Abstract
Infectious diseases exert a constant evolutionary pressure on the genetic makeup of our innate immune system. Polymorphisms in Toll-like receptor 4 (TLR4) have been related to susceptibility to Gram-negative infections and septic shock. Here we show that two polymorphisms of TLR4, Asp299Gly and Thr399Ile, have unique distributions in populations from Africa, Asia, and Europe. Genetic and functional studies are compatible with a model in which the nonsynonymous polymorphism Asp299Gly has evolved as a protective allele against malaria, explaining its high prevalence in subSaharan Africa. However, the same allele could have been disadvantageous after migration of modern humans into Eurasia, putatively because of increased susceptibility to severe bacterial infections. In contrast, the Asp299Gly allele, when present in cosegregation with Thr399Ile to form the Asp299Gly/Thr399Ile haplotype, shows selective neutrality. Polymorphisms in TLR4 exemplify how the interaction between our innate immune system and the infectious pressures in particular environments may have shaped the genetic variations and function of our immune system during the out-of-Africa migration of modern humans.
Keywords: cytokines, human migration, innate immunity, Toll-like receptor 4, sepsis
During evolution, our genetic makeup has been profoundly influenced, with important consequences for the predisposition to health and disease. In the past decades, the interest in the genetic and physiological mechanisms of predisposition to disease has led to the identification of several genetic polymorphisms that have evolved in response to specific evolutionary pressures. Among these, some of the best known are related to the susceptibility to infectious diseases: the selection of hemoglobin S, hemoglobin C, and thalassaemia α and β as protective mechanisms against malaria (1–3), the polymorphisms in CCR5 providing protection against plague or HIV infection (4).
Toll-like receptors (TLR) have been discovered as the most important class of pattern recognition receptors, involved in the host defense against bacteria, viruses, fungi, and protozoa (5). TLRs are evolutionarily conserved receptors for pathogenic microorganisms, initially described in Drosophila (6) and with homologues in plants and lower vertebrates (5). One of the best-studied TLRs is TLR4, the key receptor for the LPS component of Gram-negative bacteria and for structures of mycobacteria, fungi, and malaria parasites (7–10).
Two nonsynonymous TLR4 polymorphisms have been described, an A/G transition at SNP rs4986790 that causes an Asp/Gly polymorphism at amino acid 299 and a C/T transition at SNP rs4986791 that causes a Thr/Ile polymorphism at amino acid 399. The derived states (G and T, or Gly and Ile, respectively) have been shown to change the ligand-binding site of the receptor (11). Both these two derived states reach substantial frequencies and are found in cosegregation in Caucasian populations (12–14). Some, but not all, studies have suggested a correlation between these polymorphisms and susceptibility to infectious diseases such as Gram-negative infections or disseminated candidiasis (15–17). Despite a 98% cosegregation level of these two derived alleles in Caucasian populations, this aspect was not taken into account in susceptibility analyses: each polymorphism was treated as an independent variable in these studies (see review in ref. 13). In contrast to the situation in Caucasians, one study in West Africa identified a different TLR4 haplotype composed only of the Asp299Gly-derived allele, with no individuals bearing the Thr399Ile-derived allele (18). These differences suggest that human populations around the world differ in the prevalence of TLR4 haplotypes, which in turn may reflect particular local infectious pressure and subsequent susceptibility to these infections.
In this study, we investigated whether the differences in the TLR4 polymorphism haplotypes in various populations of the three large continental masses, Africa, Eurasia, and America, could have been the result of local evolutionary pressures by infection during or after the out-of-Africa migration of modern humans. We analyzed the prevalence of the TLR4 haplotypes formed by these two SNPs in various populations from these continents and compared the phenotype of the two most prevalent TLR4 haplotypes with the wild-type (ancestral) TLR4. These results were used to get an insight into the evolutionary pressure of infections on the TLR4 polymorphisms.
Results and Discussion
Distribution and Origin of the TLR4 Haplotypes Among the Populations.
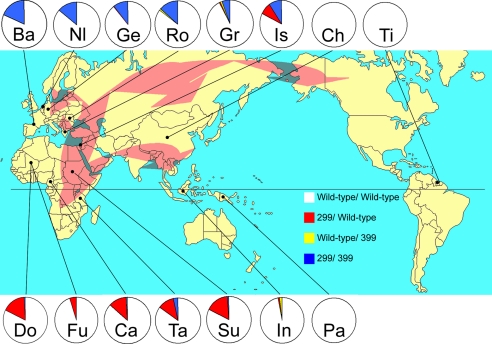
DNA from 2,491 individuals from 15 populations were included for TLR4 polymorphism analysis. The location and distribution of these populations were chosen to be representative of the original African populations before the migration and the different migration routes toward Europe, Asia, and the New World. As shown in Fig. 1, significant differences were found between the existing prevalence of the two TLR4 polymorphisms between the major continents. African populations show a high prevalence of the Asp299Gly allele (rs4986790 G). Between 10% and 18% of the individuals tested have one copy of this allele, of which only 2% are present with Thr399Ile (rs4986791 T). The Thr399Ile allele was always found in individuals with Asp299Gly. Populations from Asia (Han Chinese, Indonesian, and Papuan) and America (Trio Indians in Surinam) were practically missing TLR4 polymorphisms, with single cases of the Asp299Gly allele alone or the Thr399Ile allele alone in Indonesia. In contrast, 6–14% of Indo-European individuals were double heterozygotes for the Asp299Gly and Thr399Ile alleles (allele frequency 3–7%, respectively), a frequency that reaches 18% for Basques (allele frequency 9%) [allele frequencies, supporting information (SI) Table 3].
Fig. 1.
World distribution of the TLR4 haplotypes in human populations. Circles indicate allele frequency (red, Asp299Gly; yellow, Thr399Ile; and blue, Asp299Gly/Thr399Ile). The Asp299Gly haplotype is concentrated in Africa, and the Asp299Gly/Thr399Ile haplotype is concentrated in Europe. Arrows indicate the main out-of-Africa migration routes of modern Homo sapiens, based on refs. 19–21. Abbreviations used for the populations (and numbers): Ba, Basque, Spain (107); Nl, The Netherlands (209); Ge, Germany (632); Ro, Romania (102); Gr, Greece (162); Is, Israel (85); Ch, Han Chinese, China (100); Tr, Trio-Indians, Surinam (99); Do, Dogon tribe, Mali (241); Fu, Fulani tribe, Mali (243); Ca, Cameroon (142); Ta, Tanzania (121); Su, Sudan (101); In, Indonesia (98); and Pa, Papua New Guinea (49).
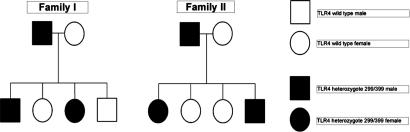
Because of the heterozygote state of most individuals bearing the Asp299Gly and Thr399Ile alleles in European populations, we analyzed whether these polymorphisms were in cosegregation on the same allele or whether the polymorphisms laid on different alleles. Several two-generation European families were investigated for the presence of these alleles to confirm that both polymorphisms were on the same haplotype and were therefore inherited in cosegregation. In all situations with a parent double heterozygote for Asp299Gly and Thr399Ile and a parent bearing wild-type TLR4 alleles, all heterozygous children had both Asp299Gly and Thr399Ile, confirming the cosegregated state of both alleles (Fig. 2). Therefore, double-heterozygous Asp299Gly/Thr399Ile individuals were assumed to contain one haplotype with the two ancestral alleles and one haplotype with the two variant alleles in further analysis. This haplotypic composition is supported by the inferred phased haplotypes for the reference HapMap populations (19).
Fig. 2.
Inheritance of the Asp299Gly/Thr399Ile heterozygote haplotype within two different families. Asp299Gly and Thr399Ile polymorphisms cosegregate in the European populations.
Statistical analysis showed that all populations were in Hardy–Weinberg equilibrium. In addition, haplotype similarity analysis between the populations confirmed that the African, West-Eurasian (European), and East-Eurasian (including the American) populations segregate into three major groups according to their TLR4 haplotypes (Table 1). Interestingly, an Israeli population was the only one showing a mixed pattern of TLR4 haplotypes. This finding could have two possible explanations: either this is a testimony of the current mosaic of the population of Israel, encompassing emigration from many parts of the world, or alternatively, this may represent a particular distribution of the TLR4 haplotypes in the Middle East. More Middle Eastern populations should be studied for clarification of these aspects.
Table 1.
TLR4 allele differentiations between populations
| Population (numbers) | Tr | Pa | Ch | In | NL | Ge | Ba | Gr | Ro | Is | Su | Ca | Ta | Do |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trio-Indians (99) | ||||||||||||||
| Papua (49) | * | |||||||||||||
| Han Chinese (100) | * | * | ||||||||||||
| Indonesia (98) | <0.120 | <0.704 | <0.123 | |||||||||||
| The Netherlands (209) | <0.001 | <0.003 | <0.001 | <0.001 | ||||||||||
| Germany (632) | <0.001 | <0.020 | <0.001 | <0.001 | <0.346 | |||||||||
| Basque (107) | <0.001 | <0.001 | <0.001 | <0.001 | <0.361 | <0.095 | ||||||||
| Greece (162) | <0.029 | <0.468 | <0.030 | <0.083 | <0.001 | <0.002 | <0.001 | |||||||
| Romania (102) | <0.001 | <0.002 | <0.001 | <0.001 | <0.403 | <0.111 | <0.547 | <0.012 | ||||||
| Israel (85) | <0.001 | <0.006 | <0.001 | <0.001 | <0.001 | <0.001 | <0.002 | <0.002 | <0.004 | |||||
| Sudan (101) | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.004 | ||||
| Cameroon (142) | <0.001 | <0.006 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.004 | <0.394 | |||
| Tanzania (121) | <0.001 | <0.012 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.156 | <0.211 | 0.337 | ||
| Dogon (241) | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.897 | 0.479 | 0.081 | |
| Fulani (243) | <0.023 | <0.235 | <0.024 | <0.021 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.008 | 0.001 | <0.001 |
Allele distribution differences between each population were tested according to the Fisher exact test. These data show a clustering of the populations into an African, West-Eurasian, and East-Eurasian group. Each asterisk indicates populations that could not be analyzed because only the wild-type allele was present. Abbreviations are as in Fig. 1.
The clustering pattern of the TLR4 haplotypes in the various populations according to their geographical location suggests, on the one hand, that the present pattern of TLR4 haplotypes may be explained by the out-of-Africa migration routes of modern humans (Fig. 1) and, on the other hand, that these differences may have been shaped by local environmental conditions (e.g., infectious pressure). Archaeological and genetic evidence suggests that anatomically modern humans originated ≈195,000 years ago in subSaharan Africa (16, 20, 21). Approximately 55,000–60,000 years ago, a first wave of migration traversed the Bab-el-Mandab strait from the Horn of Africa to the Arabic peninsula. From this West Asian source, humans migrated toward Southeast Asia, reaching Australia by 50,000 years ago (22), and toward Northern Eurasia (and subsequently to America from a Northeast Asian source) ≈40,000 years ago (ref. 23; Fig. 1).
The presence of Asp299Gly and Thr399Ile in the African populations, as well as in the Basque and Indo-European populations, suggests these are mutations that arose in Africa >60,000 years ago and migrated toward Europe through the Middle Eastern route. In addition, the much higher prevalence of the Asp299Gly haplotype compared with the Asp299Gly/Thr399Ile haplotype in Africa (whereas Thr399Ile is in linkage with Asp299Gly allele; Fig. 1 and SI Table 3), suggests the Asp299Gly mutation arose first, whereas the Thr399Ile mutation must have appeared later a chromosome bearing the Asp299Gly mutation.
Phenotypes of the Different TLR4 Haplotypes.
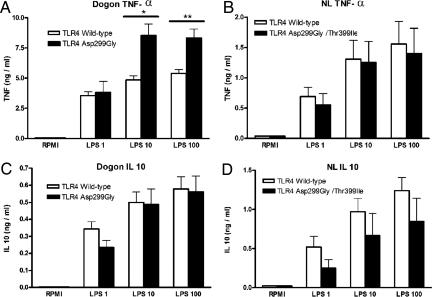
The difference in the frequency of the TLR4 haplotypes demonstrates there must have been a shift in the population frequencies of the polymorphisms during or after the migration. To answer which evolutionary or genetic force could have caused the shift responsible for the current distribution of TLR4 haplotypes in Europe and Asia, which differ from that found in Africa, we assessed whether TLR4 haplotypes result in a different functional activity of the receptor. Therefore, we studied the functionality of the two most common haplotypes, namely the ancestral 299–ancestral 399/Asp299Gly–ancestral 399 haplotypes in one African (Dogon) population and the ancestral 299–ancestral 399/Asp299Gly–Thr399Ile haplotypes in one European (Dutch) population, which both have a high prevalence of these haplotypes (Fig. 1 and SI Table 3). Whole-blood samples from Dogon individuals were exposed to increasing concentrations of bacterial LPS. Cells from Asp299Gly heterozygous individuals (homozygous for the ancestral state at 399) released significantly higher amounts of the proinflammatory cytokine TNF-α than those from donors with a double-homozygous wild-type (ancestral state) genotype TLR4 (Fig. 3A). No differences were observed in the production of the antiinflammatory cytokine IL-10 (Fig. 3B). In contrast, the presence of the double-heterozygous Asp299Gly/Thr399Ile in cosegregation in the Dutch population did not modulate cytokine production induced by LPS: no differences in TNF-α or IL-10 production were observed between individuals bearing the double-heterozygous wild-type genotype and those heterozygous for the Asp299Gly/Thr399Ile alleles (Fig. 3 C and D). Cells harvested from one individual homozygous for the Asp299Gly/Thr399Ile allele also displayed a normal production of pro- and antiinflammatory cytokines. These data demonstrate that the Asp299Gly allele is accompanied by a proinflammatory phenotype, whereas the Asp299Gly/Thr399Ile haplotype does not functionally differ from wild-type TLR4. In addition, our data represent an important argument suggesting caution in the interpretation of overexpression studies of the different forms or TLR4, which are known to be prone to artifacts due to unequal efficiency of receptor expression. In this respect, overexpression studies in cell lines have suggested that Asp299Gly and Asp299Gly/Thr399Ile TLR4 expression may lead to a defective function of TLR4 (11, 12). However, this assertion has been refuted in case of the Asp299Gly/Thr399Ile TLR4 haplotypes by many experimental and clinical studies (24–26), and our results also do not confirm a decreased function of the Asp299Gly haplotype.
Fig. 3.
Production of cytokines induced by LPS in individuals bearing the Asp299Gly or Asp299Gly/Thr399Ile haplotypes. Mean TNF-α (A and B) and IL-10 (C and D) concentrations are given as mean and SEM (nanograms per milliliter) from Asp299Gly heterozygotes (n = 4) and wild-type individuals (n = 32) with Dogon ethnicity (A and C) and Asp299Gly/Thr399Ile heterozygotes (n = 4) and wild-type individuals (n = 8) from The Netherlands (B and D). Significant difference: *, P = 0.04; **, P = 0.03 (Mann–Whitney U test).
Functional studies of the TLR4 gene neglected the TLR4 Asp299Gly allele in human primary cells; other studies to date were performed in Caucasians bearing the cosegregated Asp299Gly/Thr399Ile haplotype or used cell lines transfected with the mutated TLR4. The proinflammatory phenotype of the TLR4 Asp299Gly allele may represent the explanation of why this haplotype disappeared from Eurasia: the strongly enhanced TNF production may predispose for an increased mortality in septic shock by contributing to the systemic inflammatory response syndrome (27). In the only study that investigated the role of all TLR4 haplotypes on the susceptibility to septic shock, Lorenz et al. (16) showed that the presence of the Asp299Gly alone, but not that of the Asp299Gly/Thr399Ile haplotype, is associated with an increased mortality to septic shock. A strong body of medical literature demonstrates the deleterious effects of high cytokinemia during septic shock (28), and this most likely explains the evolutionary disadvantage of a strong proinflammatory bias in individuals with a Asp299Gly allele (and homozygous for the wild-type allele of the 399 polymorphism) who migrated into Eurasia toward northern latitudes. Pandemic infections of the past millennia such as plague (Yersinia pestis infection), typhoid fever, and influenza (with its important secondary bacterial infections) may also have been contributors to this particular geographical distribution (29). A similar scenario may have been behind the loss of the Asp299Gly allele in Asia.
Evolutionary Neutral Phenotype of Asp299Gly/Thr399Ile Haplotype.
Although these data could explain the difference in frequency of the Asp299Gly between Africa and Eurasia, they are not the reason for the variable prevalence of the cosegregated Asp299Gly/Thr399Ile haplotype in the different populations, ranging from 0% in Asian populations, 1–2% in African populations, and 5% in Dutch Indo-Europeans, to 9% in Basques. One clue may come from the observation that cells from individuals with the Asp299Gly/Thr399Ile haplotype have a response to LPS stimulation that is indistinguishable from that of cells with wild-type TLR4 alleles (Fig. 3). This is supported by data from other in vitro (17, 25, 26) and ex vivo studies (24). Thus, although the presence of Asp299Gly alone is accompanied by an increased function of the receptor, the Asp299Gly/Thr-399/Ile haplotype results in an LPS/TLR4 interaction similar to that present in wild-type individuals. To strengthen these experimental data, we investigated the effect of the Asp229Gly/Thr399Ile haplotype on the severity of sepsis. First, no significant differences were observed in the prevalence of the Asp299Gly/Thr399Ile haplotype between a healthy control population (6%) and a cohort of patients with Gram-negative sepsis (7%). Second, the severity and clinical parameters of sepsis were similar in patients bearing two wild-type haplotypes or one wild-type haplotype and one Asp299Gly/Thr399Ile haplotype (Table 2), suggesting that the later TLR4 haplotype does not increase susceptibility and mortality because of severe Gram-negative infections. Thus, these clinical data also point to a neutral effect of the Asp299Gly/Thr399Ile phenotype and suggest that the spread of this cosegregated haplotype is the consequence not of infectious pressure but more likely of functional neutrality and chance (genetic drift and/or bottlenecks).
Table 2.
Epidemiological and clinical data in 159 Indo-European patients with sepsis bearing the wild-type or heterozygote Asp299Gly/Thr399Ile haplotypes
| Wild type | Asp299Gly/Thr3999Ile | P value | |
|---|---|---|---|
| Number of patients | 150 | 9 | — |
| Age (mean ± SD, years) | 59.4 ± 18.8 | 57.4 ± 19.3 | 0.76 |
| Male/female | 112/38 | 8/1 | 0.58 |
| APACHE II (mean ± SD) | 15.9 ± 5.5 | 14.3 ± 5.1 | 0.17 |
| SOFA score (mean ± SD) | 7.87 ± 3.32 | 7.33 ± 3.00 | 0.65 |
| Lymphocytes count (mean ± SD/ml) | 13.3 ± 6.7 | 13.1 ± 3.8 | 0.93 |
| CRP (mg/liter, median IQR) | 145.0 (669) | 221.5 (260) | 0.49 |
| Septic shock, % | 67 (44.7%) | 4 (44.4%) | 0.51 |
| Mortality, % | 45 (30.0%) | 1 (11.1%) | 0.51 |
To assess possible differences in the clinical outcome between the TLR4 wild-type and Asp299Gly/Thr399Ile haplotypes, the Acute Physiology and Chronic Health Evaluation II (APACHE II) disease score, the Sepsis-Related Organ Failure Assessment (SOFA) disease severity score, C-reactive protein (CRP), and the percentage of patients with septic shock were measured. Mann–Whitney U test statistics showed no differences in the epidemiological and clinical parameters between the two groups of patients.
Another aspect concerning the phenotypic consequences of the Asp299Gly/Thr399Ile TLR4 variant refers to noninfectious diseases such as atherosclerosis (30) and nonallergic asthma (31), in which some studies (albeit not all) proposed a beneficial effect of the TLR4 polymorphisms. However, although these diseases are important for the long-term quality of life of an individual, they are very unlikely to have played any role in selection processes, because of the age at which they affect the individual and which is after the reproductive period.
In addition, to test whether the increased frequency of Asp299Gly/Thr399Ile was due to some kind of positive selection acting on this haplotype, we evaluated whether the extent of haplotype homozygosity (EHH) (EHH test; see Materials and Methods) was longer than expected under neutrality. After extensive simulations using a neutral coalescent model with recombination (see Materials and Methods), we observed that, although the EHH at the 3′ end seems to be longer than expected under neutrality (observed length until EHH < 1 = 257,122 bp; 95% expected upper limit, 242,319 bp), however, the 5′ end shows a drop in EHH already at 41,895 bp from the core, whereas the expected 95% is at least 350,000 bp. This reinforces the idea that neutrality is a compatible hypothesis to explain the evolution of this haplotype in Europeans.
Two Faces of Asp299Gly.
Why is the Asp299Gly haplotype still found in Africa? The increased mortality due to severe sepsis (i.e., severe bacterial infection) could represent an important evolutionary disadvantage in individuals bearing the Asp299Gly allele (wild-type allele homozygotes for 399) who migrated into Eurasia toward northern latitudes, whereas demography and genetic drift can be sufficient to explain the frequencies of Asp299Gly/Thr399Ile. This leaves the question why there is a high prevalence of Asp299Gly allele (associated with a wild-type allele at “399”) in subSaharan Africa. As a putative evolutionary pressure in subSaharan Africa, malaria immediately comes to mind. Indeed, it has been demonstrated that, although the Asp299Gly allele increases susceptibility to malaria in a Ghanese population, mortality was lower in individuals with Asp299Gly allele compared with “299” wild-type TLR4 patients (9). We have been able to confirm and extend this observation in a pilot study in a malaria hyperendemic region of Cameroon. In children with malaria, the presence of the Asp299Gly haplotype was associated with higher parasitemia (80% higher compared with the TLR4“299” wild type) yet lower prevalence of cerebral malaria (12% compared with 24% in the TLR4“299” wild-type group) (B.F., unpublished results). These findings strongly suggest that the higher prevalence of haplotype Asp299Gly/wild-type “399” in subSaharan Africa is due to protection against mortality from malaria. Thus, the TLR4 amino acid site 299 (more precisely, SNP rs4986790) should be added to the list of gene polymorphisms selected by malaria, next to sickle cell disease, thalassaemias, and TNF polymorphisms (1, 2, 32).
In summary, we present evidence that the TLR4“299”-“399” (rs4986790–rs4986791) haplotypes are geographically clustered. Functional data in individuals bearing the haplotype composed of the Asp299Gly allele associated to the wild-type “399” allele demonstrate a proinflammatory profile in line with an increased susceptibility to septic shock in patients bearing this haplotype (16). In contrast, this allele seems to protect against mortality in severe malaria (9), although the mechanism of protection is not yet understood. In Africa, where malaria exercises a strongly selective pressure, the beneficial effect of Asp299Gly in malaria seems to override the negative effect in sepsis. During the migration of humans into Eurasia, where malaria is less prevalent, the Asp299Gly allele was eliminated because of increased mortality in severe Gram-negative bacterial infections. The absence of the TLR4 polymorphisms in the Trio Indians from South America is a testimony that these evolutionary pressures have already eliminated the Asp299Gly/wild-type “399” haplotype in the population that migrated over the Behring strait to the New World 15,000 years ago. In contrast, the variable prevalence of the Asp299Gly/Thr399Ile haplotype in the Eurasian populations is most likely the result of genetic drift.
This study explains the geographic distribution of the TLR4 haplotypes in populations according to susceptibility to infection and provides an understanding of how our innate immune system has been molded by infectious pressures.
Materials and Methods
DNA Samples and Asp299Gly and Thr399Ile Screening.
DNA samples from the Han people came from Corriell (Camden, NY) (catalog no. HD1000CHI). All other DNA was extracted from whole blood by using the isolation kit Puregene (Gentra Sytems, Minneapolis, MN). PCR conditions, concentrations, and determinations of the Asp299Gly and Thr399Ile polymorphisms were preformed as described by Van Der Graaf et al. (17).
Cytokine Stimulation Assays.
Venous whole blood was collected into 2-ml heparin tubes (BD, Plymouth, U.K.). Blood from healthy individuals of Dogon ethnicity was diluted to a final concentration of 1:5 in RPMI medium 1640 (containing 1% glutamine, 1% pyruvate, and 1% gentamicin) and stimulated with control medium or highly purified LPS at various concentrations (0.1, 1, and 10 μg/μl). After 24 h at 37°C, supernatants were collected and stored at −80°C until cytokine measurements were done.
In the Dutch population, venous blood was collected in EDTA tubes, and peripheral blood mononuclear cells were isolated by density centrifugation on Ficoll–Hypaque (Amersham Biosciences, Diegem, Belgium). Peripheral blood mononuclear cells resuspended in RPMI medium 1640 were plated into 96-well cell culture plates at a final density of 5 × 106 cells/ml and stimulated for 24 h at 37°C with RPMI medium 1640 or LPS (0.1, 1, and 10 μg/μl). After 24 h, supernatants were collected and stored at −80°C until cytokine measurements were done.
Cytokine Measurements.
IL-10 cytokine concentrations were measured by sandwich ELISA (Sanquin, Amsterdam, The Netherlands). TNF-α production was measured by a specific ELISA, as described (33).
Sepsis Patients.
Blood was sampled from 162 patients with ventilator-associated pneumonia and sepsis. Diagnosis was made by published criteria (34). Patients were followed up for 28 days. Disease severity was assessed by Acute Physiology and Chronic Health Evaluation II (APACHE II) and Sepsis-Related Organ Failure Assessment (SOFA) scores on the first day of diagnosis, and mortality was given at the end of the observation period. C-reactive protein was estimated in serum by a nephelometric assay (Behring, Berlin, Germany).
Statistics.
The distributions between the TLR4 alleles in the different populations and the Hardy–Weinberg equilibrium were preformed with the GENEPOP program (35). The differentiation analysis is based on the unbiased estimation of the P value of a log-likelihood- (G) based exact test, which is based on the same principles as Fisher's exact test.
Cytokine and sepsis statistics were performed by using the SPSS program (Rel. 14.0.2, 2006; SPSS, Chicago, IL), and differences were tested by the Mann–Whitney U test. P < 0.05 was considered to represent a statistically significant difference.
As a neutrality test, we applied the EHH test (36) to the phased genotypic information available from HapMap Data Rel. 21a/phaseII Jan 07, on National Center for Biotechnology Information NB35 assembly, dbSNP b125. We used as the core haplotype that formed by allele G of rs4986790 and allele T of rs4986791 within TLR4 (which corresponds to haplotype Asp299Gly/Thr399Ile). We analyzed the extent to which the EHH for this core haplotype was 1 in the HapMap Caucasian population in a genomic region that extended 350 kb in both directions from the core. To test whether the distance observed was longer than expected under neutrality, we ran coalescent simulations that took into account the heterogeneity in recombination rates across these regions (obtained from the HapMap link) by means of msHOT (37). We used demographic parameters reflecting the out-of-Africa model of human evolution by slightly modifying Schaffner et al.'s model (38). Simulations were run for both the 350 kb downstream from the core and the 350 kb upstream from the core. The number of SNPs was adjusted to that observed in the HapMap sample for that region. We added three filters to the neutral simulations: (i) We considered only those simulations that resulted in a number of different haplotypes equal to that observed in the HapMap sample (as determined by the core SNPs rs4986790 and rs4986791); in the simulations, the first two SNPs were considered the core set. (ii) In addition, one of the simulated core haplotypes had to show a similar frequency to that core haplotype under examination [obtained from the HapMap Caucasian SNP information and defined by core SNPs (allele G of rs4986790 and allele T of rs4986791)]. (iii) The ancestral/derived states of the core SNPs in this preselected simulated haplotype also had to be identical to the ancestral/derived states of the core SNPs at the core haplotype under examination. By comparison with the orthologous genome regions of the chimpanzee and Rhesus macaque, both G and T were defined as “derived.” Simulations satisfying these conditions were kept, but only the information on the core haplotype was used for the EHH analysis. Finally, the extent from the core SNPs for which EHH was 1 was recorded for the observed HapMap data, and this was tested against the distribution obtained from the filtered simulations.
Supplementary Material
Acknowledgments
We thank Garrett Hellenthal (University of Oxford, Oxford, U.K.) for kindly proving a modified version of msHOT. This work was supported by a Vidi Grant of The Netherlands Organisation for Scientific Research (to M.G.N.) and an FP6 European network of Excellence (BioMalPar) fellowship (to M.B.B.M.). The Servicios Generales de Investigacion/Ikerkuntzarako Zerbitsu Orokorrak-Servicios Generales-Ikerkuntza (SGI/IZO-SGIker) UPV/EHU (supported by the National Program for the Promotion of Human Resources within the National Plan of Scientific Research, Development and Innovation–Fondo Social Europeo, McyT, and the Basque Government) is gratefully acknowledged for generous allocation of computational resources.
Abbreviations
- TLR
Toll-like receptor
- EHH
extent of haplotype homozygosity;
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704828104/DC1.
References
- 1.Allison AC. Br Med J. 1954;1:290–294. doi: 10.1136/bmj.1.4857.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flint J, Hill AV, Bowden DK, Oppenheimer SJ, Sill PR, Serjeantson SW, Bana-Koiri J, Bhatia K, Alpers MP, Boyce AJ, et al. Nature. 1986;321:744–750. doi: 10.1038/321744a0. [DOI] [PubMed] [Google Scholar]
- 3.Agarwal A, Guindo A, Cissoko Y, Taylor JG, Coulibaly D, Kone A, Kayentao K, Djimde A, Plowe CV, Doumbo O, et al. Blood. 2000;96:2358–2363. [PubMed] [Google Scholar]
- 4.Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, Goedert JJ, Buchbinder SP, Vittinghoff E, Gomperts E, et al. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 5.Akira S, Takeda K. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 6.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 7.Poltorak A, He X, Smirnova I, Liu MY, Van HC, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 8.Means TK, Wang S, Lien E, Yoshimura A, Golenbock DT, Fenton MJ. J Immunol. 1999;163:3920–3927. [PubMed] [Google Scholar]
- 9.Mockenhaupt FP, Cramer JP, Hamann L, Stegemann MS, Eckert J, Oh NR, Otchwemah RN, Dietz E, Ehrhardt S, Schroder NW, et al. Proc Natl Acad Sci USA. 2006;103:177–182. doi: 10.1073/pnas.0506803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Netea MG, Van Der Graaf CA, Vonk AG, Verschueren I, Van Der Meer JW, Kullberg BJ. J Infect Dis. 2002;185:1483–1489. doi: 10.1086/340511. [DOI] [PubMed] [Google Scholar]
- 11.Rallabhandi P, Bell J, Boukhvalova MS, Medvedev A, Lorenz E, Arditi M, Hemming VG, Blanco JC, Segal DM, Vogel SN. J Immunol. 2006;177:322–332. doi: 10.4049/jimmunol.177.1.322. [DOI] [PubMed] [Google Scholar]
- 12.Arbour NC, Lorenz E, Schutte BC, Zabner J, Kline JN, Jones M, Frees K, Watt JL, Schwartz DA. Nat Genet. 2000;25:187–191. doi: 10.1038/76048. [DOI] [PubMed] [Google Scholar]
- 13.Schroder NW, Schumann RR. Lancet Infect Dis. 2005;5:156–164. doi: 10.1016/S1473-3099(05)01308-3. [DOI] [PubMed] [Google Scholar]
- 14.Smirnova I, Hamblin MT, McBride C, Beutler B, Di RA. Genetics. 2001;158:1657–1664. doi: 10.1093/genetics/158.4.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agnese DM, Calvano JE, Hahm SJ, Coyle SM, Corbett SA, Calvano SE, Lowry SF. J Infect Dis. 2002;186:1522–1525. doi: 10.1086/344893. [DOI] [PubMed] [Google Scholar]
- 16.Lorenz E, Mira JP, Frees KL, Schwartz DA. Arch Intern Med. 2002;162:1028–1032. doi: 10.1001/archinte.162.9.1028. [DOI] [PubMed] [Google Scholar]
- 17.Van Der Graaf CA, Netea MG, Morre SA, Den HM, Verweij PE, Van Der Meer JW, Kullberg BJ. Eur Cytokine Netw. 2006;17:29–34. [PubMed] [Google Scholar]
- 18.Newport MJ, Allen A, Awomoyi AA, Dunstan SJ, McKinney E, Marchant A, Sirugo G. Tuberculosis (Edinburgh) 2004;84:347–352. doi: 10.1016/j.tube.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 19.The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 20.McDougall I, Brown FH, Fleagle JG. Nature. 2005;433:733–736. doi: 10.1038/nature03258. [DOI] [PubMed] [Google Scholar]
- 21.Cavalli-Sforza LL, Feldman MW. Nat Genet. 2003;33(Suppl):266–275. doi: 10.1038/ng1113. [DOI] [PubMed] [Google Scholar]
- 22.Bowler JM, Johnston H, Olley JM, Prescott JR, Roberts RG, Shawcross W, Spooner NA. Nature. 2003;421:837–840. doi: 10.1038/nature01383. [DOI] [PubMed] [Google Scholar]
- 23.Pitulko VV, Nikolsky PA, Girya EY, Basilyan AE, Tumskoy VE, Koulakov SA, Astakhov SN, Pavlova EY, Anisimov MA. Science. 2004;303:52–56. doi: 10.1126/science.1085219. [DOI] [PubMed] [Google Scholar]
- 24.Calvano JE, Bowers DJ, Coyle SM, Macor M, Reddell MT, Kumar A, Calvano SE, Lowry SF. Clin Immunol. 2006;121:186–190. doi: 10.1016/j.clim.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 25.Erridge C, Stewart J, Poxton IR. J Exp Med. 2003;197:1787–1791. doi: 10.1084/jem.20022078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schippers EF, van't Veer C, van Voorden S, Martina CA, le Cessie S, van Dissel JT. Cytokine. 2004;26:16–24. doi: 10.1016/j.cyto.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Beutler B, Cerami A. Nature. 1986;320:584–588. doi: 10.1038/320584a0. [DOI] [PubMed] [Google Scholar]
- 28.Hotchkiss RS, Karl IE. N Engl J Med. 2003;348:138–150. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 29.Montminy SW, Khan N, McGrath S, Walkowicz MJ, Sharp F, Conlon JE, Fukase K, Kusumoto S, Sweet C, Miyake K, et al. Nat Immunol. 2006;7:1066–1073. doi: 10.1038/ni1386. [DOI] [PubMed] [Google Scholar]
- 30.Kiechl S, Lorenz E, Reindl M, Wiedermann CJ, Oberhollenzer F, Bonora E, Willeit J, Schwartz DA. N Engl J Med. 2002;347:185–192. doi: 10.1056/NEJMoa012673. [DOI] [PubMed] [Google Scholar]
- 31.Fageras BM, Hmani-Aifa M, Lindstrom A, Jenmalm MC, Mai XM, Nilsson L, Zdolsek HA, Bjorksten B, Soderkvist P, Vaarala O. J Allergy Clin Immunol. 2004;114:561–567. doi: 10.1016/j.jaci.2004.04.050. [DOI] [PubMed] [Google Scholar]
- 32.Bayley JP, Ottenhoff TH, Verweij CL. Genes Immunol. 2004;5:315–329. doi: 10.1038/sj.gene.6364055. [DOI] [PubMed] [Google Scholar]
- 33.Grebenchtchikov N, van d V, Pesman GJ, Geurts-Moespot A, Van Der Meer JW, Sweep FC. Eur Cytokine Netw. 2005;16:215–222. [PubMed] [Google Scholar]
- 34.Giamarellos-Bourboulis EJ, Zakynthinos S, Baziaka F, Papadomichelakis E, Virtzili S, Koutoukas P, Armaganidis A, Giamarellou H, Roussos C. Intensive Care Med. 2006;32:237–243. doi: 10.1007/s00134-005-0017-1. [DOI] [PubMed] [Google Scholar]
- 35.Raymond M, Rousset F. J Hered. 1995;86:248–249. [Google Scholar]
- 36.Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, et al. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 37.Hellenthal G, Stephens M. Bioinformatics. 2007;23:520–521. doi: 10.1093/bioinformatics/btl622. [DOI] [PubMed] [Google Scholar]
- 38.Schaffner SF, Foo C, Gabriel S, Reich D, Daly MJ, Altshuler D. Genome Res. 2005;15:1576–1583. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.