Abstract
Targeted mutagenesis of Fgf9 in mice causes male-to-female sex reversal. Among the four FGF receptors, FGFR2 showed two highly specific patterns based on antibody staining, suggesting that it might be the receptor-mediating FGF9 signaling in the gonad. FGFR2 was detected at the plasma membrane in proliferating coelomic epithelial cells and in the nucleus in Sertoli progenitor cells. This expression pattern suggested that Fgfr2 might play more than one role in testis development. To test the hypothesis that Fgfr2 is required for male sex determination, we crossed mice carrying a floxed allele of Fgfr2 with two different Cre lines to induce a temporal or cell-specific deletion of this receptor. Results show that deletion of Fgfr2 in embryonic gonads phenocopies deletion of Fgf9 and leads to male-to-female sex reversal. Using these two Cre lines, we provide the first genetic evidence that Fgfr2 plays distinct roles in proliferation and Sertoli cell differentiation during testis development.
Keywords: Fgfr2, growth factor signaling, organogenesis, Sertoli cells, testis determination
The differentiation of the gonad is directed toward the testis pathway when the Y chromosome gene, Sry, is expressed in somatic progenitor cells (1–3). Precursor cells expressing Sry up-regulate the expression of a related gene, SRY-box containing gene 9 (Sox9), and initiate differentiation as Sertoli cells (4, 5). The differentiation of Sertoli cells is the pivotal event essential for testis differentiation and male sex determination. As the first testis-specific cell type, Sertoli cells coordinate early testicular morphogenesis, including testis cord formation, mesonephric cell migration, and testis-specific vascularization. Most evidence suggests that Sertoli cells induce the differentiation of other testis cell types, such as peritubular myoid cells and Leydig cells (6, 7). The male fate is established in somatic precursor cells by collective functions of transcription factors and signaling molecules. Targeted deletion of the extracellular signaling factor (Fgf9) results in disrupted Sertoli cell differentiation and male-to-female sex reversal, indicating that Fgf9 is required for testis fate determination (8).
Fgfs regulate a broad range of cellular activities, including proliferation, survival, migration, and differentiation in many organs during embryonic development. In vivo and in vitro studies suggested that Fgf9 plays several direct or indirect roles in testis development and male sex determination (8–14). However, it has been difficult to tease these roles apart.
FGF9 is required for XY-specific proliferation of cells in the coelomic epithelium, which is the earliest cellular process known to occur as XX and XY gonad development begins to diverge downstream of Sry. Cell proliferation increases the Sertoli cell population and is an obligate event for testis formation based on blocking assays (15). These data, combined with the finding that a threshold number of XY cells is required to initiate testis development in XX ↔ XY chimeric gonads (16), have led to the hypothesis that FGF9 is required for proliferation to establish numbers of Sertoli cells sufficient to block the competing female pathway and to establish testis development. However, there was no clear evidence for a direct role of FGF9 in Sertoli precursor proliferation. Furthermore, because proliferation and differentiation of Sertoli cells are so closely intertwined, it has not been clear whether loss of proliferation and reduced Sertoli cell numbers could be solely responsible for failure of the testis pathway in Fgf9 mutants or whether Fgf9 has a distinct function within Sertoli cells to maintain Sox9 during differentiation. One way to dissect the potential roles of FGF9 is to genetically address the function of Fgf receptors in the gonad.
In mammals, Fgfs bind to four high-affinity receptor tyrosine kinases, FGFR1–FGFR4, and to heparan sulfate proteoglycans in the extracellular matrix (17, 18). The expression patterns of Fgf ligands and Fgf receptors are spatiotemporally distinctive but often overlapping, and ligand–recepter interactions are possible in various combinations, complicating the understanding of their in vivo functions. Previous immunocytochemical analysis of Fgf receptors in mouse embryonic gonads indicated that all four receptors (FGFR1–FGFR4) are expressed in XX and XY gonads during early development (10). Among the four receptors, FGFR2 showed plasma membrane localization in coelomic epithelial cells, consistent with a role of Fgf9 in mediating proliferation of these cells. In contrast, it showed a nuclear localization specifically in Sertoli progenitor cells within the interior of the gonad. This expression pattern made FGFR2 a good candidate for the receptor that mediates the activity of Fgf9 in Sertoli progenitors, and suggested that it might play a role at multiple steps in the testis pathway. However, the unexpected nuclear localization of this tyrosine kinase receptor led to skepticism about the fidelity of the polyclonal antibody. To sidestep this problem we chose to take a genetic approach to test the role of Fgfr2 in testis formation.
Fgfr2 -null mice die before gonad formation (19). To overcome the embryonic lethality, we generated conditional inactivation of Fgfr2 by crossing mice carrying a Fgfr2 allele flanked with loxP sites (Fgfr2flox) (20) with two different transgenic Cre lines that direct either a temporal or a cell-specific deletion of this receptor. Phenotypic and molecular marker analysis in Fgfr2 deletion mutant gonads uncovered genetic evidence that Fgfr2 plays an essential role during testis determination. The finding that deletion of Fgfr2 phenocopies Fgf9 knockout gonads strongly suggests that Fgfr2 is the receptor for FGF9 in the gonad. Furthermore, these results, generated using two complementary Cre lines, support the hypothesis that Fgfr2 plays distinct roles in the proliferation of progenitor cells and Sertoli cell differentiation during testis development.
Results
The Expression of Fgfr2 in Gonads and Isolated Sertoli Cells.
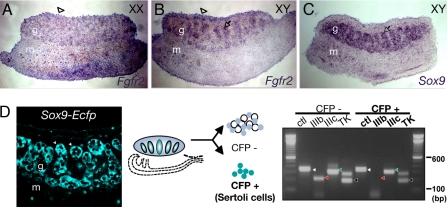
Because of concerns about the fidelity of the FGFR2 antibody, we set out to confirm the expression pattern of Fgfr2 at the mRNA level. Using mRNA in situ hybridization on sections of gonads at 12.5 days post coitum (dpc), Fgfr2 transcripts were detected in the coelomic domain of XX and XY gonads and in testis cords in a pattern similar to that of Sox9 (Fig. 1 A–C). This finding supports our previous results using an antibody against the tyrosine kinase domain of FGFR2, in which the receptor protein was detected in coelomic epithelial cells and Sertoli precursor cells (10).
Fig. 1.
Expression of Fgfr2 transcripts in embryonic gonads. (A–C) mRNA in situ hybridization of Fgfr2 and Sox9 on frozen sections of XX and XY gonads at 12.5 dpc. Fgfr2 antisense riboprobe reveals Fgfr2 expression in the coelomic domain (arrowhead) of XX and XY gonads and inside the testis cords (arrow), but not in the interstitium, of XY gonads. Sox9 is a comparative control for expression in Sertoli cells inside testis cords. (D) RT-PCR analysis of splicing isoforms of Fgfr2 in fluourescence activation-sorted Sertoli cells. A confocal image of Sox9-Ecfp XY gonad shows Sertoli cells marked with EGFP (arrowheads). EGFP+ (Sertoli) cells express mainly Fgfr2-IIIc; whereas EGFP− cells express both isoforms. PCR products are Hprt (white arrowhead), Fgfr2-IIIb (red arrowhead), Fgfr2-IIIc (green arrowhead), and the tyrosine kinase domain of Fgfr2 (black arrowhead).
Using RT-PCR analysis with two different primer sets specific to the sequences within the transmembrane domain and the intracellular tyrosine kinase domain of Fgfr2, transcripts were detected in both XX and XY gonads (data not shown), consistent with our in situ hybridization data. To validate the expression of Fgfr2 in Sertoli cells, we took advantage of transgenic mice expressing a Sertoli cell reporter, Sox9-Ecfp. ECFP-positive Sertoli cells were isolated by fluorescence-activated cell sorting, and RT-PCR of Fgfr2 was performed using primers that distinguish the expression of the two major receptor isoform variants, Fgfr2-IIIb and -IIIc. The results showed that Fgfr2-IIIc was detectable in the isolated cells, whereas Fgfr2-IIIb expression was beneath the detection limit. This finding indicates that Sertoli cells express Fgfr2 and that Fgfr2-IIIc, to which FGF9 binds with high affinity in in vitro assays (21), is the major isoform of Fgfr2 in Sertoli cells (Fig. 1D).
Conditional Deletion of Fgfr2 in Gonads Between 10.5 and 11.5 dpc Results in Disruption of Testis Morphogenesis.
Because deletion of one isoform of Fgfr2 can lead to compensation by the other isoform (refs. 22 and 23), we chose to analyze a null mutation of Fgfr2. To avoid the early embryonic lethality associated with deletion of Fgfr2, we obtained mice carrying a floxed allele (Fgfr2flox/+) to generate a conditional mutation of Fgfr2. CRE recombinase-mediated excision of the floxed allele deletes a region including the ligand-binding domain and transmembrane domain of Fgfr2 and creates a functional null allele (20). We conditionally deleted Fgfr2 by using a heat-shock-inducible Cre, Hs-Cre (24), which mediates a stage-specific deletion of the receptor.
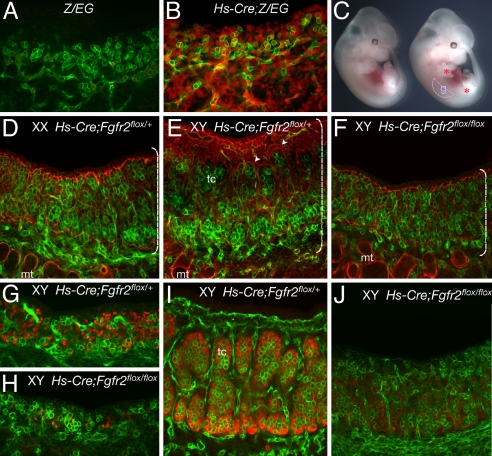
To test the efficiency of the heat-shock-activated CRE, male Hs-Cre/+ mice were crossed to female LacZ/Egfp (Z/EG) transgenic mice in which CRE-mediated loxP excision activates Egfp expression. Pregnant females were treated by heat shock at 10.5 dpc, and embryos were dissected at 11.5 dpc to examine the gonad for EGFP expression. Greater than 90% of cells in the genital ridge were positive for EGFP at the time of dissection (Fig. 2 A and B), indicating that excision by Hs-Cre is highly effective throughout the gonads. Male mice heterozygous for both Hs-Cre and Fgfr2flox (Hs-Cre/+;Fgfr2flox/+) were crossed with Fgfr2flox/+ female mice for timed matings. A heat shock was administered to pregnant females at 10.5 dpc, such that loss of Fgfr2 would affect gonads after the initial formation of the genital ridge. After the heat-shock treatment, embryos were allowed to develop for a short period, until dissection at 12.5–13.5 dpc. Conditional mutants at 13.5 dpc were normal in body size (Fig. 2C), indicating that significant deletion of Fgfr2 before heat-shock treatment was unlikely to have occurred during earlier stages, when the gene is essential for the survival and growth of embryos. However, in mutant embryos at 12.5–13.5 dpc, all XY mutant gonads failed to grow to normal sizes (Fig. 2 E and F), and limb defects (typically, a short limb with abnormal digits) were often detected (Fig. 2C and data not shown). Affected XY gonads were indistinguishable from normal XX gonads at the level of stereomicroscopy (data not shown). Heat-shock treatment alone does not impair the growth of gonads, because littermates that were wild type for either Hs-Cre or Fgfrflox appeared phenotypically normal after the treatment (data not shown). These results indicate that the deletion of Fgfr2 affected growth of the limb and the XY gonad during a temporal window during which cell proliferation increases. The small XY Hs-Cre;Fgfr2flox/flox gonads showed no sign of testis structures such as testis cords and coelomic vasculature, which are normally discernable by 12.5 dpc. Immunostaining at 13.5 dpc for laminin, a protein localized to the perimeter of testis cords, and PECAM, a marker for endothelial cells and germ cells, verified that the mutant XY gonads fail to form testis cords or the typical testicular coelomic vessel (Fig. 2 D–F).
Fig. 2.
Sex reversal caused by stage-specific conditional inactivation of Fgfr2 by Hs-Cre. (A and B) Hs-Cre efficiency in gonads was accessed in Z/EG reporter mice. Confocal scanning microscopy shows that EGFP reporter (red) is expressed in most cells throughout the gonad and mesonephros after heat shock, but expression is absent in Z/EG gonads in the absence of Hs-Cre, indicating time-specific global recombination in the genital ridge. PECAM (green) is a marker for germ cells and endothelial cells. (C) Stereomicroscopy image of control (left) and Hs-Cre/+;Fgfr2flox/flox (right) embryos 48 h after heat-shock treatment. Body shape and size is grossly normal after the treatment. However, inactivation of Fgfr2 results in defective growth in limbs and gonads. Asterisks indicate mutant limbs in heat-shock-treated animals. g, gonad, outlined. (D–F) Immunohistohemistry and confocal scanning microscopy of gonads from the heat-shock-treated embryos. Immunostaining with laminin (red) and PECAM (green) reveals that Hs-Cre/+;Fgfr2flox/flox XY gonads lack testis cords (tc), which normally form by this stage in littermate controls (arrowheads in E). Brackets highlight relative size of control and mutant XY gonads. mt, mesonephric tubules. (G–J) Control and mutant gonads immunostained for SOX9 at 11.5 dpc (G and H) or 12.5 dpc (I and J). SOX9 (red) is present in the mutant gonads at 11.5 dpc, albeit in fewer cells. By 12.5 dpc, SOX9 expression is almost absent in the mutant XY gonads.
Because Sertoli cells are critical for organizing testis cord structures, we investigated whether Sertoli cells differentiate in these mutant XY gonads by monitoring SOX9 expression between 11.5 and 12.5 dpc. SOX9 was present both in the littermate control XY gonads and in the mutant XY gonads at 11.5 dpc, but the number of SOX9-expressing cells in the mutants was reduced, often to only a few positive cells (Fig. 2 G and H). SOX9 expression was detectable in only a few cells by 12.5 dpc (Fig. 2 I and J), indicating that Sertoli differentiation was blocked. Overall, the conditional deletion of Fgfr2 by Hs-Cre at the critical stages of sex determination recapitulated the phenotype of Fgf9 null-mutant gonads: XY-specific gonad growth was blocked and testis differentiation was disrupted downstream of the initiation of SOX9 expression.
The Conditional Deletion of Fgfr2 in Pre-Sertoli Cells Rescues Gonad Growth but Blocks Sertoli Cell Differentiation.
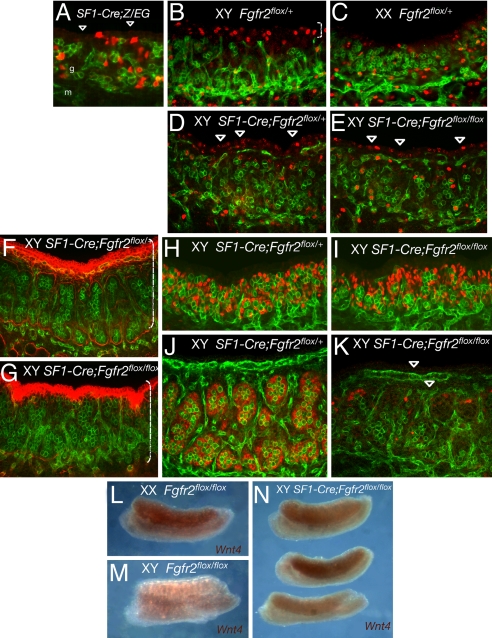
Our previous study suggested a model in which Fgf9 is required for maintenance of SOX9 and the stable commitment of precursor cells to the Sertoli cell fate (13). However, it did not distinguish between a role of Fgf9 in the proliferation/expansion of Sertoli precursors or in the differentiation/maintenance of Sertoli precursors. The model predicts that if Fgfr2 were the receptor mediating Fgf9 activity in Sertoli precursor cells, the deletion of Fgfr2 specifically in these cells would disrupt testis determination. To generate an Fgfr2 deletion specific to somatic progenitor cells, we used a different Cre transgenic line, Sf1-Cre, which express a Cre transgene driven by the regulatory sequences of steroidogenic factor 1 (SF1) (also called Nr5a1) (25). Endogenous SF1 expression is detected at high levels at 11.0–11.5 dpc in coelomic epithelial cells that proliferate and give rise to somatic progenitors within the gonad and in Sertoli precursors at the time when testis cords form (26). To determine which cells express the transgenic Sf1-Cre in gonads at bipotential stages, we crossed Z/EG females to Sf1-Cre males and examined EGFP reporter expression. In Sf1-Cre/+;Z/EG gonads at 11.5 dpc, EGFP was detected in a subset of somatic cells within the gonad and only a few coelomic epithelial cells (four of four gonad pairs examined) (Fig. 3A). Sf1-Cre/+;Fgfr2flox/+ males were mated with Fgfr2flox/flox females, directing Fgfr2 deletion to a subset of Sertoli precursor cells. In Sf1-Cre/+;Fgfr2flox/flox embryos, early gonad formation occurred normally. Mutant XY gonads were only slightly smaller than those of littermate controls (compare Fig. 3F with Fig. 3G versus the size difference in Fig. 2E and F with Hs-Cre). Consistent with this observation, immunostaining for phosphorylated histone H3 (pHH3), a mitotic cell marker, revealed little difference in proliferation in the coelomic domain of Sf1-Cre/+;Fgfr2flox/flox XY gonads and Sf1-Cre/+;Fgfr2flox/+ littermates (Fig. 3 D and E), in contrast to the difference between wild-type XY and XX at 11.5 dpc (Fig. 3 B and C and ref. 26). However, Sf1-Cre/+;Fgfr2flox/flox XY gonads showed aberrant testis cord formation based on laminin immunostaining (Fig. 3 F and G): Little detectable laminin deposition occurred, although cells aggregated as if cord formation were initiated.
Fig. 3.
Abnormal testis development caused by conditional deletion of Fgfr2 by SF1-Cre in somatic progenitor cells. PECAM1 is green throughout. (A) Confocal scanning microscopy of a SF1-Cre/+;Z/EG gonad at 11.5 dpc shows a typical example of SF1-Cre-mediated activation of the Z/EG reporter (red) in a subset of gonadal somatic progenitor cells. Arrowheads indicate coelomic domain where few cells show recombination (compare with Fig. 2B). g, gonad; m, mesonephros. (B–E) Immunostaining and confocal scanning microscopy of a mitotic marker pHH3 (red) in controls and SF1-Cre;Fgfr2flox/flox gonads at 11.5 dpc. Somatic cells in coelomic domain show increased proliferation specific to XY gonads (bracket in B). (D and E) Proliferation in the coelomic domain (arrowheads) is only slightly decreased in XY SF1-Cre;Fgfr2flox/flox gonads relative to the normal levels in XY littermates. (F–K) There is little difference in gonad size between controls and mutant XY gonads at 12.5 dpc (brackets in F and G). (G) However, immunostaining of laminin (red) reveals aberrant testis cord formation in the mutant gonad. (I and K) SOX9 expression (red) appears normal in the mutant gonads at 11.5 dpc (I), but by 12.5 dpc it is severely reduced (K). PECAM reveals fragments of the testis vasculature (arrowheads in K). (L–N) Whole-mount in situ hybridization of Wnt4 in control and mutant gonads at 12.5 dpc. (L and M) In control gonads, Wnt4 expression is specific to XX gonads. (N) In XY SF1-Cre/+;Fgfr2flox/flox gonads, Wnt4 is activated but variable between samples and across the gonad field.
Unlike Hs-Cre/+;Fgfr2flox/flox mutant gonads, in which SOX9 expression was reduced at this stage (Fig. 2 G and H), SOX9 was expressed at near-normal levels at 11.5 dpc in Sf1-Cre/+;Fgfr2flox/flox gonads (Fig. 3 H and I), suggesting that a significant population of pre-Sertoli cells is initially established, likely through the rescue of early proliferation by using Sf1-Cre as opposed to Hs-Cre to trigger deletion of Fgfr2. However, SOX9 is dramatically decreased by 12.5 dpc (Fig. 3 J and K). These results indicate that the maintenance of the Sertoli cell differentiation pathway is impaired, owing to the deletion of Fgfr2 in pre-Sertoli cells in the mutant gonads. Earlier work (13) predicted that the loss of Fgfr2 would cause the disruption of the Sox9/Fgf9 feed-forward loop and a failure to repress the ovary pathway. Accordingly, in Sf1-Cre/+;Fgfr2flox/flox XY gonads, Wnt4, an ovary-specific marker that is normally down-regulated in XY gonads by 12.5 dpc, was derepressed and expressed at variable levels (Fig. 3 L–N). This finding suggests that the competing female pathway was activated in the mutant gonads.
Ovotestis Formation in Sf1-Cre/+;Fgfr2flox/flox XY Gonads.
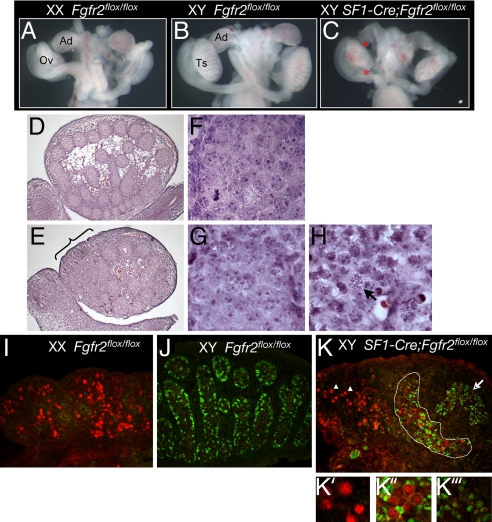
By 15.5 dpc, Sf1-Cre/+;Fgfr2flox/flox XY gonads resolved into an ovotestis structure evident at the stereomicroscopy level, in which the adrenal is also seen to be reduced or absent (Fig. 4 A–H). On the basis of histological analysis, testis cords were present in the central regions, albeit rather disrupted (Fig. 4 E and G) relative to those of XY littermate control (Fig. 4 D and F); however, they were absent from anterior (and often posterior) regions of the gonad where meiotic figures characteristic of ovarian germ cells were histologically evident in germ cell nuclei (Fig. 4H). SOX9 was detected only in domains where there was some evidence of cord-like organization, but not in the anterior region of the gonad (Fig. 4K). Instead, the meiotic marker synaptonemal complex protein 3 (SCP3) was detected in the anterior domains of the mutant XY gonads, implying that these germ cells may be under the influence of the ovarian somatic environment, as described in recent studies (27, 28). Interestingly, there was a region of overlap in which SOX9 and the meiotic marker SCP3 were simultaneously detected (Fig. 4 K and K″). This finding raises questions about the role of FGF signaling in establishing the block to meiosis that is characteristic of the male pathway.
Fig. 4.
Ovotestis formation in XY SF1-Cre/+;Fgfr2flox/flox gonads at 15.5 dpc. (A–C) Stereomicroscopy images of urogenital systems of controls and XY SF1-Cre/+;Fgfr2flox/flox mutant. In addition to gonadal defects, the adrenal glad is severely reduced or absent in these mutants. Ad, adrenal; Ov, ovary; Ts, testis; *, mutant adrenal and gonad. (D–H) Histology of wild-type XY and SF1-Cre/+;Fgfr2flox/flox XY gonads at 15.5 dpc. (E–H) Higher magnification views show testis cords in the wild-type XY gonad (F), disrupted cords from the central region in the mutant XY gonad (G), and ovarian region (H) located in the anterior of the mutant gonad (bracket in E). Arrow indicates the nucleus of a germ cell in meiosis. (I–K) Confocal scanning microscopy of sections of 15.5 dpc gonads immunostained with SOX9 (green) and SCP3 (red). (I and J) SCP3-positive germ cells are characteristic of wild-type XX gonads, but not XY gonads at this stage, whereas, SOX9 shows the reverse pattern. (K′–K‴) The ovotestis contains a region of SCP3-positive cells in the anterior domain (arrowhead in K′), a region where both markers are present (dashed line in K″), and a separated cluster of SOX9-positive cells where SCP3 is absent (arrow in K‴).
Taken together, findings in this study provide genetic evidence that Fgfr2 is necessary for testis formation and male sex determination and support a model in which paracrine signals are crucial to the establishment of testis or ovarian patterning of early gonads.
Discussion
The similarities in the phenotypes of Fgfr2 and Fgf9 mutants suggest that FGFR2 is the receptor for FGF9 in the XY gonad. The generation of conditional deletions of Fgfr2 allowed us to examine the role played by Fgfr2 in gonads during sex determination. Fgfr2 is essential for the wave of male-specific proliferation that establishes Sertoli progenitor cells in the XY gonad. We further demonstrated that Fgfr2 is required in pre-Sertoli cells to direct Sertoli cell differentiation, separating this role for Fgf signaling from the early proliferation defect characteristic of Fgf9 mutants.
The XY-specific cell proliferation is concentrated mostly at the surface of the gonad at 11.5 dpc (Fig. 3 B and C and ref. 26). This proliferation increase is not only responsible for the growth difference between XY and XX gonads by 12.5 dpc (15, 26), it is also critical for testis determination as cells dividing at the coelomic epithelium give rise to Sertoli and interstitial cells (29). Although it is evident that there is no size increase and no testicular morphogenesis in the Hs-Cre;Fgfr2flox/flox XY gonads (Fig. 2 E and F), Sf1-Cre;Fgfr2flox/flox XY gonads display near wild-type growth and improved testicular morphology (Fig. 3 F and G). Immunohistochemistry of a mitotic marker in Sf1-Cre;Fgfr2flox/flox XY gonads showed that proliferation of cells in the coelomic domain was close to normal levels, suggesting that the conditional deletion using Sf1-Cre did not affect these proliferating cells. Despite the substantial proliferation, Sf1-Cre;Fgfr2flox/flox XY gonads fail to maintain pre-Sertoli cells by 12.5 dpc (Fig. 3 J and K), suggesting that Fgfr2 is required at a step subsequent to proliferation to direct the differentiation of Sertoli cells. The partial phenotype in the mutant XY gonads may be due to incomplete deletion of Fgfr2flox alleles. The presence of ovarian regions restricted to the poles of the gonad is typical of other cases of ovotestis formation (30). The polar regions of the gonad may be more sensitive to loss of paracrine signals than the central region, where Sry is expressed at its highest levels (3, 31, 32).
Numbers of SOX9-positive cells in HS-Cre gonads are variable but usually reduced (Fig. 2H) compared with normal gonads or Fgf9−/− gonads. In normal development, the number of these precursors increases as a result of cell proliferation induced by feed-forward loops between Fgf9 and Sox9 (13, 26, 33). One possible explanation for the normal SOX9 expression in Fgf9−/− gonads at 11.5 dpc is that another Fgf can partially compensate for the loss of Fgf9. There are other Fgfs expressed at early stages in embryonic gonads (34–36). Perhaps in the absence of Fgfr2, cells are insensitive to an additional Fgf signal.
We previously reported the expression of FGFR2 in the plasma membrane of coelomic epithelial cells and in the nuclei of pre-Sertoli cells based on antibody staining patterns (10). Although this expression pattern implicated Fgfr2 in testis determination, the data were inconclusive because (i) all four FGFRs were detected in somatic progenitors of XY gonads and (ii) the specificity of the polyclonal antibody reagent detecting the unusual localization of FGFR2 in the nuclei of pre-Sertoli cells could not be conclusively verified. Our genetic approach demonstrates that Fgfr2 is required for proliferation of coelomic epithelial cells and in pre-Sertoli cells to establish the Sertoli cell fate, strongly suggesting that differential intracellular localization of the receptor has distinct functional relevance.
We show that Fgfr2-IIIc is expressed in isolated pre-Sertoli cells, whereas both Fgfr2-IIIb and Fgfr2-IIIc are expressed in other gonadal cells, including coelomic epithelial cells. It is interesting to speculate whether the regulation of alternative splicing in somatic progenitor cells is related to a switch from the proliferation of progenitors to the differentiation of Sertoli cells. Eswarakumar et al. (37) reported that homozygous Fgfr2-IIIc−/− mice were fertile, which suggests that Sertoli cells in the mutant gonads were functional. Homozygous Fgfr2-IIIb−/− mice are perinatal lethal (38), thus the fertility of the mutant animals cannot be assessed. We examined the gonads in Fgfr2-IIIb−/− embryos at 12.5–14.5 dpc and found no evidence of the disruption of Sertoli cell differentiation (data not shown). A disruption of one isoform of Fgfr2 led to the compensatory expression of another alternative splicoform in other studies (22, 23). Thus, it is not clear how alternative isoform variants of Fgfr2 underlie the regulation of Sertoli cell fate determination. This possibility should be investigated in a new system in which both alternative splicing and nuclear localization of FGFR2 can be visualized. If this regulation can account for a mechanism leading to a transition between proliferation and differentiation, it will have important implications, particularly in epithelial-to-mesenchymal transition during normal organogenesis and tumorigenesis.
Our study suggests that mutations in FGFR2 may account for an unassigned subset of human patients who show ambiguous sexual development. We hope our findings will motivate molecular analysis of FGF signaling in human syndromes associated with congenital malformations, including sexual development defects.
Methods
Mice.
The Sox9-Ecfp transgenic mice were generated in the Duke University Medical Center Transgenic Facility by using a construct provided by R.S. and R.L.-B. (unpublished manuscript). Cre transgenic lines Hs-Cre6 and Sf1-Cre, in which Hsp70–1 promoter and a regulatory region of Sf1 (also called Nr5a1) respectively drive expression of Cre recombinase, are described in refs. 24 and 25. The Fgfr2flox line was originally generated by Yu et al. (20). In the mating scheme in this study, the stage-specific inactivation of Fgfr2 was achieved by crossing Fgfr2flox/flox females to F1 generation Hs-Cre/+;Fgfr2flox/+ males. Pregnant females were heat-shocked at 10.5–11.0 dpc to delete Fgfr2 conditionally at bipotential stages in embryonic gonads. For heat shock treatment, pregnant mice were placed in a prewarmed container inside a 42° C oven for 12 min. The cell lineage-specific inactivation of Fgfr2 was generated by crossing male Sf1-Cre/+;Fgfr2flox/+ to Fgfr2flox/flox females. Mutant animals were bred on a mixed C57BL/6;129 genetic background. Genotyping for the floxed Fgfr2 allele and the Hs-Cre and SF1-Cre transgenes was determined by PCR using specific primers described in refs. 20, 24, and 25.
In Situ Hybridization and RT-PCR.
mRNA in situ hybridization was performed on frozen sections (Fgfr2) or on whole-mount preparations (Wnt4) of gonads by using a standard protocol. The plasmid template for riboprobes of Fgfr2 and Wnt4 were gifts from David Ornitz (Washington University, St. Louis, MO) (39) and Andy McMahon (Harvard University, Cambridge, MA) (40). The urogenital ridge was isolated from timed Sox9-Ecfp transgenic embryos. Gonads were separated from the mesonephros and treated with collagenase (0.025%; Sigma, St. Louis, MO) to dissociate cells, and Sertoli cells were isolated by fluorescence-activated cell sorting. Total RNA was prepared either from embryonic gonad tissues (dissected free of the mesonephros) or isolated Sertoli cells by using TRIzol reagent as instructed by the manufacturer (GIBCO/BRL, Carlsbad, CA), and quantified using a standard spectrophotometer method (Bio-Rad Laboratories, Hercules, CA). The total RNA sample was amplified with the SuperScript One-Step RT-PCR system (Invitrogen, Carlsbad, CA) using specific primer sets. The RT-PCR program was one cycle of 45° C for 15 min; followed by one cycle of 94° C for 2 min; and 32 cycles of 94° C for 30 sec, 50° C for 30 sec, and 72°C for 1 min. The primer sequences were 5′CGCCTGT G A G A G AGAAGGAGATCACG3′ and 5′AACAACGCGTCTGTCCTCAACAGC3′ to detect the tyrosine kinase domain of Fgfr2; 5′CCCATCCTCCAAGCTGGACTGCCT 3′ and 5′CAGAGCCAGCACTTCTGCATTG 3′ for Fgfr2-IIIB; 5′CCGCCGGTGTTAACACCAC3′ and 5′TGTTACCTGTCTCCGCAG 3′ for Fgfr2-IIIC; 5′CCTGCTGGATTACATTAAAGCACTG3′ and 5′GTCAAGGGCATATCCAACAACAAAC3′ for Hprt. Reaction products were resolved on a 3% agarose gel alongside a 100-bp DNA ladder.
Immunohistochemistry and Histology.
Whole-mount embryonic gonads were immunostained with antibodies directed against laminin (1:250; a gift of H. Erickson, Duke University Medical Center), SOX9 (1:1,000; a gift of F. Poulat, Institut de Genetique Humaine, Montpelier, France), phospho-histone H3 (1:250; Cell Signaling Technology, Danvers, MA), SCP3 (1:400; Novagen, San Diego, CA) and PECAM (1:250; BD BioScience, San Jose, CA). Double immunohistochemistry was detected by Cy3- and Cy5-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA), and imaged using confocal scanning microscopy (LSM510; Zeiss, Thornwood, NY). For histology, embryos were fixed in 4% paraformaldehyde in PBS, saturated through a sucrose series, and embedded in optimal cutting temperature (OCT; Sakura Rinetek, Torrence, CA). Frozen samples were sectioned at 8-μm thickness and stained with hematoxylin and eosin.
Acknowledgments
We thank Dr. Argiris Efstratiadis (Columbia University, New York, NY) for the Hs-Cre line, Drs. Sunita Verma-Kurvari and Luis Parada (both at University of Texas Southwestern, Dallas, TX) for the SF1-Cre line, Dr. Clive Dickson (Cancer Research UK, London, U.K.) for the Fgfr2-IIIb mice, and Dr. David Ornitz for the transgenic mouse line carrying a floxed allele of Fgfr2 (provided by our neighbor, Dr. Erik Meyers, Duke University Medical Center). We also thank members of the laboratory for helpful comments, especially Hao Tang for Sox9-Ecfp transgenic mice and Leo DiNapoli for establishing conditions for the use of the Hs-Cre line. This work was supported National Institutes of Health Grants HL63054 and HD39963 (to B.C.).
Abbreviations
- dpc
days post coitum
- SCP
synaptonemal complex protein.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Albrecht KH, Eicher EM. Dev Biol. 2001;240:92–107. doi: 10.1006/dbio.2001.0438. [DOI] [PubMed] [Google Scholar]
- 2.Koopman P, Munsterberg A, Capel B, Vivian N, Lovell-Badge R. Nature. 1990;348:450–452. doi: 10.1038/348450a0. [DOI] [PubMed] [Google Scholar]
- 3.Sekido R, Bar I, Narvaez V, Penny G, Lovell-Badge R. Dev Biol. 2004;274:271–279. doi: 10.1016/j.ydbio.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 4.Kent J, Wheatley SC, Andrews JE, Sinclair AH, Koopman P. Development (Cambridge, UK) 1996;122:2813–2822. doi: 10.1242/dev.122.9.2813. [DOI] [PubMed] [Google Scholar]
- 5.Morais da Silva S, Hacker A, Harley V, Goodfellow P, Swain A, Lovell-Badge R. Nat Genet. 1996;14:62–68. doi: 10.1038/ng0996-62. [DOI] [PubMed] [Google Scholar]
- 6.Polanco JC, Koopman P. Dev Biol. 2007;302:13–24. doi: 10.1016/j.ydbio.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 7.Swain A, Lovell-Badge R. Genes Dev. 1999;13:755–767. doi: 10.1101/gad.13.7.755. [DOI] [PubMed] [Google Scholar]
- 8.Colvin JS, Green RP, Schmahl J, Capel B, Ornitz DM. Cell. 2001;104:875–889. doi: 10.1016/s0092-8674(01)00284-7. [DOI] [PubMed] [Google Scholar]
- 9.Chi L, Itaranta P, Zhang S, Vainio S. Endocrinology. 2006;147:3777–3788. doi: 10.1210/en.2006-0299. [DOI] [PubMed] [Google Scholar]
- 10.Schmahl J, Kim Y, Colvin JS, Ornitz DM, Capel B. Development (Cambridge, UK) 2004;131:3627–3636. doi: 10.1242/dev.01239. [DOI] [PubMed] [Google Scholar]
- 11.Willerton L, Smith RA, Russell D, Mackay S. Int J Dev Biol. 2004;48:637–643. doi: 10.1387/ijdb.031778lw. [DOI] [PubMed] [Google Scholar]
- 12.Yoshioka H, Ishimaru Y, Sugiyama N, Tsunekawa N, Noce T, Kasahara M, Morohashi K. Dev Biol. 2005;280:150–161. doi: 10.1016/j.ydbio.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 13.Kim Y, Kobayashi A, Sekido R, DiNapoli L, Brennan J, Chaboissier MC, Poulat F, Behringer RR, Lovell-Badge R, Capel B. PLoS Biol. 2006;4:e187. doi: 10.1371/journal.pbio.0040187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El Ramy R, Verot A, Mazaud S, Odet F, Magre S, Le Magueresse-Battistoni B. J Endocrinol. 2005;187:135–147. doi: 10.1677/joe.1.06146. [DOI] [PubMed] [Google Scholar]
- 15.Schmahl J, Capel B. Dev Biol. 2003;258:264–276. doi: 10.1016/s0012-1606(03)00122-2. [DOI] [PubMed] [Google Scholar]
- 16.Palmer SJ, Burgoyne PS. Development (Cambridge, UK) 1991;112:265–268. doi: 10.1242/dev.112.1.265. [DOI] [PubMed] [Google Scholar]
- 17.Eswarakumar VP, Lax I, Schlessinger J. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Ornitz DM, Itoh N. Genome Biol. 2001;2:REVIEWS3005. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P. Proc Natl Acad Sci USA. 1998;95:5082–5087. doi: 10.1073/pnas.95.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, Towler DA, Ornitz DM. Development (Cambridge, UK) 2003;130:3063–3074. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. J Biol Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hajihosseini MK, Wilson S, De Moerlooze L, Dickson C. Proc Natl Acad Sci USA. 2001;98:3855–3860. doi: 10.1073/pnas.071586898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones RB, Wang F, Luo Y, Yu C, Jin C, Suzuki T, Kan M, McKeehan WL. J Biol Chem. 2001;276:4158–4167. doi: 10.1074/jbc.M006151200. [DOI] [PubMed] [Google Scholar]
- 24.Dietrich P, Dragatsis I, Xuan S, Zeitlin S, Efstratiadis A. Mamm Genome. 2000;11:196–205. doi: 10.1007/s003350010037. [DOI] [PubMed] [Google Scholar]
- 25.Bingham NC, Verma-Kurvari S, Parada LF, Parker KL. Genesis. 2006;44:419–424. doi: 10.1002/dvg.20231. [DOI] [PubMed] [Google Scholar]
- 26.Schmahl J, Eicher EM, Washburn LL, Capel B. Development (Cambridge, UK) 2000;127:65–73. doi: 10.1242/dev.127.1.65. [DOI] [PubMed] [Google Scholar]
- 27.Koubova J, Menke DB, Zhou Q, Capel B, Griswold MD, Page DC. Proc Natl Acad Sci USA. 2006;103:2474–2479. doi: 10.1073/pnas.0510813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowles J, Knight D, Smith C, Wilhelm D, Richman J, Mamiya S, Yashiro K, Chawengsaksophak K, Wilson MJ, Rossant J, et al. Science. 2006;312:596, 600. doi: 10.1126/science.1125691. [DOI] [PubMed] [Google Scholar]
- 29.Karl J, Capel B. Dev Biol. 1998;203:323–333. doi: 10.1006/dbio.1998.9068. [DOI] [PubMed] [Google Scholar]
- 30.Eicher EM, Beamer WG, Washburn LL, Whitten WK. Cytogenet Cell Genet. 1980;28:104–115. doi: 10.1159/000131518. [DOI] [PubMed] [Google Scholar]
- 31.Albrecht KH, Young M, Washburn LL, Eicher EM. Genetics. 2003;164:277–288. doi: 10.1093/genetics/164.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bullejos M, Koopman P. Dev Biol. 2005;278:473–481. doi: 10.1016/j.ydbio.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 33.Lovell-Badge R, Canning C, Sekido R. Novartis Found Symp. 2002;244:4–18. discussion 18–22, 35–42 253–257. [PubMed] [Google Scholar]
- 34.Cancilla B, Davies A, Ford-Perriss M, Risbridger GP. J Endocrinol. 2000;164:149–159. doi: 10.1677/joe.0.1640149. [DOI] [PubMed] [Google Scholar]
- 35.Cory AT, Boyer A, Pilon N, Lussier JG, Silversides DW. Mol Reprod Dev. 2007 doi: 10.1002/mrd.20722. in press. [DOI] [PubMed] [Google Scholar]
- 36.Nef S, Schaad O, Stallings NR, Cederroth CR, Pitetti JL, Schaer G, Malki S, Dubois-Dauphin M, Boizet-Bonhoure B, Descombes P, et al. Dev Biol. 2005;287:361–377. doi: 10.1016/j.ydbio.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 37.Eswarakumar VP, Monsonego-Ornan E, Pines M, Antonopoulou I, Morriss-Kay GM, Lonai P. Development (Cambridge, UK) 2002;129:3783–3793. doi: 10.1242/dev.129.16.3783. [DOI] [PubMed] [Google Scholar]
- 38.Revest JM, Spencer-Dene B, Kerr K, De Moerlooze L, Rosewell I, Dickson C. Dev Biol. 2001;231:47–62. doi: 10.1006/dbio.2000.0144. [DOI] [PubMed] [Google Scholar]
- 39.De Moerlooze L, Spencer-Dene B, Revest J, Hajihosseini M, Rosewell I, Dickson C. Development (Cambridge, UK) 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- 40.Stark K, Vainio S, Vassileva G, McMahon AP. Nature. 1994;372:679–683. doi: 10.1038/372679a0. [DOI] [PubMed] [Google Scholar]