Multicellular organisms face a large variety of different pathogens that can infect them. To identify and combat these potential invaders efficiently, the adaptive immune system has evolved. Its main components are T and B lymphocytes that constantly screen the body in search of unknown, and hence foreign, structures, an extremely challenging task because these structures (antigens) are unknown to the immune system and can be of diverse shape and chemical nature. Antigen recognition is accomplished by the T and B cell antigen receptors (TCRs and BCRs) that contain variable regions designed for antigen binding. The variability is generated by somatic assembly and mutations of the corresponding genes, so that each lymphocyte expresses a different receptor of random specificity. Autoreactive cells are eliminated by a selection process during the development of lymphocytes. Once the receptor on mature T or B cells has recognized (i.e., bound) an antigen with sufficient affinity, the cell is activated and can trigger an immune response against this antigen. Each single receptor has to recognize a large number of different antigens because the number of T and B cells is not sufficient to cover each potential antigen [e.g., the TCRs have to cope with >1013 possible antigens (1)]. Thus, extensive cross-reactivity is an essential characteristic of these receptors (1). Still, the TCRs exhibit a remarkable degree of specificity, capable of distinguishing minor structural differences among their ligands. These features have attracted the attention of immunologists and biophysicists.
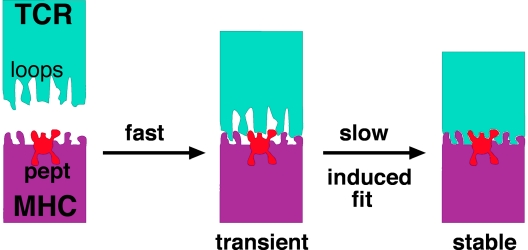
The TCR recognizes, in most cases, small peptides that are presented by MHC molecules residing on the surface of cells of its own body. MHC molecules possess a cleft in which the peptide is bound (pMHC). More than two dozen 3D structures of different TCRs and pMHCs, either individually or as TCR–pMHC complexes, have been solved (2). These structures have shown that the TCR's variable loops, which connect the β-strands of the Ig fold, undergo significant conformational changes upon binding their pMHC ligands (3, 4). In contrast, pMHC only shows very minor structural changes upon TCR binding (Fig. 1).
Fig. 1.
At least two kinetically distinct steps constitute TCR–pMHC interactions. The first step is fast, close to a diffusion-controlled association of two soluble proteins in solution. The second step is slower (kon ≈3 s−1) and reflects conformational transitions in the TCR. This induced-fit reaction determines the stability of the TCR–pMHC complex and, therefore, the outcome of the TCR stimulation signal.
In this issue of PNAS, a team of scientists (5), led by Israel Pecht, describe the energetics and kinetics of a TCR–pMHC interaction. They used a human TCR (TCRCMV) specific for a peptide derived from the human CMV presented by class I MHC. To obtain the required amounts of these proteins, soluble versions comprising only the ecto domains were expressed in bacteria and refolded under standard conditions. Surface plasmon resonance (SPR) yielded a low-affinity constant of Kd ≈8 μM, which is in the range of other TCR–pMHC interactions (6). Measurements of the temperature dependence of the TCRCMV–pMHC interaction yielded a binding enthalpy of ΔH of −3 kcal/mol and a decrease in entropy (ordered state) of TΔS of +4 kcal/mol. Thus, the TCRCMV–pMHC interaction is favored by enthalpic and entropic forces. The enthalpic contribution suggests an increase in the number of noncovalent bonds upon binding, which is consistent with the generally large contact area (≈200 Å2) between TCRs and pMHCs (2, 3). The decrease in entropy is probably caused by expulsion of bound water molecules upon complex formation that overcompensates the reduction in the conformational flexibility of the variable loops upon pMHC binding. Surprisingly, the same ΔH and TΔS values were obtained by studying a different TCR–pMHC pair (7). In contrast, in two other TCR–pMHC interactions analyzed in detail, the binding enthalpy was surprisingly high (ΔH = −23 kcal/mol) and counterbalanced by a reduction in the entropy (TΔS = −16 kcal/mol) (8, 9). The increase of the ordered state suggested that the variable loops possess conformational flexibility in the free TCR that is lost upon pMHC binding. Thus, different TCR–pMHC interactions use different enthalpic and entropic contributions to reach a similar affinity that might be necessary for activation of the TCR and, hence, the T cell.
SPR measurements have suggested that the observed low affinity was caused by a slow association rate and a fast dissociation rate. However, this method's limited time resolution did not enable the resolving of the reaction's elementary steps. Indeed, the association rates were slower than the expected diffusion-controlled reactions, already indicating that the individual steps of the association were more complicated than shown by SPR, suggesting that structural changes are also involved. Thus, the dynamics of the binding process have not been directly measured so far.
In contrast, fast kinetic measurements of antibody–antigen interactions (antibodies are the soluble form of the BCR) had been extensively performed in seminal studies by Israel Pecht's group (10, 11). These studies provided consistent kinetic evidence that conformational changes are taking place in the variable loops of the antibodies induced by antigen binding.
In this context, it is not surprising that the first report on fast kinetic measurements of TCR–pMHC interactions is published by Gakamsky et al. (5). To measure the TCR–ligand interaction by FRET, two different fluorophores were covalently attached at defined positions on the TCRCMV and MHC molecules, respectively. The reaction's time course was monitored by using the stopped-flow technique, which provides a time resolution in the milliseconds range (as opposed to SPR, which operates in the many seconds range). Interestingly, a biphasic association time course was observed in the millisecond time domain. Fitting of the binding data unequivocally supported a dynamic reaction mechanism with at least two distinct reaction phases (Fig. 1).
The first step had a calculated fast rate constant (kon ≈106 mol−1·s−1) that is close to the diffusion-controlled limit for the binding of two macromolecules in solution. The rate constant for the second step was slow (kon ≈3 s−1), clearly indicating the operation of an assumed conformational change (Fig. 1). Although this study did not distinguish between changes at the TCR or pMHC, the many cases of reported crystallographic structures strongly support that it is the TCR that undergoes an induced fit transition in its variable loops to optimally accommodate the antigen. Importantly, the induced fit type of interaction might explain the known promiscuity of TCR–ligand interactions because the variable loops could adapt differently to distinct antigens. Because these changes at the TCR's variable regions are not transmitted to the constant regions of
TCRs exhibit a remarkable degree of specificity.
the TCR (2), they most likely do not cause the rearrangements of the TCR's quartery structure that lead to transmembrane signaling (12, 13). Nevertheless, the second kinetic step determines the final complex stability and thus the outcome of TCR–ligand binding and T cell activation.
This landmark study by Gakamsky et al. (5) should be a basis for future work to unravel in further detail the interactions between TCRs and their ligands. TCRs always bind pMHC in a similar, relative diagonal orientation (2). One open and interesting controversy is whether it is a germ-line-encoded structure of all TCRs that predisposes the diagonal MHC binding or whether it is a result of positive and negative selection (14). Very recently, structural evidence has suggested that TCRs possess germ-line-encoded structures in their variable regions that recognize MHC molecules independent of the bound peptide (15). These germ-line-encoded TCR–MHC interactions would allow a fast screening of many different pMHCs presented on the target cell by a TCR (16) and might be reflected by the first fast association of the TCRCMV with MHC, as observed by Gakamsky et al. Systematic mutations of the TCR and pMHC combined with structural and kinetic measurements could resolve this issue. One implication is that the TCR and MHC coevolved to recognize each other. In contrast, the BCR, which can potentially recognize any molecular entity, and its ligand could not be directly fitted to each other during evolution (17).
Even more intriguing and important, the study by Gakamsky et al. (5) could provide a quantitative understanding of the difference between agonistic and antagonistic peptides presented by the same MHC. Another challenge for the future would be to measure the TCR–pMHC interactions on the surface of living cells because that is where they are expressed and act in physiological situations.
Footnotes
The authors declare no conflict of interest.
See companion article on page 16639.
References
- 1.Mason D. Immunol Today. 1998;19:395–404. doi: 10.1016/s0167-5699(98)01299-7. [DOI] [PubMed] [Google Scholar]
- 2.Rudolph MG, Stanfield RL, Wilson IA. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 3.Garcia KC, Degano M, Pease LR, Huang M, Peterson PA, Teyton L, Wilson IA. Science. 1998;279:1166–1172. doi: 10.1126/science.279.5354.1166. [DOI] [PubMed] [Google Scholar]
- 4.Reiser JB, Gregoire C, Darnault C, Mosser T, Guimezanes A, Schmitt-Verhulst AM, Fontecilla-Camps JC, Mazza G, Malissen B, Housset D. Immunity. 2002;16:345–354. doi: 10.1016/s1074-7613(02)00288-1. [DOI] [PubMed] [Google Scholar]
- 5.Gakamsky DM, Lewitzki E, Grell E, Saulquin X, Malissen B, Montero-Julian F, Bonneville M, Pecht I. Proc Natl Acad Sci USA. 2007;104:16639–16644. doi: 10.1073/pnas.0707061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis MM, Boniface JJ, Reich Z, Lyons D, Hampl J, Arden B, Chien Y. Annu Rev Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- 7.Ely LK, Beddoe T, Clements CS, Matthews JM, Purcell AW, Kjer-Nielsen L, McCluskey J, Rossjohn J. Proc Natl Acad Sci USA. 2006;103:6641–6646. doi: 10.1073/pnas.0600743103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boniface JJ, Reich Z, Lyons DS, Davis MM. Proc Natl Acad Sci USA. 1999;96:11446–11451. doi: 10.1073/pnas.96.20.11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willcox BE, Gao GF, Wyer JR, Ladbury JE, Bell JI, Jakobsen BK, van der Merwe PA. Immunity. 1999;10:357–365. doi: 10.1016/s1074-7613(00)80035-7. [DOI] [PubMed] [Google Scholar]
- 10.Lancet D, Pecht I. Proc Natl Acad Sci USA. 1976;73:3549–3553. doi: 10.1073/pnas.73.10.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zidovetzki R, Blatt Y, Pecht I. Biochemistry. 1981;20:5011–5018. doi: 10.1021/bi00520a030. [DOI] [PubMed] [Google Scholar]
- 12.Gil D, Schamel WW, Montoya M, Sanchez-Madrid F, Alarcon B. Cell. 2002;109:901–912. doi: 10.1016/s0092-8674(02)00799-7. [DOI] [PubMed] [Google Scholar]
- 13.Minguet S, Swamy M, Alarcon B, Luescher IF, Schamel WW. Immunity. 2007;26:43–54. doi: 10.1016/j.immuni.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 14.Housset D, Malissen B. Trends Immunol. 2003;24:429–437. doi: 10.1016/s1471-4906(03)00180-7. [DOI] [PubMed] [Google Scholar]
- 15.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Nat Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 16.Wu LC, Tuot DS, Lyons DS, Garcia KC, Davis MM. Nature. 2002;418:552–556. doi: 10.1038/nature00920. [DOI] [PubMed] [Google Scholar]
- 17.Reth M, Wienands J, Schamel WW. Immunol Rev. 2000;176:10–18. doi: 10.1034/j.1600-065x.2000.00610.x. [DOI] [PubMed] [Google Scholar]