Abstract
We have applied a comparative phylogenomic analysis to study the evolutionary relationships between GC content, CpG-dinucleotide content (CpGs), potential nuclear factor I (NFI) binding sites, and potential Z-DNA forming regions (ZDRs) as representative structural and functional GC-rich genomic elements. Our analysis indicates that CpG and NFI sites emerged with a general accretion of GC-rich sequences downstream of the eukaryotic transcription start site (TSS). Two distinct classes of ZDRs are observed at different locations proximal to the eukaryotic TSS. A robust CA/TG class of ZDRs was seen to emerge upstream of the TSS and independently of GC content, CpGs, and NFIs, whereas a second, weaker CG type appears to have evolved along with these downstream GC-rich elements. Taken together, the results provide a model for how GC-rich structural and functional eukaryotic markers emerge relative to each other, and indicate two distinct transition points for their occurrence: the first at the pro/eukaryotic boundary, and the second at or near the amniotic boundary.
Keywords: evolution, genomic analysis, Z-DNA
GC-rich regions of genomic DNA sequences are located at or near eukaryotic genes, serving as structural and functional “punctuation marks” for transcription (1). Analysis of the prevalence and locations of GC-rich elements across a large number of prokaryotic and eukaryotic genomes allows us to now trace their initial emergence and continued evolution in the eukaryotic genome and decipher the phylogenomic relationships between various transcription-related elements.
The GC content of a genome varies locally and regionally (1, 2). Enrichment of GC-rich regions has been implicated in mutational bias, gene conversion bias, increased thermostability of the DNA duplex in thermophilic prokaryotes and warm-blooded eukaryotes, and structural plasticity associated with transcription (3–7). High GC content has also been correlated with short introns and elevated levels of gene transcription and recombination, whereas low GC content has been correlated with, among other things, tissue specificity and chromatin condensation (7–11). Increased GC content of sequences at and around the transcriptional start sites (TSSs) of genes suggests a functional relevance for GC-rich elements in higher eukaryotes (1). Indeed, GC-rich mammalian genes exhibit up to 100-fold greater transcription rates than orthologous GC-poor genes (12). Variations in GC content distribution may be a general property of the genes or may be associated with the emergence of GC-rich structural and functional transcriptional elements that contribute to the increased GC content. For example, CpG islands, defined as regions with GC content >50% and observed ratios of CpG dinucleotides ≥60% (13), have been shown to accumulate coincidentally with GC enrichment at the TSS of human genes and are used to predict genes in higher eukaryotes (14, 15). Other examples of GC-rich functional and structural elements include the CAAT-box sequence recognized by the nuclear factor I (NFI) transcription factor (16) and CG-rich alternating pyrimidine-purine sequence regions with the potential to form left-handed Z-DNA (ZDRs) (17). Z-DNA has been implicated in several biological functions (17), including gene activation (18) and chromatin remodeling (19), and large-scale deletions in mammalian cells (20). Distributions of both NFI binding sites and ZDRs are correlated with the distribution of known and predicted genes across human chromosome 22 (21), accumulating around the TSS of human genes in a manner generally similar to those of GC content and CpG islands (21, 22). Here, we survey the patterns of occurrence of four GC-rich elements (GC content, potential CpG islands as reflected in the CpG-dinucleotide content, potential NFI bindings sites, and ZDRs) across a broad representation of genomic sequences to establish their phylogenomic relationships. Because CpG islands are not expected in prokaryotes, we do not directly count their occurrence, per se, but analyze for potential CpG islands by monitoring the percent of CpG dinucleotides across sequences. The patterns of distribution observed for these functional and structural elements result in models for their emergence through divergence from a common GC-rich element and/or convergence of disparate elements.
Results
In the current study, we survey the annotated genomes of nine eukaryotic and seven prokaryotic representative organisms. The genomes were placed in approximate order of increasing evolutionary complexity (Fig. 1A): cyanobacteria (generally recognized as one of the most primitive prokaryotic organisms; ref. 23), other eubacteria, archaea, simple eukaryotes, and higher eukaryotes (including invertebrates and vertebrates, with chicken and mammals representing the amniotic organisms).
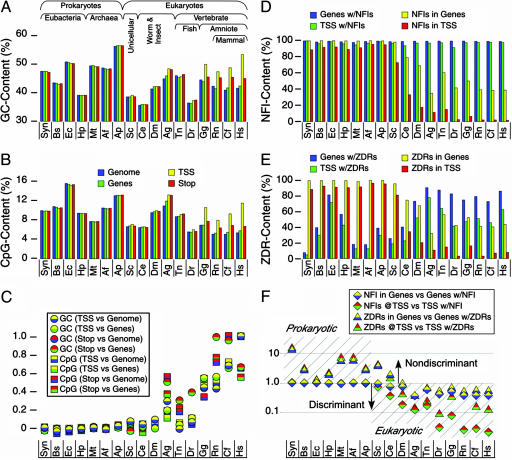
Fig. 1.
Occurrence of GC-rich elements across organisms. (A and B) The abundance of GC-rich and CpG sites are shown for the genomes (blue) and genes (green) and at the transcription start (TSS, yellow) and termination (stop, red) sites of genes across prokaryotic and eukaryotic organisms. The TSS and stop sites are defined as the 40-bp bin that is centered at the respective gene marker. Gene sequences include the ORFs as well as the promoter and termination sequences both upstream of the TSS and downstream of the termination sequence, respectively. Representative genomes are arranged by approximate increasing complexity in the following order: cyanobacterium Synechocystis sp. (Syn); eubacteria Bacillus subtilis (Bs), Escherichia coli (Ec), and Helicobacter pylori (Hp); archaea Methanothermobacter thermoautotrophicus (Mt), Archaeoglobus fulgidus (Af), Aeropyrum pernix (Ap); unicellular eukaryote Saccharomyces cerevisiae (Sc); the invertebrate worm Caenorhabditis elegans (Ce); fruit fly Drosophila melanogaster (Dm); mosquito Anopheles gambiae (Ag); fugu fish Tetraodon nigroviridis (Tn), and zebrafish Danio rerio (Dr); chicken Gallus gallus (Gg); the mammals rat Rattus norvegicus (Rn), dog Canis familiaris (Cf) and human Homo sapiens (Hs). (C) Analysis of GC content (circles) and CpG content (squares) at the TSS and stop sites relative to those of the genome and in genes. The two colors in each point represent the ratio of content in the TSS versus the overall genome (yellow over blue), TSS versus genes (yellow over green), the stop site versus genome (red over blue), or stop versus genes (red over green). These ratios increase with increasing organismal complexity, indicating a general accumulation of these global elements at the beginning and end of genes. (D and E) The contents of NFI and ZDR sites were analyzed as the percent of genes (blue) and TSS (green) with these elements, and as the percent of NFI and ZDR sites within genes (yellow) and within TSS (red) sequences. (F) The ratio of NFI (diamonds) and ZDRs (triangles) in all genes (yellow) versus genes with either NFI or ZDR (blue) and at the TSS of all genes (red) versus TSS of genes with either NFI or ZDR (green) drops one order of magnitude with increasing complexity, reflecting an increasing discrimination in the location of these functional and structural elements. A ratio ≥1.0 was taken as evidence for no discrimination in localization of each element within genes and at the TSS of genes, but a ratio <1.0 indicates discrimination in localization of the elements.
We initially compared the global GC and CpG content across the genomes, within genes, and at the TSS and termination (Stop) sites of genes (Fig. 1 A and B). This analysis shows that eukaryotic genomes are generally GC-poor (significantly less than 50%), as expected, whereas prokaryote genomes vary ≈50% (3). Neither overall GC nor CpG content across genomes shows any particular pattern that correlates to the order of organismal complexity. Compared with genomic and genetic levels, however, one distinguishing feature is the significant enrichment of both GC content and CpG dinucleotides at the TSS and, to a lesser extent, at the stop sites, starting at the amniotic boundary between fish and birds. This enrichment is already evident in early eukaryotes (Fig. 1C), although it is not as striking.
This boundary becomes better defined from the analyses of the functional NFI and structural ZDR elements (Fig. 1 D–F). Global distributions were analyzed for their general occurrences in genes (as opposed to intergenic regions) and at the TSS, whereas more specific distributions were determined from the number of genes and TSS that include these elements. Nearly all genes and TSS sequences across the genomes have at least one potential NFI binding sequence. However, the total numbers of overall NFI sites within genes and at the TSS decrease significantly starting at S. cerevisiae, and continue to drop precipitously with increased eukaryotic complexity. ZDR content shows parallel decreases within genes and TSS sequences, but a gradual increase is observed in the number of genes and TSS with ZDRs, consistent with the developing functionality of this structural element. Unlike the more general GC and CpG content that shows increasing accumulation at the TSS relative to their distributions in genes and across genomes (Fig. 1C), NFIs and ZDRs actually appear to become discriminated against in genes and at the TSS. Because NFI binding has no known function in prokaryotes, the sequences recognized by this eukaryotic activator should occur randomly (i.e., there would be no discrimination as to when and where NFIs might occur in prokaryotes). High ratios of NFIs in genes vs. genes with NFIs (Fig. 1F) indicate that there are many potential binding sites among the genes that have such sites and thus an indiscriminant distribution of such sites across the gene. Low ratios, on the other hand, reflect a more discriminant localization with fewer sites within genes. For prokaryotes, this high ratio indicates very little discrimination for these potential binding sites in their genes. In eukaryotes, this ratio is reduced, reflecting an increase in discrimination for NFIs as a functional element within each gene. The same trends are observed for ZDRs in genes and for NFIs and ZDRs at the TSS of genes, suggesting that, as these elements assume specific functions, their localization along the genome becomes more explicit. The global analyses of these GC-rich elements indicate a distinct phylogenomic boundary at the lower eukaryotes (yeast and worms).
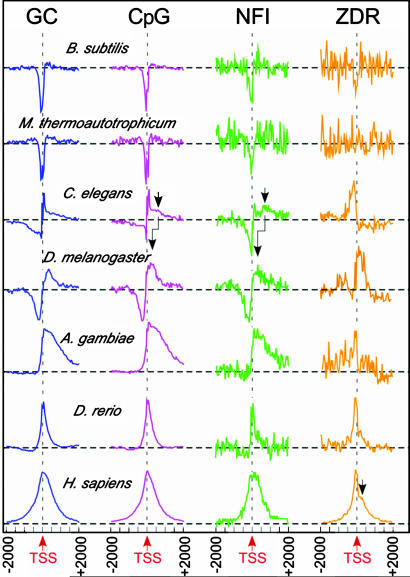
Detailed analyses of the distribution profiles of GC content, CpGs, and NFIs around the TSS (Fig. 2) suggest that these eukaryotic elements are closely related. The distributions show sharp dips immediately upstream of the TSS in eubacteria and archaea, which can be attributed to the localization of AT-rich promoters upstream of prokaryotic genes. In lower eukaryotes (C. elegans), the distributions show a broad negative peak upstream of the TSS, followed by a sharp spike immediately 5′ of the TSS and a weak positive shoulder further downstream of the TSS. This general pattern is sustained, but broadened in D. melanogaster. Interestingly, this negative–positive pattern around the TSS mirrors that for H3 localization in D. melanogaster genes (24). In A. gambiae, the upstream negative distribution is lost, but the downstream distribution becomes a broad, highly asymmetric positive peak. In vertebrates (D. rerio and H. sapiens), the positive distribution becomes sharper, more symmetric, and centered at the TSS. Thus, distribution of GC content in eukaryotic genomes starts as asymmetric (being both positive and negative in lower eukaryotes) and becomes more symmetric and positive with increasing organismal complexity. The broad positive downstream shoulder seen in C. elegans becomes more distinct as a separate peak in the CpG and NFI distributions. This pattern suggests that the highly asymmetric positive downstream distributions in A. gambiae result from either migration toward or enhancement of this broad downstream peak around the TSS. A similar analysis of these distributions around termination sites showed no regular patterns across the phyla (data not shown).
Fig. 2.
Distribution of GC-rich elements around the TSS of genes. The distributions (normalized for peak height) of GC content (GC) and CpG, NFI, and ZDR sites are plotted from −2,000 bp upstream to +2,000 bp downstream of the TSS for a set of representative genomes. Dashed horizontal lines indicate the average number of each element seen in the genomes. Arrows indicate positions of secondary (weaker) shoulders or peaks that are identified in these distributions.
The phylogenomic pattern of ZDR distributions shows a weak suppression just upstream of the TSS of eubacteria, and no discernible pattern in archaea. A sharp positive peak is seen to emerge upstream of the TSS in C. elegans. A second, broader positive distribution is seen to emerge downstream of the TSS, weakly in D. rerio, but more distinctly in H. sapiens. Thus, the emergence of ZDRs appears to be distinct from that of GC content, CpGs, and NFIs.
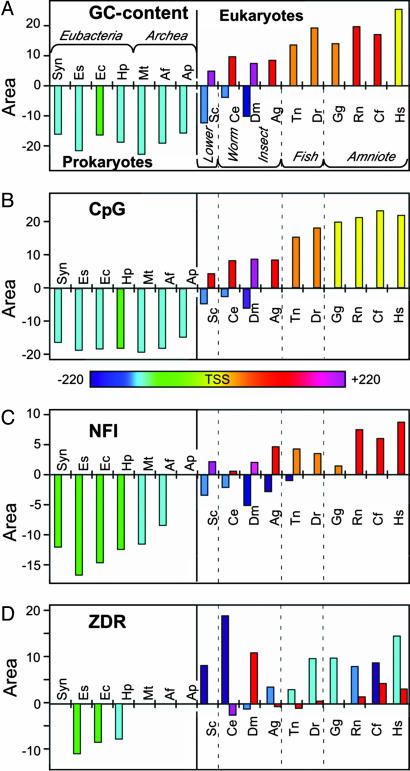
The distributions in Fig. 2 were quantified by first calculating their first derivative, which allowed us to identify the centers and the boundaries of all peaks in each distribution [supporting information (SI) Fig. 5]. The intensity of each peak was calculated by summing the data between the peak boundaries. This analysis provides an overall picture across all organisms in the study of how each GC-rich element emerges and positions itself relative to the TSS. The patterns for GC content and CpGs are similar, both showing a negative peak 5′ of the TSS in prokaryotes that evolutionarily migrates further upstream in lower eukaryotes (Fig. 3 A and B). Within eukaryotes, the peaks are positive and start downstream of the TSS, but migrate toward the TSS with increasing organismal complexity. The CpGs, however, are centered at the TSS for all amniotic organisms (recapitulating the amniotic boundary seen in Fig. 1), whereas GC-content distributions are centered at the TSS only in H. sapiens, S. cerevisae, and C. elegans show both upstream suppression and downstream accumulation of GC content, indicative of the lower eukaryotes serving as a transitional boundary with features of both kingdoms. The NFI pattern displays features similar to GC content and CpGs, but with the pro/eukaryotic boundary extended into insects.
Fig. 3.
Phylogenomic patterns of enrichment or suppression of GC-rich transcriptional elements. The intensity of GC content (A), CpG (B), NFI (C), and ZDR (D) distributions are shown for various representative genomes. The positions of the centers of each distribution are shown relative to the TSS (red arrow in Fig. 2) of the genes in the genomes, with those centered increasingly upstream shown as green to blue to violet and those increasingly downstream shown as orange to red to magenta (the color scale for positions of centers is shown between the panels).
The phylogenomic pattern of ZDR distributions is weakly suppressed immediately upstream of the TSS of eubacterial genes, but shows no discernible pattern in archaea. A sharp positive peak is seen to emerge upstream of the TSS in lower eukaryotes and remains ≈100–200 bp upstream of the TSS in all eukaryotes. This class of strong, upstream Z-DNA elements (ZDR1) clearly arose independently of the other GC-rich elements. A second, weaker distribution of ZDRs (ZDR2) starts as a negative peak far downstream of the TSS in C. elegans, which becomes weakly positive in R. norvegicus and centered closer to the TSS in H. sapiens. Thus, there are apparently two distinct classes of ZDRs that emerge: the first is distinct from and the second appears correlated to GC enrichment and the emergence of CpGs and NFIs across the phylogenomic spectrum.
Discussion
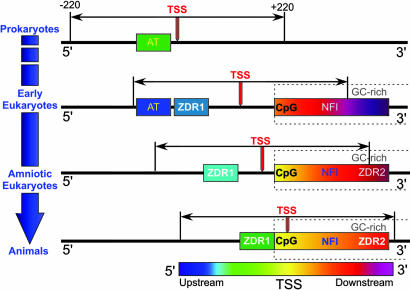
When considered together, the phylogenomic patterns suggest a model for the emergence of GC-rich structural and functional elements for eukaryotic genes. It is clear from these patterns that CpG dinucleotides (and, by extension, CpG islands) and the GC-rich NFI transcriptional activator binding sites both emerged coincidentally with the increasing GC content just 3′ of the eukaryotic TSS. The starting and ending positions for the centers of these distributions suggests that CpG islands evolved with a subset of high GC-content regions that are relatively close to the TSS, and NFI sequences started further downstream of the TSS. All three elements migrate toward the TSS in parallel, as one would expect for elements that become increasingly important for transcriptional regulation, but the migration stops once it reaches the TSS. An attractive alternative model is that, rather than the transcriptional elements migrating relative to the TSS, both TA- and GC-rich elements are relatively fixed across the various organisms, but the TSS migrates evolutionarily in the 3′ direction (Fig. 4). This would be consistent with the dramatic increase in the size of the transcriptosome (the proteins of the transcription machinery) at the pro/eukaryotic boundary (the number of subunits of RNA polymerase triples from E. coli to yeast) and the increased numbers of transcriptional regulatory elements in the higher eukaryotes. The increase in size and complexity of the transcriptosome that accompanies evolutionary complexity would provide a physical rationale for the downstream migration of the TSS away from the primordial TA-rich transcriptional elements.
Fig. 4.
Model for the emergence of GC-rich transcriptional elements and migration of the transcription start site (TSS, red arrow) of genes from prokaryotes to early eukaryotes to amniotic eukaryotes, and, finally, to higher eukaryotes. In this model, the prokaryotic AT-rich promoters and the GC-rich eukaryotic elements are seen to be fixed, whereas the TSS and the analysis window (±220 bp relative to the TSS) migrate in the 3′ direction as the size and complexity of the transcriptosome increases. In prokaryotes, the primary transcriptional control elements are AT-rich upstream promoters, which account for the strong suppression of GC-rich elements that are characteristic of eukaryotes. Early eukaryotes show both the persistence of localized upstream AT-rich promoters that are characteristic of prokaryotes, as well as the accumulation of the eukaryotic GC-rich elements (CpG and NFI elements) as the GC content (dashed boxes) downstream of the TSS increases. The first Z-DNA regions (ZDR1) also emerge at this point, independently of the other GC-rich elements. In progressing toward amniotic eukaryotes, the TSS migrates further in the 3′ direction, followed by the ZDR1 elements. This is in concert with AT-rich promoters becoming less distinct (and, therefore, their locations are not specified in this figure), the emergence of a second class of CG-rich Z-DNAs (ZDR2) accumulated downstream of the TSS, and convergence of the upstream ZDR1 sites with the GC-rich elements.
The results of the phylogenomic analysis suggest that the stronger ZDR1-type structural elements emerged independently of GC and CpG content, even though Z-DNA is characteristic of alternating GC-rich sequences. ZDR1s are most likely alternating CA/TG-type Z-DNA sequences, as opposed to the prototypical alternating GC motif. ZDR1 sequences, however, are not simply repeats of CA/TG, as seen in the repetitive regions of eukaryotic chromosomes, but are similar to the CA/TG-rich sequences characteristic, for example, of the promoters in rat genes (25). The convergence of ZDR1s toward the downstream GC-rich elements such as NFI may reflect the emergence of the more complex mechanism of structural/nuclear factor coactivation, as seen in the human CSF-1 promoter (17).
The lower intensity ZDR2 class follows the general trend of the GC-rich elements, suggesting that these are the prototypical GC-type Z-DNA sequences and they arose perhaps as a consequence of GC content and CpG islands rather than as a distinct element in itself. The emergence of GC-rich isochores has been proposed to be associated with Z-DNA, as well as thermal stability and helix bendability (10). The emergence of two distinct classes of ZDRs may reflect the plurality of functions now recognized for Z-DNA in various genomes (17).
When viewed as a whole, the phylogenomic relationships seen here suggest that GC-rich transcriptional elements evolved gradually rather than abruptly across organisms, but with two distinct boundaries. The lower eukaryotes can be perceived as the pro/eukaryotic transition, showing characteristics of both types, consistent with a continuity across this transition. The second interface is at or near the amniotic transition, where the GC content changes from a broad asymmetric to a sharper symmetric distribution, CpG dinucleotides have fully localized at the TSS, and ZDR2-type sequences are enriched rather than suppressed. Thus, these GC-rich elements are a means to decipher phylogenomic relationships at the gene level, even without knowing their specific functions. What remains unclear at this level of analysis is whether patterns of emergence of these punctuation elements are entirely organismal or related to the emergence of specific genes or gene functions in each class of organism.
Materials and Methods
Genome Analyses.
Sequences and annotations of prokaryotic genomes were accessed from the National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov/genomes/lproks.cgi) and eukaryotic genomes from the Ensembl database (www.ensembl.org) (26) as their December 2005 releases. The current analyses include the genomes from 16 organisms (Table 1), representing four eubacteria, three archaea, yeast, worm, mosquito, two fish, chicken, and three mammals (rat, dog, and human). The particular eukaryotic genomes were chosen for analysis because of the consistency in their annotations in the Ensembl database and methylation of their genomes.
Table 1.
Analyses of prokaryotic and eukaryotic genomes for GC-rich transcriptional elements (percentage GC content, percentage CpG dinucleotides, number of NFI binding sites, and number of Z-DNA sequences)
| Genome size, Mbp (no. of chromosomes) | No. of genes | GC content, % | CpG, % | Total no. of NFIs | Total no. of ZDRs | |
|---|---|---|---|---|---|---|
| Prokaryotes (Complete Microbial Genomes, www.ncbi.nlm.nih.gov/genomes/lproks.cgi) | ||||||
| Eubacteria | ||||||
| Synechocystis sp. PCC6083 (Syn) | 3.57 | 3,218 | 47.72 | 10.05 | 15,982 | 118 |
| B. subtilis subsp. subtilis str.168 (Bs) | 4.21 | 4,226 | 43.52 | 10.92 | 7,827 | 2,143 |
| E. coli K12 (Ec) | 4.64 | 4,915 | 50.79 | 15.75 | 12,472 | 10,424 |
| H. pylori J99 (Hp) | 1.64 | 1,496 | 39.19 | 9.54 | 3,077 | 1,282 |
| Archaea | ||||||
| M. thermoautotrophicus Δ H (Mt) | 1.75 | 1,921 | 49.54 | 7.80 | 2,791 | 206 |
| A. fulgidus DSM 4304 (Af) | 2.18 | 2,487 | 48.58 | 10.60 | 4,019 | 292 |
| A. pernix K1 (Ap) | 1.67 | 1,894 | 56.53 | 13.17 | 4,154 | 603 |
| Eukaryotes (Ensembl v36-Dec 2005, www.ensembl.org/index.html) | ||||||
| S. cerevisiae (Sc) | 12 (16) | 6,652† | 38.42 | 6.73 | 20,729 | 1902 |
| C. elegans (Ce) | 100 (6) | 19,723† | 35.47 | 6.48 | 165,940 | 39,734 |
| D. melanogaster (Dm)‡ | 118 (6) | 13,733§ | 41.30 | 9.47 | 165,940 | 148,116 |
| A. gambiae (Ag) | 278 (5) | 12,500§ | 44.73 | 11.06 | 388,446 | 693,596 |
| T. nigroviridis (Tn) | 402 (21) | 15,357† | 45.99 | 8.81 | 363,806 | 475,219 |
| D. rerio (Dr) | 1,688 (25) | 18,009§ | 36.42 | 5.63 | 1,715,635 | 1,285,246 |
| G. gallus (Gg) | 1,054 (30) | 15,348§ | 44.47 | 7.02 | 1,644,258 | 231,151 |
| R. norvegicus (Rn) | 2,719 (21) | 21,939§ | 42.25 | 5.13 | 4,468,986 | 2,526,023 |
| C. familiaris (Cf) | 2,385 (39) | 17,861§ | 40.90 | 5.14 | 4,555,797 | 1,061,843 |
| H. sapiens (Hs) | 3,272 (24¶) | 20,121† | 41.53 | 5.44 | 5,693,028 | 1,065,255 |
†Number of annotated known RNA polymerase II (Pol II) transcribed genes.
‡Ensembl v42-Dec 2006.
§Number of annotated known and novel RNA Pol II transcribed genes.
¶Includes both the X and Y chromosomes.
GC contents were calculated as the percent of G+C within 40-bp bins. Transcription start and stop sites for eukaryotic genes were as annotated in the Ensembl database (according to experimental transcripts). The near-identical distributions of GC content around the TSS and stop sites for human genes seen here, and as reported by Zhang et al. (1), indicate that the annotations for these transcriptional markers are consistent with previous analyses. Potential CpG islands (reflected in the CpG content) were analyzed similarly. The accepted definition for CpG islands (13) are long stretches of sequence (≥200 bp) with observed CpG occurrence versus expected occurrence (1/16 = 12.5%) ≥0.6; therefore, an actual CpG island would be five or more contiguous 40-bp bins having CpG contents ≥0.6, as calculated here. Because no CpG islands are expected in prokaryotic genomes, we calculated the potential for this element for all genomes (prokaryotic and eukaryotic) as the percent of CpG dinucleotides within a 40-bp window, rather than actual CpG islands, per se. NFI binding sites were analyzed according to affinity constants reported by Roulet et al. (16), with a binding score of 65 (of a possible 100) representing a nonrandom and potentially functional binding site (enhancing gene expression in vitro) as the criteria for identifying an NFI site for this study, as described in ref. 21. With the threshold set at a lower binding score, the distributions around the TSSs approach the distributions of random sequences (i.e., become flat), whereas, at higher scores, the distributions become noisier (because of smaller numbers), but retain the general shapes reported here. ZDRs were defined as contiguous stretches of DNA with a PZ score ≥500 bp, as defined in ref. 21, using the program Z-hunt (27, 28).
Quantitative Analysis of Distributions.
The occurrence of each class of CpG, NFI, and ZDR elements were counted in 40-bp bins starting from −2 kb to +2 kb relative to the TSS of identified genes. Note that, for prokaryotic and lower eukaryotic genomes, this 4-kb window for analysis encompasses primarily coding sequences, whereas, for higher eukaryotes, it would include mostly noncoding sequences of introns, the 5′-untranslated regions, and intergenic DNA. In addition, for the compact prokaryotic and yeast genomes, there would be significant overlapping of transcriptional start and stop sites from multiple genes within this analysis window. However, these overlapping sites occur randomly across the windows, and any suppression or accumulation of elements is expected to contribute to the overall background levels within the windows and not contribute significantly to the patterns of positive and negative spikes identified at the reference TSS for the analysis of a particular genome. Indeed, the features observed for these compact genomes distribute over a very narrow range (≤200 bp). The magnitudes of each element within the 40-bp bins are normalized as the number of standard deviations from the mean of the average count for that organism, allowing us to directly compare the uniqueness of the GC-rich elements across all organisms regardless of the background levels. The center and boundaries of each peak in the normalized distributions were determined by calculating the first derivative of the distribution (SI Fig. 5). Centers were defined as points where the first derivative crosses zero, and boundaries were defined as the positive and negative peaks of the derivative. Although these boundaries do not account for the entire area within each peak, they provide a consistent definition that is independent of shifting baselines.
Supplementary Material
Acknowledgments
We thank Drs. C. K. Mathews, G. Merrill, and M. Freitag at Oregon State University for reviewing the manuscript and providing comments before submission. This work was funded by National Institutes of Health Grant R01GM62957A and the Medical Research Foundation of Oregon.
Abbreviations
- CpG
cytosine-phosphate-guanine dinucleotide
- NFI
potential nuclear factor I binding site
- TSS
transcriptional start site
- ZDR
potential Z-DNA regions.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707203104/DC1.
References
- 1.Zhang L, Kasif S, Cantor CR, Broude NE. Proc Natl Acad Sci USA. 2004;101:16855–16860. doi: 10.1073/pnas.0407821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eyre-Walker A, Hurst LD. Nat Rev Genet. 2001;2:549–555. doi: 10.1038/35080577. [DOI] [PubMed] [Google Scholar]
- 3.Basak S, Ghosh TC. Biochem Biophys Res Commun. 2005;330:629–632. doi: 10.1016/j.bbrc.2005.02.134. [DOI] [PubMed] [Google Scholar]
- 4.Bernardi G. Gene. 2000;241:3–17. doi: 10.1016/s0378-1119(99)00485-0. [DOI] [PubMed] [Google Scholar]
- 5.Hurst LD, Williams EJ. Gene. 2000;261:107–114. doi: 10.1016/s0378-1119(00)00489-3. [DOI] [PubMed] [Google Scholar]
- 6.Galtier N. Trends Genet. 2003;19:65–68. doi: 10.1016/s0168-9525(02)00002-1. [DOI] [PubMed] [Google Scholar]
- 7.Vinogradov AE. Nucleic Acids Res. 2003;31:5212–5220. doi: 10.1093/nar/gkg699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montoya-Burgos JI, Boursot P, Galtier N. Trends Genet. 2003;19:128–130. doi: 10.1016/S0168-9525(03)00021-0. [DOI] [PubMed] [Google Scholar]
- 9.Versteeg R, van Schaik BD, van Batenburg MF, Roos M, Monajemi R, Caron H, Bussemaker HJ, van Kampen AH. Genome Res. 2003;13:1998–2004. doi: 10.1101/gr.1649303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vinogradov AE. Nucleic Acids Res. 2003;31:1838–1844. doi: 10.1093/nar/gkg296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vinogradov AE. Trends Genet. 2005;21:639–643. doi: 10.1016/j.tig.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Kudla G, Lipinski L, Caffin F, Helwak A, Zylicz M. PLoS Biol. 2006;4:e180. doi: 10.1371/journal.pbio.0040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardiner-Garden M, Frommer M. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 14.Bird A. Trends Genet. 1987;3:342–348. [Google Scholar]
- 15.Antequera F. Cell Mol Life Sci. 2003;60:1647–1658. doi: 10.1007/s00018-003-3088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roulet E, Bucher P, Schneider R, Wingender E, Dusserre Y, Werner T, Mermod N. J Mol Biol. 2000;297:833–848. doi: 10.1006/jmbi.2000.3614. [DOI] [PubMed] [Google Scholar]
- 17.Rich A, Zhang S. Nat Rev Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 18.Liu R, Liu H, Chen X, Kirby M, Brown PO, Zhao K. Cell. 2001;106:309–318. doi: 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- 19.Liu H, Mulholland N, Fu H, Zhao K. Mol Cell Biol. 2006;26:2550–2559. doi: 10.1128/MCB.26.7.2550-2559.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang G, Christensen LA, Vasquez KM. Proc Natl Acad Sci USA. 2006;103:2677–2682. doi: 10.1073/pnas.0511084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Champ PC, Maurice S, Vargason JM, Camp T, Ho PS. Nucleic Acids Res. 2004;32:6501–6510. doi: 10.1093/nar/gkh988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schroth GP, Chou PJ, Ho PS. J Biol Chem. 1992;267:11846–11855. [PubMed] [Google Scholar]
- 23.Taylor TE. The Biology and Evolution of Fossil Plants. Englewood Cliffs, NJ: Prentice Hall; 1993. [Google Scholar]
- 24.Mito Y, Henikoff JG, Henikoff S. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 25.Rothenburg S, Koch-Nolte F, Rich A, Haag F. Proc Natl Acad Sci USA. 2001;98:8985–8990. doi: 10.1073/pnas.121176998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Birney E, Andrews D, Caccamo M, Chen Y, Clarke L, Coates G, Cox T, Cunningham F, Curwen V, Cutts T, et al. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schroth GP, Chou PJ, Ho PS. J Biol Chem. 1992;267:11846–11855. [PubMed] [Google Scholar]
- 28.Ho PS, Ellison MJ, Quigley GJ, Rich A. EMBO J. 1986;5:2737–2744. doi: 10.1002/j.1460-2075.1986.tb04558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.