Abstract
Schizophrenia is a complex multifactorial brain disorder with a genetic component. Convergent evidence has implicated oxidative stress and glutathione (GSH) deficits in the pathogenesis of this disease. The aim of the present study was to test whether schizophrenia is associated with a deficit of GSH synthesis. Cultured skin fibroblasts from schizophrenia patients and control subjects were challenged with oxidative stress, and parameters of the rate-limiting enzyme for the GSH synthesis, the glutamate cysteine ligase (GCL), were measured. Stressed cells of patients had a 26% (P = 0.002) decreased GCL activity as compared with controls. This reduction correlated with a 29% (P < 0.001) decreased protein expression of the catalytic GCL subunit (GCLC). Genetic analysis of a trinucleotide repeat (TNR) polymorphism in the GCLC gene showed a significant association with schizophrenia in two independent case-control studies. The most common TNR genotype 7/7 was more frequent in controls [odds ratio (OR) = 0.6, P = 0.003], whereas the rarest TNR genotype 8/8 was three times more frequent in patients (OR = 3.0, P = 0.007). Moreover, subjects with disease-associated genotypes had lower GCLC protein expression (P = 0.017), GCL activity (P = 0.037), and GSH contents (P = 0.004) than subjects with genotypes that were more frequent in controls. Taken together, the study provides genetic and functional evidence that an impaired capacity to synthesize GSH under conditions of oxidative stress is a vulnerability factor for schizophrenia.
Keywords: genetic association, glutamate cysteine ligase, oxidative stress, GAG trinucleotide repeat polymorphism, skin fibroblasts
Schizophrenia is a multifactorial disease with a strong heritable component. Although schizophrenia has begun to be studied on the level of molecular genetics, knowledge about genetically based functional alterations is sparse (1–4). Recent gene-expression analysis, genetic studies, and quantifications of brain glutathione (GSH) levels in vivo and on postmortem tissues led to the hypothesis that a dysregulation of the GSH metabolism is involved in the pathogenesis of schizophrenia (5–9). GSH levels were reduced by 27% in cerebrospinal fluid and by 52% in medial prefrontal cortex of schizophrenia patients (6). Similarly, GSH levels were decreased by 40% in the caudate region of postmortem-brain tissue from schizophrenia patients, as compared with control subjects (7).
GSH plays a crucial role as a cellular antioxidant scavenger of reactive oxygen species (ROS), and it maintains intracellular redox potential, detoxifies xenobiotics, and protects cells from oxidative stress (10). Several biological and psychological factors can increase oxidative stress in the brain. For example, environmental risk factors of schizophrenia such as viral infections, inflammations, or obstetrical complications are known to increase oxidative stress (11). Psychological stress can increase oxidative stress via the hypothalamic-pituitary-adrenal axis, especially in hormone-sensitive or dopamine-innervated brain regions (12). In rats, for example, stress induced by one hour restrain can decrease GSH brain levels (13). The human brain is metabolically very active and thus particularly sensitive to an impaired capacity to react against oxidative stress (14).
Genetic polymorphisms or mutations that cause a deficit in GSH synthesis have been associated to various pathological processes or disorders, including oxidative stress (15), myocardial infarction (16), hemolytic anemia (17), neurological alterations, or mental retardation (18). Interestingly, an increased risk for cardiovascular morbidity has been described for schizophrenia (19, 20).
Cellular GSH levels are highly regulated (21), and several substances known to produce oxidative stress have been shown to increase GSH synthesis (22). GSH is synthesized in two consecutive enzymatic reactions: the first is catalyzed by the enzyme glutamate cysteine ligase (GCL) and the second by the GSH synthetase (GSS). GCL consists of a catalytic (GCLC) and a modulatory subunit (GCLM) (23). We recently reported a decrease in GCLM and GSS gene expression in cultured skin fibroblasts derived from schizophrenia patients, as compared with controls (8). The same study revealed a genetic association between allelic variants of the GCLM gene and schizophrenia.
The aim of the present study was to test whether schizophrenia is associated with a deficit in GSH synthesis. As a model, we selected fibroblasts that were obtained by skin biopsy. We supposed that a deficit in GSH synthesis is more pronounced under conditions of oxidative stress, and we thus treated cultured fibroblasts with tert-butylhydroquinone (t-BHQ), a substance known to increase the expression of phase II genes (including GCLM and GCLC), to increase GSH synthesis, and to increase GSH content (10, 24). We measured GSH content, GCL activity, as well as protein expression of GCLM and GCLC under baseline (untreated) and t-BHQ- treated conditions. We compared the regulation of GCLM and GCLC protein expression in relation to GCL activity and GSH content in patients and controls.
As GCLC protein expression and GCL activity were reduced in patients, the GCLC gene became our focus of interest. This gene was reported to contain a GAG trinucleotide repeat (TNR) polymorphism in the 5′-untranslated region (25). We compared the genotype distribution of this polymorphism in a Swiss sample of 66 schizophrenia patients and 48 control subjects and a Danish sample of 322 schizophrenia patients and 331 control subjects. Finally, we analyzed the GAG TNR polymorphism for functional relevance on GCLC protein expression, GCL activity, and GSH content.
Results
GCL Activity and GSH Content.
GCL activity and GSH content were quantified in the Swiss sample (Fig. 1A and Table 1). The ability to increase GCL activity after t-BHQ treatment was significantly reduced in fibroblasts of patients (P = 0.001) as compared with controls. GCL activity after t-BHQ treatment increased by a factor of 3.45 in controls, whereas it only increased by a factor of 2.95 in patients. Thus, GCL activity under t-BHQ-treated conditions was 26% (P = 0.002) lower in patients than in controls. The increase in GSH content induced by t-BHQ treatment was very similar in patients and controls and amounted to a factor of 2.53 (Table 1).
Fig. 1.
GCL activity and GCLC protein expression in patients and controls under untreated and t-BHQ-treated conditions. (A) GCL activity of 25 patients and 25 controls, expressed as nanomoles of GSH per minute per milligram of protein. Each box describes 25%, 75%, and median values. The whisker bars show values in the 1.5 box lengths range. The open circle depicts an outlier value. (B) The plot shows GCLC protein levels of 26 patients and 26 controls, determined as α-tubulin corrected arbitrary units. **, P < 0.01; ***, P < 0.001 vs. the respective controls were calculated by using ANOVA test (two-tailed). (C) Representative Western blots of GCLC, GCLM, and α-tubulin of two controls and two patients under untreated and t-BHQ-treated conditions.
Table 1.
GCL activity, GSH contents, GCLM, and GCLC protein expression in patients vs. controls in the Swiss sample
| Variable | N | Untreated |
t-BHQ |
||
|---|---|---|---|---|---|
| Controls | Patients | Controls | Patients | ||
| GCL activity | 25 | 0.250 ± 0.08 | 0.215 ± 0.06 | 0.847 ± 0.26 | 0.627 ± 0.16** |
| GSH content | 25 | 56.7 ± 8.0 | 53.6 ± 6.4 | 144.9 ± 21.5 | 136.3 ± 19.1 |
| GCLM | 26 | 62.3 ± 16.3 | 61.7 ± 11.3 | 108.0 ± 24.0 | 107.6 ± 24.1 |
| GCLC | 26 | 44.4 ± 12.3 | 34.5 ± 10.9** | 64.1 ± 17.5 | 45.4 ± 15.8*** |
N, number of tested subjects. Values are given as average and SD under untreated or t-BHQ-treated conditions. GSH content is expressed as nanomoles of GSH per milligram of protein, and GCL activity is expressed as nanomoles of GSH per minute per milligram of protein. GCLM and GCLC protein expressions are presented as α-tubulin corrected arbitrary units.
**, P < 0.01;
***, P < 0.001 vs. the respective controls were calculated using ANOVA test (two-tailed).
GCLC and GCLM Protein Expression.
Parallel to GCL activity and GSH content, protein levels of GCLM and GCLC were quantified (Fig. 1 B and C and Table 1). GCLC protein expression was lower in patients than in controls by 22% (P = 0.009) under baseline conditions and by 29% (P < 0.001) under conditions of t-BHQ treatment. GCLC protein expression was increased in controls after t-BHQ treatment by a factor of 1.44, but significantly less (P = 0.005) in patients by a factor of 1.32. GCLM protein expression did not differ between patients and controls, and the increase induced by t-BHQ treatment was similar: 73% in control subjects and 74% in patients.
Regulation of GSH Synthesis in Patients and Controls.
GSH content, GCL activity, and GCLC and GCLM protein expression are parameters of the GSH synthesis. To understand interactions among these parameters in response to t-BHQ treatment, we performed more detailed statistical analysis. We assumed that the ratio of the two GCL subunits may reflect the enzymatic activity of the GCL holoenzyme. Using two-way ANOVA, we tested the influence of the three independent variables “disease status” (patient vs. control), “treatment” (with or without t-BHQ), and “ratio of GCLM/GCLC protein expression” on the two dependent variables “GSH content” and “GCL activity.” There was a significant interaction between treatment and disease status (P = 0.029). Moreover, GSH content and GCL activity were influenced by the disease status (P = 0.018), the GCLM/GCLC protein ratio (P = 0.021), and the treatment (P < 0.001).
In a second step, we performed multivariate analysis of variances (MANOVA) for the variables “GCLM protein expression,” “GCLC protein expression,” “GCL activity,” and “GSH content.” The regulation of the GSH synthesis after t-BHQ treatment was different between patients and controls (P = 0.019). Univariate analysis (ANOVA) of the same parameters revealed the following effects: (i) GCLC protein expression (P < 0.001) and GCL activity were differently regulated (P < 0.001) in patients vs. controls; (ii) GCL activity after t-BHQ treatment was influenced (P = 0.025) by the degree of GCLC protein expression present before the treatment; (iii) in patients and controls, the GSH content after t-BHQ treatment was influenced by the baseline protein expression of GCLC (P = 0.032); and (iv) the level of GCLC protein expression after t-BHQ treatment was influenced (P = 0.011) by the level of GCLM protein expression before t-BHQ treatment.
Aside from the decreased GCLC protein expression, we consistently observed in patients a different regulation of GCLM and GCLC protein expression as compared with controls. In control subjects protein expressions of GCLM and GCLC were correlated under untreated and under t-BHQ-treated conditions (Fig. 2A). This correlation was lost in patients (Fig. 2B). The reduced GCLC protein expression in patients under t-BHQ-treated conditions correlated with GCL activity (Fig. 2D). This correlation underlined a limiting effect of the decreased GCLC protein expression on GSH synthesis and on GSH content (R = 0.435; P = 0.034). Such correlations were not observed in controls (Fig. 2C). Finally, analysis of correlations between the ratio of GCLM/GCLC with GCL activity or GSH content under t-BHQ-treated conditions revealed a different regulation between patients and controls. In patients, both GCL activity (Fig. 2F) and GSH content (R = −0.481; P = 0.017) were inversely correlated with the GCLM/GCLC ratio, whereas in controls there was no correlation (Fig. 2E).
Fig. 2.
Different regulation of GSH synthesis in patients and controls. The plots show GCLM and GCLC protein expression under untreated (gray diamonds) and t-BHQ-treated (black triangles) conditions of 26 controls (A) and 26 patients (B). Note that the correlation between GCLM and GCLC is lost in patients. GCLC protein expressions compared with GCL activities under untreated (gray diamonds) and t-BHQ-treated (black triangles) conditions of 24 controls (C) and 24 patients (D). Note that, in patients under oxidative stress, the GCL activity depends on GCLC protein levels. Shown are ratios of GCLM vs. GCLC protein amounts under untreated conditions (gray squares) and after t-BHQ treatment (black triangles) compared with GCL activities of 24 controls (E) and 24 patients (F). Note that, in patients, GCL activity is inversely correlated with the GCLM/GCLC ratio. R is the Spearman correlation; P is the probability of type I error.
GAG TNR Polymorphism in the GCLC Gene: Genetic Analysis.
The GCLC gene contains a TNR polymorphism 10 base pairs upstream of the start codon, with 7, 8, or 9 GAG repeats. We investigated this polymorphism in a Swiss and a Danish sample [Tables 2–4 and supporting information (SI) Table 5]. In the Swiss sample, 50% of the controls had a 7/7 genotype, 44% a 7/9, 4% a 7/8, and 2% an 8/9 (Table 3). Forty-five of 48 controls had no 8 TNR allele and genotypes 8/8 and 9/9 were absent. In contrast, 24 of 66 (36%) schizophrenia patients had a genotype with an 8 TNR allele or had genotype 9/9, resulting in a significant inter-group difference (P = 0.012) regarding the overall genotype distribution.
Table 2.
Demographic characteristics of samples of patients and controls
| Group | Sample | Material | Study | Subjects | N | Age | Sex, m/f | Diagnostic |
|---|---|---|---|---|---|---|---|---|
| A | Swiss | Skin fibroblasts ± | [GSH], GCL activity, | Controls | 26 | 36.6 ± 12.4 | 1.2 | DIGS |
| t-BHQ treatment | protein | Patients | 26 | 36.7 ± 11.1 | 3.3 | DSM-IV, DIGS | ||
| B | Swiss | Blood samples | Genotyping | Controls | 48 | 34.3 ± 11.6 | 1.2 | DIGS |
| Patients | 66 | 33.9 ± 10.7 | 2.8 | DSM-IV, DIGS | ||||
| C | Swiss | Skin fibroblasts | Baseline [GSH] | Controls | 39 | 35.7 ± 10.4 | 1.1 | DIGS |
| Patients | 33 | 35.3 ± 13.2 | 2.5 | DSM-IV, DIGS | ||||
| D | Danish | Blood samples | Genotyping | Controls | 331 | 40.2 ± 10.5 | 1.5 | Random selection |
| Patients | 322 | 38.8 ± 12.1 | 1.5 | ICD-10 |
Groups A and C are both part of group B. Age is presented as mean ± SD; sex as the ratio between male and female. DSM-IV, Diagnostic System Manual IV; DIGS, Diagnostic Interview for Genetic Studies; ICD-10, International Classification for Diseases.
Table 3.
GCLC GAG TNR genotypes in the Swiss sample
| Genotypes | Controls |
Patients |
OR | 95% CI | P value | ||
|---|---|---|---|---|---|---|---|
| N | % | N | % | ||||
| 7/7 | 24 | 50.0 | 20 | 30.3 | 0.44 | 0.20–0.94 | 0.033 |
| 7/8 | 2 | 4.2 | 13 | 19.7 | 5.64 | 1.21–26.32 | 0.015 |
| 7/9 | 21 | 43.7 | 22 | 33.3 | 0.64 | 0.30–1.38 | 0.257 |
| 8/8 | 0 | — | 2 | 3.0 | 0.57 | 0.48–0.67 | 0.333 |
| 8/9 | 1 | 2.1 | 5 | 7.6 | 3.9 | 0.43–34.1 | 0.195 |
| 9/9 | 0 | — | 4 | 6.1 | 0.56 | 0.48–0.66 | 0.108 |
| Controls vs. patients: (24, 2, 21, 0, 1, and 0 vs. 20, 13, 22, 2, 5, and 4)* | 0.012 | ||||||
N, number of genotypes in a group; %, percentage of genotypes in a group. Data are given together with OR and 95% confidence interval (CI). P values were calculated using χ 2 test (two-tailed), or Fisher exact test (two-tailed) if cells had an expected count of <5.
*The frequency of genotypes between patients and controls was compared using Fisher exact test (two-tailed) with a 2 × 6 contingency table (controls and patients × genotypes 7/7, 7/8, 7/9, 8/8, 8/9, and 9/9).
Table 4.
GCLC GAG TNR genotypes in the Danish sample
| Genotypes | Controls |
Patients |
OR | 95% CI | P value | ||
|---|---|---|---|---|---|---|---|
| N | % | N | % | ||||
| 7/7 | 125 | 37.8 | 87 | 27.0 | 0.61 | 0.43–0.85 | 0.003 |
| 7/8 | 65 | 19.6 | 64 | 19.9 | 1.02 | 0.69–1.49 | 0.939 |
| 7/9 | 86 | 26.0 | 103 | 32.0 | 1.34 | 0.95–1.88 | 0.091 |
| 8/8 | 8 | 2.4 | 22 | 6.8 | 2.96 | 1.30–6.75 | 0.007 |
| 8/9 | 27 | 8.2 | 19 | 5.9 | 0.71 | 0.38–1.30 | 0.260 |
| 9/9 | 20 | 6.0 | 27 | 8.4 | 1.42 | 0.78–2.59 | 0.247 |
| Controls vs. patients: (125, 65, 86, 8, 27, and 20 vs. 87, 64, 103, 22, 19, and 27)* | 0.004 | ||||||
N, number of genotypes in a group; %, percentage of genotypes in a group. Data are given together with OR and 95% confidence interval (CI). P values were calculated using χ2 test (two-tailed).
*The frequency of genotypes between patients and controls was compared using χ 2 test (two-tailed) with a 2 × 6 contingency table (controls and patients × genotypes 7/7, 7/8, 7/9, 8/8, 8/9, and 9/9).
The Danish sample analysis also showed genotype distribution differences (P = 0.004) between patients and controls (Table 4). Patients of the Danish sample and those of the Swiss sample had a very similar genotype distribution. In particular, the results in the Danish sample revealed that genotype 8/8 was three times more frequent in patients (OR = 2.96, P = 0.007), whereas genotype 7/7 was more often present in controls (OR = 0.61, P = 0.003). Combining the two samples resulted in a slightly more pronounced (P = 0.002) genotype separation between patients and controls (SI Table 5). Logistic regression analysis also rejected the global null hypothesis (P < 0.001) and stepwise selection revealed significant effects for the genotype 7/7 (P < 0.001), the genotype 8/8 (P = 0.019), and the sample (P = 0.048).
The GCLC GAG TNR allele distribution of the Danish and the Swiss control samples was compared with that of a white population from New York (25) (SI Table 6). The Danish and the New York control samples had a similar allele distribution. In contrast, the allele distribution among the Swiss controls was characterized by an overrepresentation of the “protective” allele 7 (74% vs. 64%) and an underrepresentation of the “risk” allele 8 (3% vs. 15%).
Functional Evidence for the GCLC GAG TNR Polymorphism.
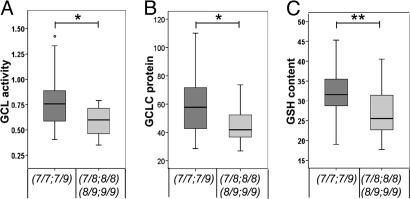
Genetic analysis in the Swiss sample showed an overrepresentation of genotypes 7/7 and 7/9 in controls and an overrepresentation of genotypes 7/8, 8/8, 8/9, and 9/9 in patients. Genetic analysis in the Danish sample supported the findings from the Swiss sample regarding the overrepresentation of genotype 7/7 (OR = 0.61) among controls and the overrepresentation of genotype 8/8 among schizophrenia patients (OR = 2.96). Based on these observations, the Swiss sample was split according to the genotypes into a “low- risk” group (genotypes 7/7 and 7/9) and a “high-risk” group (genotypes 7/8, 8/8, 8/9, and 9/9). GCL activity and GCLC protein expression (both under conditions of t-BHQ treatment) were compared between groups (Fig. 3 A and B). Subjects of the low-risk group had higher GCL activity (0.784 ± 0.246 vs. 0.584 ± 0.141; P = 0.017) and higher GCLC protein expression (58.9 ± 19.0 vs. 45.6 ± 13.3; P = 0.037) than those of the high-risk group. Similar findings were also obtained regarding GSH contents. We have measured fibroblast GSH contents of 39 patients and 33 controls (Table 2). Subjects with the high-risk genotypes had a higher GSH content (32.1 ± 5.8 vs. 27.0 ± 6.0; P = 0.004) than subjects with the low-risk genotypes (Fig. 3C). Analyzing the functional effect of the GCLC GAG polymorphism in patients and controls showed that it was independent of the disease (SI Fig. 4).
Fig. 3.
Functional relevance of the GCLC GAG TNR polymorphism in the Swiss sample. Because genotypes 7/7 and 7/9 were present more often in controls, whereas genotypes 7/8, 8/8, 8/9, and 9/9 were present more often in patients, data of GCL activity, GCLC protein expression, and GSH content were regrouped according their genotypes (7/7 and 7/9 vs. 7/8, 8/8, 8/9, and 9/9). The plots show GCL activity (t-BHQ) (A) and GCLC protein expression (t-BHQ) (B) of 38 subjects with genotypes 7/7 and 7/9 and 11 subjects with genotypes 7/8, 8/8, 8/9, and 9/9. (C) The plot shows GSH content (baseline) of 56 subjects with genotypes 7/7 and 7/9 and 16 subjects with genotypes 7/8, 8/8, 8/9, and 9/9. Each box describes 25%, 75%, and median values. The whisker bars show values in the 1.5 box lengths range. *, P < 0.05; **, P < 0.01 vs. the respective controls was calculated by using Mann–Whitney U test (two-tailed).
In a previous study, we measured GCLC mRNA levels in skin fibroblast cultures of patients and controls (8). A trend (P = 0.064) of lower GCLC mRNA levels was found in patients as compared with controls. In the 72 Swiss subjects of the present study, we again tested a potential association between risk genotypes and GCLC gene expression. There was no association (SI Fig. 5).
No Correlation Between the Disease-Associated Polymorphisms of the GCLC and GCLM Genes.
In the preceding study, we found an association between genetic variants of GCLM and schizophrenia (8). In the Danish sample, we tested whether an interaction between the GAG TNR polymorphism of the GCLC gene and the disease-associated single nucleotide polymorphisms in the GCLM gene (rs2301022 and rs3170633) would affect the risk of schizophrenia. No association was found pointing to an independent contribution of GCLC and GCLM polymorphisms to the vulnerability for schizophrenia.
Discussion
We report evidence that schizophrenia patients have a decreased capacity to synthesize GSH that is most likely of genetic origin. The major findings can be summarized as follows. (i) GCL activity in patients, as compared with controls, was impaired in skin fibroblast cultures under conditions of oxidative stress. (ii) This reduced GCL activity correlated with decreased GCLC protein expression. (iii) Two independent case-control studies showed a significant association between schizophrenia and a GAG TNR polymorphism in the GCLC gene. The most common TNR genotype 7/7 was more frequent in controls, whereas the rarest TNR genotype 8/8 was three times more frequent in patients. (iv) The disease-associated genotypes of this polymorphism correlated with a decrease in GCLC protein expression, GCL activity, and GSH content.
Aside from GCLC and GCLM, the protein expressions of three additional phase II genes were determined: NQO1, NQO2, and NRF2 (data not shown). GCLC was the only protein with an impaired expression in patients, strengthening the hypothesis that this deficit was due to a polymorphism in the GCLC gene. GCLC is located on chromosome 6p12. Several genetic studies have shown an association between schizophrenia and markers or genes on chromosome 6p, mostly between 6p23 and 6p21 (26). It is not clear whether these linkage signals were in fact mediated across the region through linkage with the GCLC GAG TNR polymorphism, or whether the different markers were independently associated with schizophrenia. Further studies are required to address this aspect.
Patients of the Swiss and the Danish sample had comparable genotype distribution of the GCLC GAG TNR polymorphism. However, the genotype distribution differed between the controls of the two samples. The allele distribution of the Danish control sample was very similar to the one of randomly selected Caucasian subjects in New York (25). In contrast, controls of the Swiss sample (where subjects with a history of a major psychiatric disorder have been excluded) had a more different genotype distribution as compared with schizophrenia patients. The fact that all but one control subject in the Swiss sample had a genotype containing at least one 7 allele suggests that subjects containing a genotype without a 7 allele have an increased vulnerability for schizophrenia. Indeed, genetic analysis within both the Swiss and the Danish sample showed a protective effect for the genotype 7/7 and a vulnerable effect for the genotype 8/8. Among Swiss control subjects the genotypes 7/7 and 7/9 were overrepresented, whereas schizophrenia patients in both the Swiss and the Danish sample had a shift toward genotypes 8/8 and 9/9 pointing to a GAG TNR allele rank order for risk of 7 < 9 < 8.
Comparing genetic and functional data for GCLC in the Swiss sample revealed an association between disease-associated GCLC GAG TNR genotypes and decreased protein levels, but not with decreased mRNA levels. In accordance with this observation, a study performed on 60 tumor cell lines found no correlation between the GAG TNR polymorphism and GCLC gene expression (25). As the GAG TNR is present in the mRNA of the GCLC gene (NM_001498), it is likely to influence either mRNA transport or translation rather than mRNA expression.
The GAG TNR polymorphism in the GCLC gene was not sufficient to explain all observed variation in the GCL activity. Aside from patients with disease-associated genotypes (7/8, 8/8, 8/9, and 9/9), there are subjects with low GCL activity, although they had genotypes 7/7 or 7/9. It is possible that additional polymorphisms in the GCLC and/or GCLM gene can affect GCL activity. Moreover, alterations in stress-induced signaling pathways could lead to decreased GSH synthesis.
Aside from the genetic findings, our results provide evidence in a non-tumor cell line for a functional effect of the GCLC GAG TNR polymorphism on GCLC protein expression, GCL activity, and GSH contents. The effect of the polymorphism on GSH contents observed in skin fibroblasts was different from the associations observed in tumor cell lines (25). In tumor cell lines allele 7 was associated with lower GSH content than alleles 8 and 9, suggesting an allele rank order for GSH content of 7 < 8 < 9. In contrast, in skin fibroblasts genotypes 7/7 and 7/9 were associated with higher GSH contents than genotypes containing an 8 allele or genotype 9/9, pointing to an allele rank order for GSH content of 8 < 9 < 7. The divergent associations may be due to the fact that the 60 tumor cell lines originated from a variety of tissues with considerable differences in GSH contents (21).
Consequences of oxidative stress have been reported in patients suffering from schizophrenia, although the causes of this redox imbalance remained unclear. In contrast to neurodegenerative diseases (such as Alzheimer's disease or Parkinson's disease) in which GSH synthesis was not impaired (27) and GSH deficit is considered to be secondary to an excess of ROS, our studies (present study and refs. 8 and 9) provide evidence that the GSH deficit observed in schizophrenia was of primary origin, because of a genetic impairment of GSH synthesis. It is suggested that a GSH deficit when combined with a brain specific insult such as dopamine excess (an important generator of ROS) could play a crucial role in the development of abnormal synaptic connections leading, in turn, to the known perceptive, cognitive, and behavioral dysfunctions of schizophrenia (9). Interestingly, experimental models with a GSH deficit showed morphological, electrophysiological, and behavioral alterations (28–33) that are analogous to anomalies observed in schizophrenia (34, 35). GSH-deficient rats with an excess of dopamine during the development exhibited a decreased GABA-parvalbumin immunoreactivity selectively in the anterior cingulate cortex (31) and had an impairment in object recognition (29). Moreover, a GSH deficit in rat hippocampal slices led to impaired NMDA-receptor (NMDA-R) function and NMDA-dependent synaptic plasticity (30).
Taken together, the results of the present study provide evidence for a genetic source of the redox dysregulation in schizophrenia. The GCLC GAG TNR polymorphism may serve as a marker to identify individuals at risk. This marker may contribute to gather a complete picture of genetic risk factors of schizophrenia in the GSH and oxidative stress associated pathways. These attempts may help to identify critical period and specific brain location during development where such deficits are important to the emergence of the disease. Such knowledge is important to be able to select and target (e.g., pharmaceutical) preventive or even curative interventions.
Materials and Methods
Chemicals and Reagents.
Western blots.
Anti-human α-tubulin antibody was from Santa Cruz Biotechnology. Antibodies for human GCLC and GCLM were obtained from P. Vliet (University of Washington, Seattle, WA). Protease inhibitor tablets were from Roche Diagnostics (Mannheim, Germany). Protein concentration was determined by BCA protein assay (Pierce, Rockford, IL). The enhanced chemiluminescence (ECL) detection system was purchased from Amersham International (Little Chalfont, U.K.).
GSH content and GCL activity.
2,3-Naphthalenedicarboxyaldehyde GSH, γ-glutamylcysteine, ATP, 5-sulfosalicylic acid, l-glutamic acid, l-cysteine, t-BHQ, and other common reagents were purchased from Sigma–Aldrich (Buchs, Switzerland); we purchased the GSH assay kit from Calbiochem (San Diego, CA).
Subjects.
Subjects of the Swiss sample (66 patients and 48 controls; Table 2) were assessed by using the Diagnostic Interview for Genetic Studies (36, 37). Control subjects with a major mood, psychotic, or substance-use disorder or a first-order relative with a psychotic disorder were excluded. Subjects were of Caucasian origin and matched for age. Sex was not matched, but all parameters were analyzed with sex as covariate for its influences on variances (ANCOVA). No sex specific influence was observed. Patients met criteria for schizophrenia or schizoaffective disorder of the Diagnostic and Statistical Manual of Mental Disorders-Fourth Edition (DSM-IV). The subjects were recruited with fully informed written consent according to ethical guidelines of Lausanne University. The Danish sample included 322 schizophrenia patients from the Danish Psychiatric Biobank and 331 unrelated anonymous blood donors serving as controls (Table 2). Danish patients met criteria for schizophrenia or schizoaffective disorder of the International Classification of Diseases (ICD-10). Danish subjects were of Caucasian origin and had signed an informed consent before participation. The Danish Data Protection Agency and the Danish Scientific Ethics Committee approved the study.
Cell Cultures and Treatment.
Fibroblast cultures were established from skin biopsies as described in Tosic et al. (8). Fibroblasts after five passages near confluency were treated for 18 h with 50 μM t-BHQ in 0.05% dimethyl sulfoxide (DMSO) or 0.05% DMSO alone.
Western Blot Analysis.
Cells were scraped with lysis buffer [2.37 ml of buffer (50 mM Tris·HCl, pH 7.2/150 mM NaCl/10 mM NaF/2 mM EDTA/2 mM EGTA/1% Triton X-100); plus 100 μl of proteinase inhibitor (one tablet per 2 ml of H2O); 25 μl of 100 mM PMSF; 2.5 μl of 1 mM DTT]. Lysates were homogenized and centrifuged (20,000 × g, 14 min, 4°C). Protein concentration was determined by BCA protein assay. Equal amounts of protein (40 μg) were separated by 12% SDS/PAGE and electro-transferred to a polyvinylidene difluoride Immobilion-P membrane. Membranes were blocked (milk powder 3%) for 1 h. After incubation with primary and secondary antibodies, protein was visualized by using ECL detection system, and protein abundance was quantified by densitometry analysis (Multi Genius, Bio Imaging System, Syngene). The results were normalized to α-tubulin.
GSH Content and GCL Activity.
Cells were trypsinized, pelleted, and frozen in 0.5 ml of PBS. GSH content and GCL activity were determined in protein extracts from fibroblast cultures, with a fluorescence-based microtiter plate assay according to White et al. (38). GCL activity was determined as the difference between GSH synthesis in unblocked and GSH synthesis in BSO-treated wells per minute and per milligram of protein. GSH contents were also routinely measured in all participants that had spent skin fibroblasts (33 controls and 39 patients) by a colorimetric GSH assay kit (Calbiochem, Darmstadt, Germany).
Genotyping.
DNA was purified from blood samples by using the NucleonBACC3 system (Amersham Pharmacia Biotech, Buckinghamshire, U.K.). GCLC GAG TNR polymorphism was assessed as originally described (25) with the following modifications. PCR was performed in reaction mixtures of 25 μl containing 40 ng of each primer, 200 μM each dNTP, and 0.3 units of EuroTaq DNA polymerase (Euroclone, U.K.). Temperature cycling of the PCR was as originally described. PCR products were separated on an 8% polyacrylamide gel (6 × 8 cm) for 5 h with 40 V and stained by using SilverXpress Silver Staining Kit (Invitrogen, Carlsbad, CA).
Statistical Analysis.
Statistical analysis was performed by using the SAS statistical package version 9 (SAS Institute, Cary, NC). The distribution of the protein levels was tested for normality (Univariate procedure, SAS Users Guide: Base, 1989), and data were transformed with an exponential function to obtain normally distributed values. A two-way ANOVA was used to test the effect of disease status and treatment (GLM procedure, SAS/STAT User's Guide, version 6, 1989). The treatment effect was studied also with a MANOVA model in which the baseline values were taken as covariables. Correlations between protein levels were studied by using the CORR procedure. In the Swiss and Danish sample, genotype distribution between patients and controls was tested by using a χ2 test (two-tailed) or, if cells had an expected count <5, the Fisher exact test (two-tailed). Genotype distribution of both samples together was first analyzed by using a χ2 test (two-tailed), and their contribution to the disease and the national differences were analyzed by using a logistic regression. The Mann–Whitney U test (two-tailed) was used to compare GCL activity, GCLC protein expression, and GSH content between the groups of low- and high-risk TNR polymorphisms.
Supplementary Material
Acknowledgments
We thank all patients and controls for their participation; Nathalie Ballanfat, Valérie Perroud, and Christel Reuterswaerd for technical help; and Dr. Portia Vliet for the GCLM and GCLC antibodies. This work was supported by the “Loterie Romande.”
Abbreviations
- GCL
glutamate cysteine ligase
- GCLC
GCL catalytic subunit
- GCLM
GCL modulatory subunit
- GSH
glutathione
- t-BHQ
tert-butylhydroquinone
- TNR
trinucleotide repeat
- ROS
reactive oxygen species
- OR
odds ratio.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706778104/DC1.
References
- 1.Owen MJ, Craddock N, O'Donovan MC. Trends Genet. 2005;21:518–525. doi: 10.1016/j.tig.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 2.Harrison PJ, Weinberger DR. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- 3.Lewis DA, Levitt P. Annu Rev Neurosci. 2002;25:409–432. doi: 10.1146/annurev.neuro.25.112701.142754. [DOI] [PubMed] [Google Scholar]
- 4.Ross CA, Margolis RL, Reading SA, Pletnikov M, Coyle JT. Neuron. 2006;52:139–153. doi: 10.1016/j.neuron.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 5.Do KQ, Lauer CJ, Schreiber W, Zollinger M, Gutteck-Amsler U, Cuenod M, Holsboer F. J Neurochem. 1995;65:2652–2662. doi: 10.1046/j.1471-4159.1995.65062652.x. [DOI] [PubMed] [Google Scholar]
- 6.Do KQ, Trabesinger AH, Kirsten-Kruger M, Lauer CJ, Dydak U, Hell D, Holsboer F, Boesiger P, Cuenod M. Eur J Neurosci. 2000;12:3721–3728. doi: 10.1046/j.1460-9568.2000.00229.x. [DOI] [PubMed] [Google Scholar]
- 7.Yao JK, Leonard S, Reddy R. Dis Markers. 2006;22:83–93. doi: 10.1155/2006/248387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tosic M, Ott J, Barral S, Bovet P. Am J Hum Gen. 2006;79:586–592. doi: 10.1086/507566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Do KQ, Bovet P, Cabungcal JH, Conus P, Gysin R, Lavoie S, Steullet P, Cuenod M. In: Schizophrenia, Handbook of Neurochemistry and Molecular Neurobiology. 3rd Ed. Lajtha A, editor. New York: Springer; 2007. in press. [Google Scholar]
- 10.Soltaninassab SR, Sekhar KR, Meredith MJ, Freeman ML. J Cell Physiol. 2000;182:163–170. doi: 10.1002/(SICI)1097-4652(200002)182:2<163::AID-JCP4>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 11.Robertson GS, Hori SE, Powell KJ. J Psychiatry Neurosci. 2006;31:157–167. [PMC free article] [PubMed] [Google Scholar]
- 12.Piazza PV, Le Moal ML. Annu Rev Pharmacol Toxicol. 1996;36:359–378. doi: 10.1146/annurev.pa.36.040196.002043. [DOI] [PubMed] [Google Scholar]
- 13.Chakraborti A, Gulati K, Banerjee BD, Ray A. Behav Brain Res. 2007;179:321–325. doi: 10.1016/j.bbr.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 14.Sokoloff L. Neurochem Res. 1999;24:321–329. doi: 10.1023/a:1022534709672. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, Dieter MZ, Chen Y, Shertzer HG, Nebert DW, Dalton TP. J Biol Chem. 2002;277:49446–49452. doi: 10.1074/jbc.M209372200. [DOI] [PubMed] [Google Scholar]
- 16.Koide S, Kugiyama K, Sugiyama S, Nakamura S, Fukushima H, Honda O, Yoshimura M, Ogawa H. J Am Coll Cardiol. 2003;41:539–545. doi: 10.1016/s0735-1097(02)02866-8. [DOI] [PubMed] [Google Scholar]
- 17.Beutler E, Gelbart T, Kondo T, Matsunaga AT. Blood. 1999;94:2890–2894. [PubMed] [Google Scholar]
- 18.Dahl N, Pigg M, Ristoff E, Gali R, Carlsson B, Mannervik B, Larsson A, Board P. Hum Mol Genet. 1997;6:1147–1152. doi: 10.1093/hmg/6.7.1147. [DOI] [PubMed] [Google Scholar]
- 19.Curkendall SM, Mo J, Glasser DB, Rose SM, Jones JK. J Clin Psychiatry. 2004;65:715–720. doi: 10.4088/jcp.v65n0519. [DOI] [PubMed] [Google Scholar]
- 20.Enger C, Weatherby L, Reynolds RF, Glasser DB, Walker AM. J Nerv Ment Dis. 2004;192:19–27. doi: 10.1097/01.nmd.0000105996.62105.07. [DOI] [PubMed] [Google Scholar]
- 21.Dringen R, Gutterer JM, Hirrlinger J. Eur J Biochem. 2000;267:4912–4916. doi: 10.1046/j.1432-1327.2000.01597.x. [DOI] [PubMed] [Google Scholar]
- 22.Satoh T, Okamoto SI, Cui J, Watanabe Y, Furuta K, Suzuki M, Tohyama K, Lipton SA. Proc Natl Acad Sci USA. 2006;103:768–773. doi: 10.1073/pnas.0505723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meister A. Methods Enzymol. 1995;252:26–30. doi: 10.1016/0076-6879(95)52005-8. [DOI] [PubMed] [Google Scholar]
- 24.Prestera T, Holtzclaw WD, Zhang Y, Talalay P. Proc Natl Acad Sci USA. 1993;90:2965–2969. doi: 10.1073/pnas.90.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walsh AC, Feulner JA, Reilly A. Toxicol Sci. 2001;61:218–223. doi: 10.1093/toxsci/61.2.218. [DOI] [PubMed] [Google Scholar]
- 26.Schwab SG, Hallmayer J, Albus M, Lerer B, Eckstein GN, Borrmann M, Segman RH, Hanses C, Freymann J, Yakir A, et al. Mol Psychiatry. 2000;5:638–649. doi: 10.1038/sj.mp.4000791. [DOI] [PubMed] [Google Scholar]
- 27.Schulz JB, Lindenau J, Seyfried J, Dichgans J. Eur J Biochem. 2000;267:4904–4911. doi: 10.1046/j.1432-1327.2000.01595.x. [DOI] [PubMed] [Google Scholar]
- 28.Cabungcal JH, Preissmann D, Delseth C, Cuenod M, Do KQ, Schenk F. Neurobiol Dis. 2007;26:634–645. doi: 10.1016/j.nbd.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Castagne V, Rougemont M, Cuenod M, Do KQ. Neurobiol Dis. 2004;15:93–105. doi: 10.1016/j.nbd.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Steullet P, Neijt HC, Cuenod M, Do KQ. Neuroscience. 2006;137:807–819. doi: 10.1016/j.neuroscience.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 31.Cabungcal JH, Nicolas D, Kraftsik R, Cuenod M, Do KQ, Hornung JP. Neurobiol Dis. 2006;22:624–637. doi: 10.1016/j.nbd.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 32.Grima G, Benz B, Parpura V, Cuenod M, Do KQ. Schizophr Res. 2003;62:213–224. doi: 10.1016/s0920-9964(02)00405-x. [DOI] [PubMed] [Google Scholar]
- 33.Castagne V, Cuenod M, Do KQ. Neuroscience. 2004;123:821–834. doi: 10.1016/j.neuroscience.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 34.Kolluri N, Sun Z, Sampson AR, Lewis DA. Am J Psychiatry. 2005;162:1200–1202. doi: 10.1176/appi.ajp.162.6.1200. [DOI] [PubMed] [Google Scholar]
- 35.Lewis DA, Hashimoto T, Volk DW. Nat Rev Neurosci. 2005;6:312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- 36.Nurnberger JI, Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T. Arch Gen Psychiatry. 1994;51:849–859. doi: 10.1001/archpsyc.1994.03950110009002. [DOI] [PubMed] [Google Scholar]
- 37.Preisig M, Fenton BT, Matthey ML, Berney A, Ferrero F. Eur Arch Psychiatry Clin Neurosci. 1999;249:174–179. doi: 10.1007/s004060050084. [DOI] [PubMed] [Google Scholar]
- 38.White CC, Viernes H, Krejsa CM, Botta D, Kavanagh TJ. Anal Biochem. 2003;318:175–180. doi: 10.1016/s0003-2697(03)00143-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.