic histology of WT and Ppargldi/+ mice. Ppargldi/+ hepatocytes are hypertrophic, and contain large unstained vacuoles, but do not display the prototypic lipid droplet accumulation of steatotic livers. (L and M) Dox treatment circumvents hypertriglyceridemia and hyperinsulinemia in Ppargldi/+ mice. Values were measured in untreated 5.5-wk-old WT and Ppargldi/+ cohorts (−) compared with matched cohorts exposed to dox from midgestation onwards (+). (N) Aldehyde Fuchsin-stained pancreas from a 16-month-old Ppargldi/+ male demonstrates rich insulin content in β-cells of aged mutants. Tissue specimens were collected from 10-wk-old male cohorts, except in N. (Scale bars: F, G, and N, 100 μm; I, 1 cm; J and K, 50 μm.)
Despite dyslipidemia and severe insulin resistance throughout life, Ppargldi/+ mice developed neither bona fide steatosis nor type II diabetes. Mice as old as 12–18 months exhibited hyperplastic yet intact and insulin-laden pancreatic islets (Fig. 3N), with hyperinsulinemia and normoglycemia (data not shown). These observations suggest that steatosis and diabetes are not automatic complications of chronic hyperlipidemic or hyperinsulinemic burdens, respectively, and the development of each requires additional physiological factors.
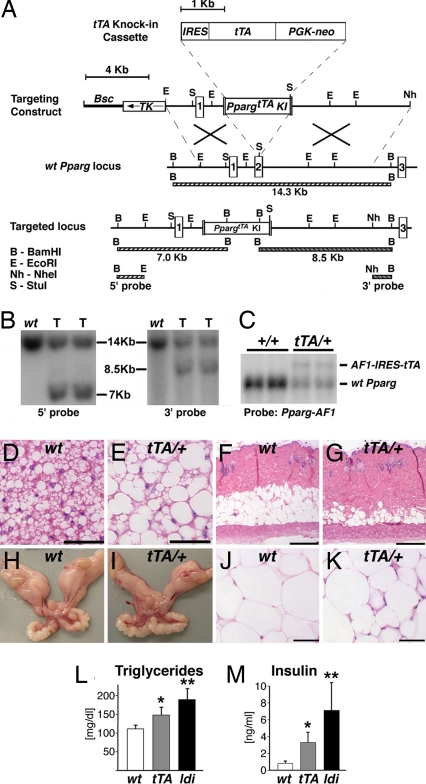
Pathogenic Determinants of the Ppargldi Allele.
To dissect the causes of the Ppargldi phenotype, we generated and analyzed a targeted allelic variant, PpargtTA, which expressed the exact IRES-tTA knockin configuration as in Ppargldi but lacked the TetO-driven FlagPparg1 module (Fig. 4 A–C). Surprisingly, mice derived from two independent PpargtTA/+ ES clones manifested histological and metabolic characteristics of lipodystrophy, albeit substantially more modest than Ppargldi/+ mice. Specifically, (i) brown adipocytes were pale, unilocular, and hypertrophic (Fig. 4 D and E), yet not to the overt extent of buffalo humps; (ii) s.c. fat was reduced, but not eliminated (Fig. 4 F and G); (iii) the gonadal fat pads were smaller than those of WT mice (Fig. 4 H and I), although not as severely as in Ppargldi/+ mice, and exhibited neither adipocyte hypertrophy nor fibrosis and almost no leukocyte infiltration (Fig. 4 J and K); (iv) triglyceride and insulin levels were elevated, albeit to a significantly lesser extent than in Ppargldi/+ mice (Fig. 4 L and M); and (v) hepatomegaly was modest (data not shown). Dox treatment completely suppressed these pathologies (data not shown), suggesting that basal lipodystrophy results from promiscuous transcriptional activity of tTA, rather than generalized transcription cofactor titration by its VP16 moiety (17). Most critically, the markedly attenuated PpargtTA/+ phenotype indicated that the Flag-Pparg1 transgene is responsible for the dramatically exacerbated pathology of Ppargldi/+ mice. Resequencing of the Ppargldi targeting construct confirmed that apart from the engineered Flag epitope in its N terminus, this cDNA was 100% identical to WT Pparg1, and, thus, its pathogenic activity is not due to accidental mutation.
Fig. 4.
tTA causes mild lipodystrophy with modest metabolic anomalies. (A) The PpargtTA allele contains an exact replica of the IRES, tTA, Pgk-neo, and synthetic polyA modules of the Ppargldi allele, targeted to the same location within the second coding exon of the Pparg gene. (B) Southern blots of BamHI-digested genomic DNA hybridized to 5′ and 3′ external probes. Correctly targeted PpargtTA/+ ES cell clones (T) exhibit the digestion patterns predicted in A. (C) A Northern blot of two WT (+/+) and two PpargtTA/+ EFPs was hybridized the 5′ Pparg cDNA probe, as described in Fig. 1C. PpargtTA/+ WAT expresses the same AF1-IRES-tTA fusion transcript seen in Ppargldi/+ fat. (D–K) BAT (D and E), skin (F and G), EFPs (H and I), and WAT (J and K) from 10-wk-old WT (D, F, H, and J) and PpargtTA/+ (E, G, I, and K) males. BAT exhibits approximately the same phenotype of Ppargldi/+ mice (see Fig. 2C), whereas subdermal fat and WAT exhibit a markedly attenuated phenotype (compare with Fig. 2 D, I, and J. (Scale bars: D, E, J, and K, 50 μm; F and G, 250 μm.) (L and M) Fasting triglyceride (L) and insulin (M) levels in 5.5-wk-old WT, PpargtTA/+, and Ppargldi/+ males (n = 5–6 mice). All cohorts were born within the same week and assayed simultaneously. Notice the intermediate hypertriglyceridemic and hyperinsulinemic phenotypes of PpargtTA/+ compared with Ppargldi/+ mice. * and **, levels of both analytes are significantly different in each strain compared with the other two (P < 0.05).
Ppargldi/+ and Sr Fibroblasts Undergo Normal Adipogenesis.
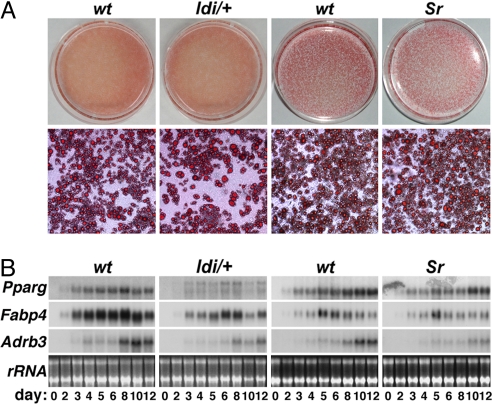
Congenital lipodystrophy is commonly ascribed to defective adipocyte differentiation (1, 2, 5, 7, 8). Adipogenic disorders clearly underlie lipodystrophy in both A-ZIP/F-1 transgenic mice (5) and Lpin1-deficient Fld mouse (8). However, such an etiology is harder to reconcile with the normal elaboration of Ppargldi/+ brown adipose tissue (BAT), as hypertrophic and dysfunctional as it eventually becomes, or with the presence of WAT in these mice, as residual as it may be. Similar considerations apply to the etiologically distinct yet phenotypically similar Sr mice (7). Indeed, in vitro adipogenesis of both Ppargldi/+ and Sr embryonic fibroblasts was consistently normal, as judged by both lipid accumulation and expression of adipogenic markers in at least five independent preparations of each (Fig. 5). These observations suggest that lipodystrophy in these two mouse models may not be the result of perturbed adipocyte differentiation per se.
Fig. 5.
Unperturbed adipogenesis of Ppargldi/+ and Sr embryonic fibroblasts. Primary fibroblasts from embryonic day 13.5 WT, Ppargldi/+, and Sr embryos were subject to adipogenic differentiation. (A) On the 12th day of differentiation, cultures were stained with Oil- red-O and were photographed at low (Upper) and high magnification (Lower), revealing comparable lipid accumulation between both Ppargldi/+ and Sr cells and the corresponding WT cultures. (B) RNA was extracted from cultures throughout differentiation and was analyzed by Northern blot hybridization to Pparg, Fabp4 (aP2), and Adrb3 (β3-adrenergic receptor). Pparg levels are more substantially decreased in Ppargldi/+ fibroblasts, reflecting gene dosage reduction to a single WT copy. Although nominal marker expression levels are slightly reduced in the mutant fibroblasts, induction kinetics is similar between WT and mutant cells, demonstrating normal progress of adipogenesis. Ribosomal RNA (rRNA) levels are shown for normalization.
Overlapping Gene Expression Aberrations in Ppargldi/+ and Sr Adipose Tissue.
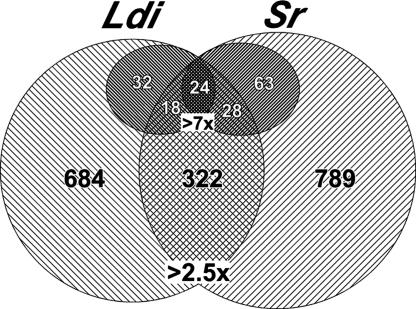
To better define the Ppargldi/+ mouse in relation to other models of lipodystrophy, we compared the gene expression profiles of Sr and Ppargldi/+ gonadal fat pads. These profiles combine genuine changes in gene regulation with changes that arise from the skewed cellular demographics of lipodystrophic fat pads, i.e., richer stromal cell populations and a decrease in the relative adipocyte fraction. By averaging the reduction in multiple adipocyte-specific genes and the increase in prototypic markers of endothelial cells and leukocytes we estimated that adipocyte abundance is reduced by ≈2.5-fold in the mutant tissues, whereas different stromal cell types are enriched 2.5- to 4-fold. Differentials that exceed these values are therefore more likely to represent regulatory changes than aberrant tissue composition. A staggering 1,006 and 1,111 transcripts changed by >2.5-fold in WAT of Ppargldi/+ and Sr mice, respectively, relative to their WT counterparts (Fig. 6). Of these, 322 differentials were shared by both mouse strains. Moreover, 74 and 115 genes changed by >7-fold in Ppargldi/+ and Sr fat, respectively, 24 of which were shared by both strains, and an additional 46 genes changed by >7-fold in one strain and between 2.5- and 7-fold in the other. This substantial overlap in robust gene expression aberrations points to widespread molecular similarities between the two strains and reveals a gene expression signature of lipodystrophic fat. Most genes and pathways that comprise this shared signature are new in the context of adipose tissue biology. Recurrent functional themes include up-regulation of genes regulating immunity, inflammation, and chemotaxis, and down-regulation of imprinted genes and secreted and membranal components of diverse extracellular signaling pathways [supporting information (SI) Table 1].
Fig. 6.
Overlapping gene expression aberrations in Ppargldi/+ and Sr fat. Shown is a Venn diagram depicting the numbers of overlapping and strain-specific genes that are differentially expressed by >2.5-fold in Ppargldi/+ (Ldi) and/or Sr mice. (Inset) The number of genes that change by at least 7-fold in each strain partitioned according to their overlap with the reciprocal strain (7-fold, 2.5- to 7-fold, <2.5-fold). See SI Table 1 for the full list of genes in the 7-fold inset.
Discussion
We report here the generation and detailed analysis of a new mouse model of lipodystrophy, which confers the disease dominantly through cooperative unanticipated effects of the synthetic transcription factor tTA and a minutely expressed Flag-PPARγ1 transgene. Although most mouse models of lipodystrophy exhibit indiscriminate degeneration of all adipose tissue depots, the Ppargldi/+ phenotype is strikingly similar to that of aP2-nSREBP1c (Sr) mice in the unique combination of hypoplastic WAT and hypertrophic, isoplastic BAT. This trait and its metabolic comorbidities resemble the lipodystrophic syndrome of HIV patients, in which some fat depots degenerate whereas others expand (3). This analogy raises the hypothesis that all three cases of lipodystrophy arise from insults that converge on similar cellular processes and renders the Ppargldi/+ mouse and its milder PpargtTA/+ counterpart attractive animal models for severe and mild grades of this serious complication. The ability to fully suppress lipodystrophy by doxycycline in both strains makes them unique models of conditional lipodystrophy, further expanding their utility.
Ppargldi/+ mice teach us several valuable lessons, both in general and with regard to PPARγ and lipodystrophy. The cautionary tale of tTA-induced lipodystrophy is the first reported deleterious activity of the system in vitro or in vivo throughout its rather extensive history (18). The complete suppression of the PpargtTA/+ phenotype by doxycycline suggests that its molecular mechanism of action likely entails promiscuous transcriptional induction of endogenous genes rather than generalized transcription factor titration by the VP16 moiety (17). This finding is surprising in light of the bacterial origin and complex response element of the Tet repressor but is strongly supported by follow-up studies that identify at least one cellular gene, Gpr56, as a robust, dox-repressible cellular target of tTA (S.K., L.-W.H., and Y.B., unpublished work).
The strong pathogenic impact of Flag-PPARγ1 provides the second surprise. The flag-tagged receptor recapitulates the DNA-binding, transactivation potential and adipogenic activity of native PPARγ1 (ref. 19 and data not shown) and was expected to benefit adipocyte functions and systemic metabolism. However, not only does it markedly exacerbate lipodystrophy in Ppargldi/+ over PpargtTA/+ mice, it does so with minute expression levels relative to the companion WT PPARγ1 allele. Possible explanations to this confounding effect include the following: (i) The N-terminal blockade might generate a defective PPARγ variant with strong dominant-negative effect on the product of the companion WT PPARγ allele. This idea is based on the lipodystrophy of patients with dominant negative PPARG mutations but is harder to reconcile with the difference between N-terminal blockade here and mutations in the ligand-binding domain of PPARγ in patients (20–22). (ii) The N-terminal modification may underlie a gain-of-function, which either exaggerates normal deleterious functions of PPARγ or elicits altogether new off-target pathogenic interactions. (iii) The transgene may have non-protein-mediated functions at the level of either its RNA product or allele structure. This possibility gains some support from the dramatic decline in PPARγ2 protein levels in Ppargldi/+ adipose tissue (Fig. 1F). However, endogenous PPARγ1 levels are intact and PPARγ2 is not repressed in fibroblast-derived Ppargldi/+ adipocytes in vitro (data not shown), suggesting that its down-regulation is likely not a direct effect of Flag-PPARγ1.
The common contention that heritable lipodystrophy results from defective adipocyte differentiation has been proven in several instances (5, 8). However, the adipogenic potential of both Ppargldi/+ and Sr fibroblasts is unperturbed. Although we cannot rule out an adipogenic defect that is manifested only in vivo, the most plausible interpretation is that lipodystrophy in these two mouse models is independent of and subsequent to adipogenesis. This conclusion should not be surprising in the broader sense, as congenital and acquired degenerative diseases of other tissues, such as muscle, heart, brain, and kidney, frequently arise in properly differentiated tissues. Moreover, the phenotypically analogous HIV-associated lipodystrophy clearly evolves in mature adipose tissue.
Last, the gene expression profiles of Ppargldi/+ and Sr fat pads recapitulate their high phenotypic similarity and imply a putative molecular signature of lipodystrophic fat. The vast majority of genes involved in the regulation of adipogenesis and classical lipid metabolism are glaringly unchanged in both strains, with the obvious exception of up-regulated cholesterol-repressed genes in Sr mice. This observation is consistent with the conclusion that neither phenotype arises from defective adipogenesis and suggests that aberrant lipid metabolism is also not at fault. Instead, several genes associated with immunity and inflammation, most conspicuously Mhc2q10 and Ccl8 (see SI Table 1), are up-regulated in the two strains, hinting at potential involvement of immune and inflammatory processes in both phenotypes. A broader array of inflammation-related genes is up-regulated in Ppargldi/+ mice, suggesting that these processes are not general attributes of lipodystrophy, but rather strain-specific events that may contribute to discrete aspects of the phenotype. Most of these genes are not changed in PpargtTA/+ mice (data not shown), suggesting that inflammation and immunity are involved primarily in severe, but not basal lipodystrophy. The substantial leukocytic infiltrate in adipose tissue of Ppargldi/+ and Sr, but not PpargtTA/+ mice is consistent with this interpretation. Multiple imprinted genes and genes encoding cell surface receptors and secreted factors from diverse signaling pathways are highly down-regulated in adipose tissue of both strains, two phenomena whose biological significance remains to be determined. On the whole, most of the acutely affected genes are previously unrecognized in the context of adipocyte biology, hindering current understanding of these phenotypes, but offering fresh research directions. Importantly, a preliminary screen revealed significant similarities to gene expression anomalies of adipose tissue from HIV patients (F. Villarroya, personal communication). This observation buttresses the functional analogy between HIV-associated lipodystrophy and the two mouse strains and validates their utility as models for studying this poorly understood syndrome.
Experimental Procedures
Ppargldi/+ and PpargtTA/+ Mice.
Targeting vectors were constructed by standard recombinant DNA technology and introduced into the Pparg locus by homologous recombination as described in ref. 23. Correct integration was validated by Southern blots by using probes external to the homology arms of the vectors, as shown. Germ-line chimeras were established from three distinct Ppargldi/+ and two distinct PpargtTA/+ ES clones. Mice carrying either allele were subsequently propagated on the closely related 129S1/SvImJ (129S) background. All histological, metabolic, and gene expression analyses were performed on F1 hybrid progeny of 129S-Ppargldi/+ or -PpargtTA/+ sires and WT C57BL/6J (B6) dams. This configuration ensured genetic homogeneity of the cohorts and was required because of poor fecundity of Ppargldi/+ mice after introgression onto B6. Where indicated, doxycycline was administered continuously in drinking water at 0.5 mg/ml. All mouse studies were approved by The Jackson Laboratory's Animal Care and Use Committee.
Nucleic Acid and Protein Detection.
Northern and Western blot analyses were performed using standard methods (24). tTA was visualized with an anti-VP16 antibody (Clontech, Mountain View, CA). Custom PPARγ antiserum was raised in rabbits (QCB, Hopkinton, MA) against a bacterially expressed, 6X-His-tagged polypeptide comprising the 98 N-terminal amino acids and was affinity-purified using the same antigen.
Histology and Metabolic Phenotyping.
All tissues were fixed in Bouin's fixative, except liver, which was fixed in formalin. Fixed tissues were embedded in paraffin, sectioned, and stained with hematoxylin and eosin. Pancreas sections were also stained with aldehyde fuchsin to detect insulin granules. Plasma analytes were determined in retroorbital blood collected from animals subject to 5–6 h of fasting. Glucose, triglycerides, free fatty acids, and cholesterol were measured in a Beckman (Fullerton, CA) Synchron CX5 Delta Clinical Chemistry Analyzer. Insulin was measured using a rat insulin RIA kit (LINCO, St. Charles, MO). For glucose tolerance tests, retroorbital blood was collected from 6 h-fasted animals, followed by i.p. injection of 2 g glucose per kilogram of body weight, collection of retroorbital blood samples 30, 60, 90, and 120 min thereafter, and determination of glucose concentrations in the Synchron Analyzer.
Adipogenesis of Embryonic Fibroblasts.
To prepare fibroblasts carcasses of embryonic day 13.5 embryos were individually minced by 4–5 passages through an 18-guage syringe needle. Cells were plated on 6-cm dishes, brought to complete confluence, and 2 days later were treated for 72 h with insulin (10 μg/ml), 5 μM dexamethazone, 0.2 mM isobutyl-methyl-xanthine, and 1 μM rosiglitazone. Cultures were fed with fresh insulin-containing medium at 72-h intervals thereafter. Lipid content was assessed by Oil-red-O staining at the end of differentiation. In other experiments, RNA was extracted from cultures at different stages of differentiation and analyzed.
Affymetrix Microarrays and Statistical Analysis.
RNA from epididymal fat pads of three 10-wk-old mutant males (Ppargldi/+ or Sr) and three litter-matched WT controls for each strain was treated with DNase I and probes were generated according to manufacturer's instructions (Affymetrix, Santa Clara, CA). Each sample hybridized to one MOE430 version 2.0 GeneChip array in a Fluidics Station 450 instrument (Affymetrix). Arrays were imaged with a GeneChip 3000 Laser Confocal Slide Scanner, were quantified using GeneChip Operating Software version 1.2 (Affymetrix), and expression values were summarized using the robust multichip average (RMA) function. P values were assigned using ANOVA methodology (25). False discovery rates (FDR) were assigned by computing q values from test statistics (26).
Supplementary Material
Acknowledgments
We thank Dr. Ed Leiter for discussions and encouragement; Dr. Francesc Villarroya for sharing unpublished microarray data; Drs. Sue Ackerman, Jürgen Naggert, and Ron Evans for helpful comments on the manuscript; The Jackson Laboratory's diagnostic laboratory, microinjection, histology, and gene expression services for expert technical assistance; and Pat Cherry for administrative assistance. Support for this work was received through pilot funding from National Institute of Diabetes and Digestive and Kidney Diseases Grant 5 P30 DK32520 (to Y.B.), Grant CA34196 (to The Jackson Laboratory), an American Heart Association Postdoctoral Fellowship (to S.K.), and private gifts from the Fraternal Order of Eagles.
Abbreviations
- BAT
brown adipose tissue
- dox
doxycycline
- EFP
epididymal fat pad
- PPARγ
peroxisome proliferator-activated receptor γ
- Sr
aP2-nSrebp1c
- tTA
tetracycline-regulated transactivator
- WAT
white adipose tissue.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Microarray data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE9132).
This article contains supporting information online at www.pnas.org/cgi/content/full/0707797104/DC1.
References
- 1.Garg A. N Engl J Med. 2004;350:1220–1234. doi: 10.1056/NEJMra025261. [DOI] [PubMed] [Google Scholar]
- 2.Capeau J, Magre J, Lascols O, Caron M, Bereziat V, Vigouroux C, Bastard JP. Biochem Soc Trans. 2005;33:1073–1077. doi: 10.1042/BST0331073. [DOI] [PubMed] [Google Scholar]
- 3.Carr A, Samaras K, Burton S, Law M, Freund J, Chisholm DJ, Cooper DA. AIDS. 1998;12:F51–F58. doi: 10.1097/00002030-199807000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Ross SR, Graves RA, Spiegelman BM. Genes Dev. 1993;7:1318–1324. doi: 10.1101/gad.7.7b.1318. [DOI] [PubMed] [Google Scholar]
- 5.Moitra J, Mason MM, Olive M, Krylov D, Gavrilova O, Marcus-Samuels B, Feigenbaum L, Lee E, Aoyama T, Eckhaus M, et al. Genes Dev. 1998;12:3168–3181. doi: 10.1101/gad.12.20.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pajvani UB, Trujillo ME, Combs TP, Iyengar P, Jelicks L, Roth KA, Kitsis RN, Scherer PE. Nat Med. 2005;11:797–803. doi: 10.1038/nm1262. [DOI] [PubMed] [Google Scholar]
- 7.Shimomura I, Hammer RE, Richardson JA, Ikemoto S, Bashmakov Y, Goldstein JL, Brown MS. Genes Dev. 1998;12:3182–3194. doi: 10.1101/gad.12.20.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peterfy M, Phan J, Xu P, Reue K. Nat Genet. 2001;27:121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- 9.Cutler DA, Sullivan T, Marcus-Samuels B, Stewart CL, Reitman ML. Biochem Biophys Res Commun. 2002;291:522–527. doi: 10.1006/bbrc.2002.6466. [DOI] [PubMed] [Google Scholar]
- 10.Pendas AM, Zhou Z, Cadinanos J, Freije JM, Wang J, Hultenby K, Astudillo A, Wernerson A, Rodriguez F, Tryggvason K, et al. Nat Genet. 2002;31:94–99. doi: 10.1038/ng871. [DOI] [PubMed] [Google Scholar]
- 11.El-Haschimi K, Dufresne SD, Hirshman MF, Flier JS, Goodyear LJ, Bjorbaek C. Diabetes. 2003;52:1340–1346. doi: 10.2337/diabetes.52.6.1340. [DOI] [PubMed] [Google Scholar]
- 12.Laustsen PG, Michael MD, Crute BE, Cohen SE, Ueki K, Kulkarni RN, Keller SR, Lienhard GE, Kahn CR. Genes Dev. 2002;16:3213–3222. doi: 10.1101/gad.1034802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He W, Barak Y, Hevener A, Olson P, Liao D, Le J, Nelson M, Ong E, Olefsky JM, Evans RM. Proc Natl Acad Sci USA. 2003;100:15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones JR, Barrick C, Kim KA, Lindner J, Blondeau B, Fujimoto Y, Shiota M, Kesterson RA, Kahn BB, Magnuson MA. Proc Natl Acad Sci USA. 2005;102:6207–6212. doi: 10.1073/pnas.0306743102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imai T, Takakuwa R, Marchand S, Dentz E, Bornert JM, Messaddeq N, Wendling O, Mark M, Desvergne B, Wahli W, et al. Proc Natl Acad Sci USA. 2004;101:4543–4547. doi: 10.1073/pnas.0400356101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tasset D, Tora L, Fromental C, Scheer E, Chambon P. Cell. 1990;62:1177–1187. doi: 10.1016/0092-8674(90)90394-t. [DOI] [PubMed] [Google Scholar]
- 18.Zhu Z, Zheng T, Lee CG, Homer RJ, Elias JA. Semin Cell Dev Biol. 2002;13:121–128. doi: 10.1016/s1084-9521(02)00018-6. [DOI] [PubMed] [Google Scholar]
- 19.Hummasti S, Tontonoz P. Mol Endocrinol. 2006;20:1261–1275. doi: 10.1210/me.2006-0025. [DOI] [PubMed] [Google Scholar]
- 20.Barroso I, Gurnell M, Crowley VE, Agostini M, Schwabe JW, Soos MA, Maslen GL, Williams TD, Lewis H, Schafer AJ, et al. Nature. 1999;402:880–883. doi: 10.1038/47254. [DOI] [PubMed] [Google Scholar]
- 21.Agarwal AK, Garg A. J Clin Endocrinol Metab. 2002;87:408–411. doi: 10.1210/jcem.87.1.8290. [DOI] [PubMed] [Google Scholar]
- 22.Hegele RA, Cao H, Frankowski C, Mathews ST, Leff T. Diabetes. 2002;51:3586–3590. doi: 10.2337/diabetes.51.12.3586. [DOI] [PubMed] [Google Scholar]
- 23.Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM. Mol Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 24.Shalom-Barak T, Nicholas JM, Wang Y, Zhang X, Ong ES, Young TH, Gendler SJ, Evans RM, Barak Y. Mol Cell Biol. 2004;24:10661–10669. doi: 10.1128/MCB.24.24.10661-10669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Churchill GA. BioTechniques. 2004;37:173–177. doi: 10.2144/04372TE01. [DOI] [PubMed] [Google Scholar]
- 26.Storey JD, Tibshirani R. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.