Abstract
Uropathogenic Escherichia coli (UPEC) cause most uncomplicated urinary tract infections (UTIs) in humans. Because UTIs are considered to occur in an ascending manner, flagellum-mediated motility has been suggested to contribute to virulence by enabling UPEC to disseminate to the upper urinary tract. Previous studies from our laboratory and others have demonstrated a modest yet important role for flagella during ascending UTI. To better understand the role of flagella in vivo, we used biophotonic imaging to monitor UPEC infection and temporospatial flagellin gene expression during ascending UTI. Using em7-lux (constitutive) and fliC-lux transcriptional fusions, we show that flagellin expression by UPEC coincides with ascension of the ureters and colonization of the kidney. The patterns of fliC luminescence observed in vitro and in vivo were also validated by comparative quantitative PCR. Because fliC expression appeared coincident during ascension, we reassessed the contribution of fliC to ascending UTI using a low-dose intraurethral model of ascending UTI. Although wild-type UPEC were able to establish infection in the bladder and kidneys by 6 hours postinoculation, fliC mutant bacteria were able to colonize the bladder but were significantly attenuated in the kidneys at this early time point. By 48 hours postinoculation, the fliC mutant bacteria were attenuated in the bladder and kidneys and were not detectable in the spleen. These data provide compelling evidence that wild-type UPEC express flagellin and presumably utilize flagellum-mediated motility during UTI to ascend to the upper urinary tract and disseminate within the host.
Keywords: biophotonic imaging, urinary tract infection, pyelonephritis, motility, fliC
It has been hypothesized that flagella, organelles required for motility, facilitate the establishment and spread of infection by microbial pathogens within the host (1, 2). Studies suggest that up to 95% of urinary tract infections (UTIs) develop in an ascending manner (3) beginning with periurethral colonization by bacteria such as uropathogenic Escherichia coli (UPEC), followed by migration to the bladder to establish infection, and if left untreated, ascension to the upper urinary tract or ureters and kidneys (4). Once in the kidneys, UPEC can gain access to the bloodstream, causing bacteremia and sometimes death (5). Because UTIs caused by UPEC are ascending infections involving multiple organs, we reasoned that monitoring flagella expression during UTI in real time would be useful as a model system to examine the connection between motility and the establishment and spread of bacterial infection.
The bacterial flagellum is a long helical surface appendage composed of polymerized subunits of flagellin encoded by fliC. Mutation of fliC in UPEC leads to loss of flagellation and motility. Recently, our laboratory and others showed that fliC mutants were out-competed by motile wild-type strains during experimental cochallenge of mice, demonstrating that flagella contribute to the efficient colonization of the urinary tract (6, 7). The limitation of these challenge experiments was the inability to define the precise location and time flagella are expressed in vivo to provide the wild-type strain with the fitness advantage. Using a DNA microarray for UPEC strain CFT073, our laboratory previously determined the transcriptional profile of CFT073 during ascending UTI in CBA/J mice (8). We demonstrated that, overall, genes involved in flagellum-mediated motility and chemotaxis were down-regulated in vivo when compared with in vitro growth conditions (8). Although this study provided important insight into the in vivo transcriptome of UPEC during ascending UTI, data represented averages of expression over a 10-day period. Thus, using transcriptional profiling, it is possible neither to assess the level of transient expression for flagellin or any given gene nor to determine the specific tissue in which this expression occurred.
For some bacterial pathogens such as pandemic Vibrio cholerae strains, motility mutants are highly attenuated in vivo (9). In others, such as Salmonella enterica serovar Typhimurium, fliC is expressed during invasion through Peyer's Patches but not in the mesenteric lymph nodes or spleen to promote escape from immune responses and to spread to systemic sites of replication (10). Because much attention has been focused on immune recognition of bacterial flagellin via Toll-like receptor 5 (11–13), and it is unknown how flagella contribute to ascending infections, it would be advantageous to assess the spatial and temporal expression of UPEC flagellin in real time during UTI. Noninvasive biophotonic imaging has been used for real-time monitoring of organism density and spread in various animal models of infection, including UTI caused by Pseudomonas aeruginosa and Proteus mirabilis (14–16). This technology has also been used to measure gene-specific real-time expression of a global virulence regulator, agr, during Staphylococcus aureus s.c. abscess formation (17).
Here we used noninvasive biophotonic imaging to assess the level and site of flagellar gene expression during ascending infection and to resolve the fitness advantage flagella provide UPEC in vivo. A luminescent fliC reporter plasmid was constructed that was stably maintained in vivo and demonstrated flagellin-specific patterns of luminescence during swimming and swarming in vitro. Using this construct, we found that fliC expression coincided with bacterial ascension to the upper urinary tract, showing that motility is involved in the migration of the infection from the bladder to the kidneys. The in vivo expression of fliC was verified by quantitative PCR (qPCR), and the contribution of fliC to ascending infection was reassessed by using a low-dose intraurethral inoculation model (18). Although wild-type UPEC were able to ascend the urethra to the bladder and migrate to the kidneys, fliC mutant bacteria were significantly less able to reach the upper urinary tract.
Results
Construction of a Bioluminescent fliC Reporter and Correlation of Expression with Motility.
All bioluminescence reporters were expressed in UPEC strain CFT073 (O6:K2:H1), which was isolated from the blood of a hospitalized patient with acute pyelonephritis (19). The luxCDABE gene cluster encoding bacterial luciferase [isolated from pUT-Tn5-EM7-lux-Km1 (20), a generous gift from C. Contag, Stanford University, Palo Alto, CA] was cloned into the low-copy plasmid pGEN222 [ref. 21; a gift from J. Galen (University of Maryland, Baltimore] [supporting information (SI) Fig. 4 and SI Table 1]. The resulting plasmid, pGEN-luxCDABE, carries the synthetic em7 promoter that drives constitutive expression of the luxCDABE operon and is referred to as Pem7-lux. Pem7-lux retains the novel plasmid maintenance system from pGEN222, with the hok sok postsegregation killing system and two par (partitioning) loci to enhance plasmid stability over multiple bacterial generations (ref. 21; SI Fig. 4 and SI Table 1). The maintenance of Pem7-lux was assessed in vitro in the absence of antibiotic pressure by examining the loss of antibiotic resistance of CFT073 cultured statically at 37°C and subcultured every 24 h for 3 days. After 1 and 3 days of passaging, the plasmid appeared relatively stable in the absence of antibiotics in that 97.8% and 86.6% of the CFT073 population, respectively, retained Pem7-lux (SI Fig. 5).
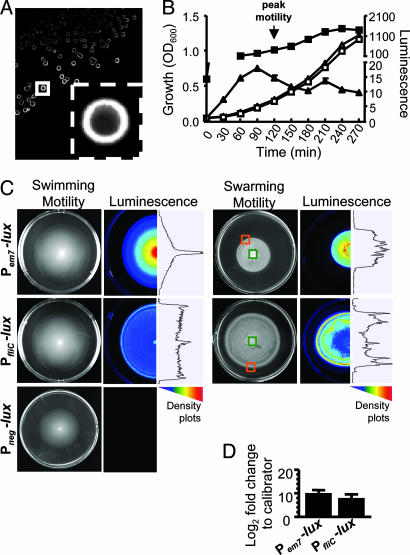
To create the bioluminescent fliC reporter, the em7 promoter of Pem7-lux was replaced with a 244-bp fragment that includes the fliC promoter of CFT073 (SI Fig. 4) (7, 22). The resulting construct, PfliC-lux, was transformed into CFT073. Unlike the Pem7-lux transformants, CFT073 transformants containing the PfliC-lux reporter expressed lux as a halo concentrated at the periphery of colonies on LB agar (Fig. 1A). In vitro growth rates of CFT073 containing the Pem7-lux or PfliC-lux reporters were comparable and lux expression from the Pem7-lux reporter was constitutive (Fig. 1B). Levels of lux expression from the PfliC-lux reporter, however, corresponded to changing levels of motility observed by phase-contrast microscopy (Fig. 1B). As predicted, peak PfliC-lux activity preceded the maximal motility of the culture at early exponential phase (OD600 = 0.3) (Fig. 1B, arrow), presumably because of the length of time required to assemble the complex flagellar apparatus after transcription of the class III genes.
Fig. 1.
In vitro characterization of wild-type CFT073 PfliC-lux and Pem7-lux reporters. (A) Luminescence of wild-type CFT073 transformants containing the PfliC-[infi]lux reporter. (Inset) Enlargement of boxed colony. (B) Growth and luminescence expression of PfliC-lux (▵ and ▴, respectively) and Pem7-lux (□ and ■, respectively). Average (n = 3) optical densities and luminescence were plotted. Arrow indicates the highest level of motility as observed by phase-contrast microscopy. (C) Motility and luminescence images of PfliC-lux, Pem7-lux, and Pneg-lux reporters swimming within 0.25% tryptone agar (Left) or swarming atop 0.45% Eiken agar (Right). Cultures of each reporter were stabbed into soft agar plates or spotted atop swarm plates and incubated for 16 h at 30°C or 37°C, respectively. (D) qPCR analysis of fliC transcription within the inside and outer edge of swarms formed by the PfliC-lux and Pem7-lux reporters. Black bars represent the average changes (log2-fold; n = 3) in expression of fliC between the inside (green boxes in C) and outer edge (orange boxes in C) of the swarms. Error bars represent the standard error of the mean.
To further correlate motility and expression of the PfliC-lux reporter, we examined the pattern of luminescence displayed by the fliC and em7 reporters while swimming in 0.25% agar or swarming atop 0.45% agar (Fig. 1C). To demonstrate the level of background luminescence in vitro, we also measured the luminescence of CFT073 containing Pneg-lux, a promoterless control reporter, during motility in 0.25% agar. Overnight cultures of the PfliC-lux, Pem7-lux and Pneg-lux reporters were recultured in fresh LB, standardized to an OD600 = 1.0, and then either stabbed into 0.25% tryptone agar or spotted (5 μl) onto 0.45% Eiken agar. The swimming and swarming plates were incubated for 16 h at 30°C and 37°C, respectively. Although all three reporters formed similar motility patterns, the patterns of luminescence displayed were distinct (as demonstrated by the density plots to the right of the luminescence images in Fig. 1C). As expected, the level of in vitro lux expression by Pneg-lux was not detectable by the ChemiDoc XRS system (Bio-Rad, Hercules, CA) (Fig. 1C). The pattern of luminescence exhibited by the Pem7-lux reporter correlated with the concentration of the bacterial population; the more bacteria present, the brighter the luminescence (which was expected with the constitutive lux reporter) (Fig. 1C). In contrast, the pattern of PfliC-lux luminescence was brightest at the outer ring of motility in 0.25% agar and along the outer edge of the swarm atop 0.45% agar (Fig. 1C). This phenomenon is consistent with previous observations that FliC expression, and overall flagella production are maximal along the edge of E. coli swarm fronts (23). Here, we also demonstrated by comparative qPCR that the amount of fliC transcription along the outer edge of the swarms produced by PfliC-lux and Pem7-lux were approximately log2 8.0-fold (256-fold) and log2 10.1-fold (1,098-fold) greater, respectively, than the bacteria residing in the center of the swarms (Fig. 1D). It is important to note, however, that only the luminescence profile of the PfliC-lux reporter correlates with the pattern of fliC transcription observed in the swarm assays (Fig. 1 C and D).
Temporal and Spatial Expression of fliC in Vivo.
Before examining the temporospatial expression of fliC, the progression of UPEC infection was monitored by imaging mice infected with the constitutive CFT073 Pem7-lux reporter using the in vivo imaging system, IVIS 200 (Xenogen, Hopkinton, MA). Preliminary studies conducted in our laboratory demonstrated that, on average, 99.3% of the CFT073 population retained the Pem7-lux reporter plasmid after 48 h in vivo (from both the bladder and kidneys; data not shown). Once the in vivo stability of the Pem7-lux reporter was confirmed, we monitored the progression of ascending UTI by biophotonic imaging and demonstrated that the luminescence observed is indicative of bacterial infection within the lower (bladder) and/or upper (kidneys) urinary tract. Indeed, the luminescence emitted from regions of interest (ROIs) positively correlated (P < 0.0001) with the number of viable bacteria harvested from the kidneys at 48 hpi for the constitutive em7 reporter (data not shown). Additionally, as we monitored the progression of infection, we were able to demonstrate that the bladders and kidneys, which appeared to be bioluminescent in vivo, were also bioluminescent ex vivo (SI Fig. 6). The enzymatic activity of bacterial luciferase required oxygen, as demonstrated by the disappearance of the luminescent signals emitted from the ROIs within 5 min after death [most likely due to the loss of circulating oxygenated blood to the tissues as described by Wiles et al. (24) (data not shown)]. Furthermore, we observed luminescence from the kidney ex vivo only when it was minced into small pieces allowing oxygenation (SI Fig. 6). It is not surprising that the bladder is extremely luminescent ex vivo, because after harvesting the organ, it acquired a large opening that exposed the lumen of the bladder to air (SI Fig. 6).
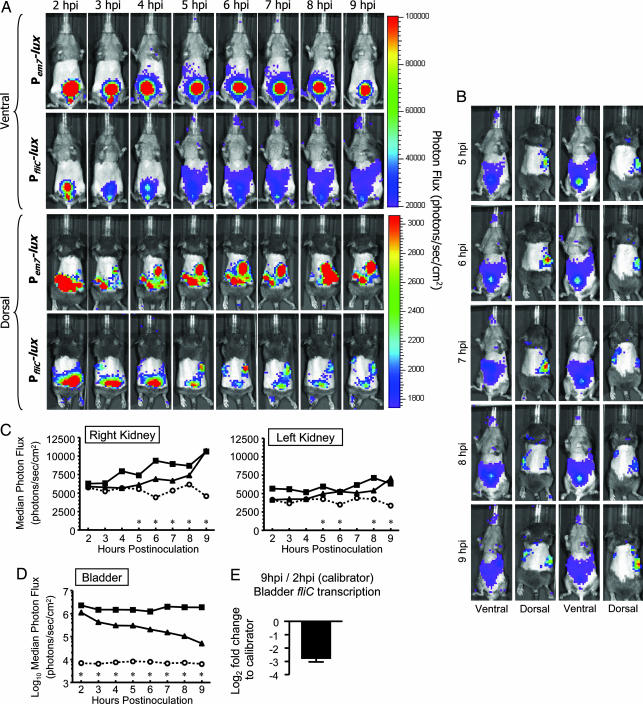
To more closely monitor the progression of CFT073 infection during the earliest time points of UTI ascension, 109 cfu of CFT073 containing Pem7-lux were inoculated into the bladders of 20 CBA/J mice, respectively. To demonstrate that em7 reporter luminescence was significantly higher than background, an additional 14 CBA/J mice were monitored that were infected with CFT073 containing Pneg-lux. From 2 to 9 h postinoculation (hpi), mice were imaged hourly by using the IVIS 200, and luminescence was acquired and quantified in the bladder and kidneys using the Living Image software (Xenogen). Images of a representative mouse infected over time with CFT073 containing the constitutive Pem7-lux reporter are presented in Fig. 2A. In Fig. 2A, the scale is demonstrating luminescence emission per individual pixel and not total luminescence of a region of interest as indicated by the scatter plots in Fig. 2 C and D. The amount of luminescence emitted from the bladder from 2 to 9 hpi by CFT073 containing the constitutive em7 reporter remained for the most part unchanged and significant over background (Fig. 2 A Ventral and D), suggesting that the colonization of bladder by UPEC remains at a steady state during early time points of infection. In contrast, the amount of luminescence emitted from the kidneys steadily increased from 2 to 9 hpi (it was, however, significant over background at all early time points with the exception of the 2-h time point in the right kidney) (Fig. 2 A Dorsal and C). Moreover, during some of the early time points on the ventral side of the mice, it is possible to observe a trail of luminescent signal with the constitutive reporter leading from the bladder to the right kidney on the ventral side of the mice (Fig. 2A Ventral). Altogether, these data illustrate that, although the bladder retains a steady state of UPEC colonization, there is a gradual ascension of UPEC from the bladder to the kidneys, which causes an increase in luminescence signal as UPEC colonizes the kidneys. Because we have been able to culture UPEC from the ureters during ascending murine infection (data not shown), it is likely that the route of ascension from the bladder to the kidneys is through the ureters.
Fig. 2.
Biophotonic imaging of wild-type CFT073 Pem7-lux and PfliC-lux reporter luminescence during ascending UTI and confirmation of fliC expression in vivo by qPCR. (A) Dorsal and ventral sides of mice (n = 2) transurethrally inoculated with 109 cfu of wild-type CFT073 containing either Pem7-lux or PfliC-lux. Mice were imaged from 2 to 9 hpi by using the IVIS 200, and luminescence was quantified in the bladder and kidneys by using Living Image software. (B) Additional images of mice (n = 2) inoculated with 109 cfu of wild-type CFT073 containing PfliC-lux. Images are shown between 5 and 9 hpi to further demonstrate the trail of signal produced by the fliC reporter during ascension to the upper urinary tract. The scales for ventral and dorsal luminescence are the same as those shown for A. (C and D) Scatter plots depicting luminescence over time for PfliC-lux (n = 19 mice), Pem7-lux (n = 20 mice), and Pneg-lux (n = 15 mice). Each data point represents either the median or log10 median luminescence emitted from the bladder (D) and individual kidneys (C) of 14–20 CBA/J mice during specific times after inoculation. Significance of fliC gene expression over background at each time point was assessed by using a Mann–Whitney test. *, P < 0.05; ■, Pem7-lux; ▴, PfliC-lux; and ○, Pneg-lux. (E) qPCR analysis of in vivo fliC transcription by the PfliC-lux reporter. The black bar represents the log2-fold change in fliC expression between 2 and 9 hpi during bladder colonization (n = 20 mice). The error bar represents the standard error of the mean.
To determine the temporospatial expression of fliC during early bladder colonization and ascension to the upper urinary tract, 19 additional CBA/J mice were transurethrally inoculated with 109 cfu of CFT073 containing PfliC-lux and imaged every hour from 2 to 9 hpi. It is important to note that wild-type CFT073 containing each of the Pem7-lux, Pneg-lux, and PfliC-lux equally colonized the urinary tract at 9 hpi, because the cfus per gram of bladder or kidney tissue did not significantly differ (data not shown). Images of a representative mouse infected over time with CFT073 containing the constitutive PfliC-lux reporter are presented in Fig. 2A. In contrast to the constitutive luminescence observed over time in the bladder with the em7 reporter, the fliC reporter luminescence decreases considerably between 2 and 9 hpi (Fig. 2D). At all time points, however, the level of fliC reporter luminescence remains significant above background (Fig. 2D). These data appear to be consistent with the observation made by Snyder et al. (8) in the in vivo transcriptome study, where, over the course of a 10-day UPEC infection, fliC transcription is down-regulated. To further substantiate these findings, we isolated total RNA from the bladders of mice that were infected for 2 and 9 h with CFT073, prepared gene-specific (fliC and gapA) cDNA, and then determined the differences in fliC transcription using comparative qPCR. As expected, the level of fliC transcription was log2 2.8-fold (7.0-fold) less at 9 hpi compared with 2 hpi and directly correlated with the drop in fliC reporter luminescence from 2 to 9 hpi (Fig. 2E).
Despite the drop in total fliC reporter luminescence in the bladder between 2 and 9 hpi, a more pronounced trail of luminescence leading from the bladder to the kidneys emerged at 5 hpi and was maintained up to 9 hpi (similar to the trail observed with the em7 reporter during ascension and early colonization of the kidneys). This phenomenon was also observed with the remaining 18 infected mice (for at least one time point between 5 and 9 hpi) and is illustrated with two additional representative mice in Fig. 2B. Because the trail of signal emerged at 5 hpi, it also correlated with a significant increase in fliC reporter luminescence over background in one or both kidneys (Fig. 2 A and B). In fact, the amount of luminescence at all time points after 5 hpi in the kidneys (with the exception of the 7-h time point in the left kidney) is significantly greater than background (Fig. 2C). Altogether, these data demonstrate the temporospatial expression of flagellin (and presumably flagella) during ascending UPEC infection, and that the expression of fliC coincides with the ascension of UPEC to the upper urinary tract.
Reassessment of the Contribution of Flagellin During Ascending Murine UTI Using an Intraurethral Challenge Method.
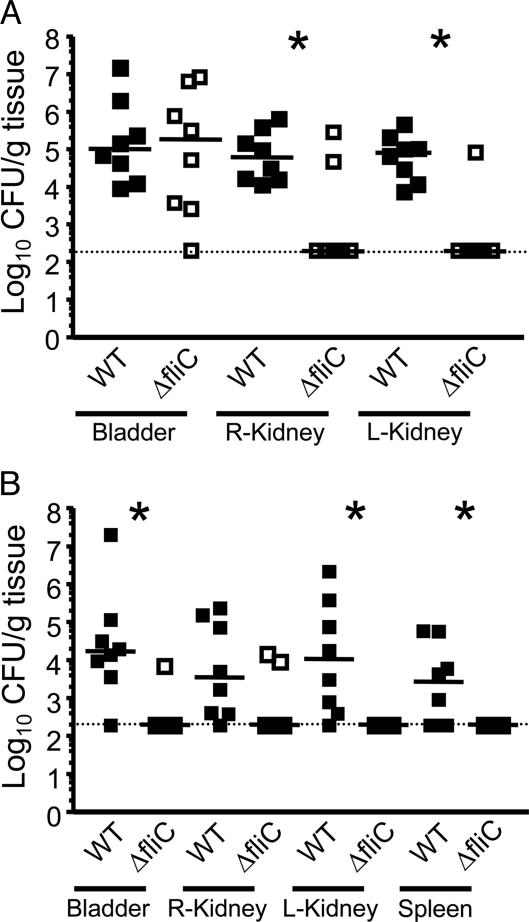
Because fliC expression appeared to coincide with the ascension of UPEC from the bladder to the kidneys, we hypothesized that flagella must be playing a more important role during urinary tract ascension than what has been previously observed by our laboratory and others. It has been debated that the current methods of transurethral inoculation (using a 20- to 50-μl volume) used by our laboratory and several others results in minimal to substantial vesicoureteral reflux (that is, leakage of bacteria from the bladder into the ureters and kidneys due to the pressure or volume of the inoculum) (25). In fact, we confirmed that our method of transurethral inoculation results in minimal to no vesicoureteral reflux, as determined by viable counts present in the kidneys and biophotonic imaging after 5-min postinoculation (SI Fig. 7). Because flagella are presumed to be important for ascension of the bladder through the ureters to the kidneys, we then wanted to rule out the effect of inoculation on development of kidney infection. Therefore, we reassessed the contribution of flagellin during ascending UTI by inoculating a smaller volume (10 μl) and concentration (107 cfu) directly into the urethra (instead of the bladder). For comparison, the ID50 for transurethral inoculation is 106 cfu per mouse. Hopkins et al. (18) previously demonstrated that with this method of intraurethral challenge, vesicoureteral reflux could be avoided. Specifically, 107 cfu of wild-type CFT073 and CFT073 ΔfliC were independently inoculated into the urethras of eight mice per each time point. After 6 and 48 hpi, cfu/gram of bladder, kidney, and spleen tissue (for the 48-h time point only) were determined. Both the wild-type and fliC mutant were recovered in equal numbers from the bladder at 6 hpi, suggesting that both strains were equally able to colonize the bladder at early time points of infection (Fig. 3A). In contrast, the fliC mutant was recovered in significantly lower numbers than wild-type in both the left and right kidneys at 6 hpi (P = 0.002 and 0.021, respectively), thus demonstrating that the fliC mutant was less able to colonize the kidneys (Fig. 3A). At 48 hpi, the fliC mutant was also recovered in significantly lower numbers than wild-type from the bladder, left kidney, and spleen (P = 0.006, 0.004, and 0.037, respectively) and tended to be less well recovered than wild-type from the right kidney (Fig. 3B; P = 0.051). The 48-hpi data demonstrate that fliC is not only important for colonization of the kidneys but is also required for maintenance of bladder colonization and dissemination into the bloodstream.
Fig. 3.
Independent challenges of mice inoculated with a 10-μl suspension of 107 cfu/ml of either wild-type CFT073 or CFT073 ΔfliC directly into the urethra. Eight mice were challenged for each strain and time point. At 6 hpi (A) and 48 hpi (B), bladders, kidneys, and/or spleens were harvested to determine bacterial concentrations. Each data point represents the log10 cfu per gram of tissue collected from one mouse. Horizontal bars represent the median values of the populations. The limit of detection of this assay is 2 × 102 cfu per gram, as indicated by the dotted line. Significant differences in colonization between the wild-type and fliC mutant were determined by using a Mann–Whitney test. *, P < 0.05; ■, wild type (WT); □, ΔfliC.
Discussion
Here, we describe the use of biophotonic imaging for visualization of the bioluminescent UPEC inoculum in vivo, for monitoring the spread of infection, and to assess the contribution of motility for this movement of the bacteria within the host. This was accomplished by measuring the level and localization of fliC expression in vivo during ascending UTI in real time. Using this technology, fliC expression was easily detectable over background luminescence produced by the promoterless Pneg-lux construct. The early decrease observed for fliC expression in the bladder and subsequent peak of fliC expression in the kidneys is intriguing, because it is coincident with the time that UPEC strain CFT073 is known to reach and colonize the kidneys (6, 26). Although it has been speculated that flagellum-mediated motility contributes to the movement of infection within the host (1, 2), the data presented in this study provide direct evidence that flagella of UPEC strain CFT073 are expressed at both the time (4–6 hpi) and location (ureters) at which UPEC ascend from the bladder to the kidneys.
Previously, our group determined the transcriptional profile of UPEC during ascending UTI by collecting urine from infected mice over an extended period (8). In that study, urine was pooled from multiple infected mice over time to allow isolation of sufficient mRNA to measure the UPEC transcriptome (8). Although this study served as an excellent tool for identifying the transcriptional profile of the CFT073 genome during ascending UTI, it did not allow us to identify periods of transient gene expression or localize site-specific gene expression. Despite fliC being down-regulated in the in vivo transcriptome, results from cochallenge experiments conducted by our laboratory and others have demonstrated that UPEC flagella contribute significantly to fitness during colonization of the urinary tract (6, 7). Resolving this apparent paradox, biophotonic imaging for the fliC-luciferase transcriptional fusion revealed that wild-type UPEC express flagellin while migrating from the bladder to the kidneys. This flagellar expression during ascension to the upper urinary tract likely represents the nature of the fitness advantage seen for wild-type bacteria during the cochallenge experiments in that UPEC mutants deficient for flagellin are unable to migrate throughout the urinary tract as efficiently as bacteria expressing flagella.
To better understand the contribution of flagellin for the establishment and movement of the infection within the host, a low-dose intraurethral UTI model was used. Although the bacterial inoculum for this technique is not sufficient to generate suitable bioluminescent signal in vivo for localizing fliC expression, it was useful to evaluate the impact that deletion of the gene has on the ability of UPEC to colonize the urinary tract. Although wild-type UPEC were able to establish infection in the bladder and kidneys by 6 hpi, fliC mutant bacteria were able to colonize the bladder but were significantly attenuated in the kidneys at this early time point. By 48 hpi, the fliC mutant bacteria were attenuated in the bladder and kidneys and were not detectable in the spleen. These data provide compelling evidence that wild-type UPEC express flagellin, and presumably use flagellum-mediated motility during UTI to ascend to the upper urinary tract and disseminate within the host.
The sharp decrease in fliC signal within the bladder and concomitant increase in expression within the ureters and kidney that dissipates after a short time demonstrates that flagellin expression is transient in vivo. Consistent with our findings, Wright et al. (7) observed robust expression of class I flagellar genes but only minimal fliC (class III flagellar gene) expression within intracellular bacterial communities during development within the bladder. Recently, much attention has been focused on pattern recognition receptors that are important for proper innate immune function. Toll-like receptor 5 is known to recognize flagellated bacteria (13). Thus, bacterial pathogens such as UPEC would benefit by tightly controlling flagellar expression within the host to limit its exposure to immune defenses. It is important to know what signals are driving UPEC to switch on/off flagella expression to avoid immune responses or to ascend to the upper urinary tract and colonize the kidneys. Therefore, one could speculate that a two-component signal transduction system, such as the quorum-sensing regulators of EHEC, QseB, and QseC [involved in the transcriptional regulation of flhDC (27)] or other regulatory mechanisms, exists to strictly control the complex flagellar transcriptional hierarchy. Future studies investigating the effect that environmental cues and bacterial signal transduction have on flhDC may provide insight as to the nature of the ability for UPEC to tightly control flagellin expression in vivo.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions.
Bacterial strains and plasmids used in this study are listed in SI Table 1. All strains were cultured on LB agar plates with appropriate antibiotics (100 μg/ml ampicillin or 25 μg/ml kanamycin) and incubated at 37°C for 18 h. Overnight broth cultures were obtained by inoculating a single colony into LB broth containing appropriate antibiotics and incubating at 37°C for 18 h with aeration.
In Vitro Growth and Motility Assays.
To assess growth in vitro, bacteria from overnight cultures were diluted 1:100 into 40 ml of tryptone broth. Suspensions were cultured with aeration at 37°C in 125-ml flasks. For both the Pem7-lux and PfliC-lux reporter constructs, the OD600 was measured, and luminescence was quantified at various time points by using a Synergy HT plate-reader operating KC4 software (Bio-Tek, Winooski, VT). Luminescent emission spectra were collected from 100-μl samples of culture contained within triplicate wells of a 96-well one-half area white polystyrene plate (Corning, Corning, NY) using a 530-nm filter with a bandwidth of 25 nm. For swimming and swarming motility studies, 50 μl of overnight cultures of wild-type CFT073 containing Pem7-lux or PfliC-lux were reinoculated into 5 ml of sterile LB broth and incubated at 37°C with aeration to an OD600 ≅ 1.0. At that point, cultures were stabbed into 0.25% tryptone agar plates [1% tryptone/0.5% NaCl, 0.25% Difco (BD, Sparks, MD) agar] by using a sterile inoculating needle or spotted (5 μl) onto the surface of modified 0.45% Eiken agar [per liter: 10.49 g of Eiken agar (Eiken Chemical, Tokyo, Japan)/3.50 g of yeast extract/7.00 g of tryptone/3.50 g of NaCl/40% d-glucose] containing 100 μg/ml ampicillin. Swimming and swarming plates were incubated for 16 h at 30°C or 37°C, respectively. After incubation, luminescence emitted from the swimming and swarming bacteria were captured by using the ChemiDoc XRS system (Bio-Rad). Further analysis was conducted to obtain density plots of the luminescence using the Discovery Series Quantity One software (Bio-Rad).
CBA Mouse Model of Ascending UTI.
For biophotonic imaging, all mice were infected as described, with modification (6, 28). Female CBA/J mice (Harlan Sprague–Dawley, Indianapolis, IN) were anesthetized with ketamine/xylazine and inoculated transurethrally over a 30-s period with a 50-μl bacterial suspension delivering 109 cfu per mouse. To prepare the inoculum, overnight cultures (grown in the absence of antibiotic selection) were adjusted to deliver 109 cfu per mouse. Dilutions of each inoculum were spiral-plated by using an Autoplate 4000 (Spiral Biotech, Norwood, MA) to determine the input cfu per milliliter. For the low-dose independent challenge studies, a similar procedure was followed; however, mice were inoculated into the proximal end of the urethra over a 6-s period with a 10-μl bacterial suspension delivering 107 cfu per mouse. Six and 48 hpi, mice were killed by overdose with isoflurane, and the bladder, kidneys, and spleen were aseptically removed, weighed, and homogenized in sterile culture tubes containing 3 ml of PBS using an OMNI mechanical homogenizer (OMNI International, Marietta, GA). Appropriate dilutions of the homogenized tissue were then spiral-plated onto plain LB plates with or without antibiotic to determine the output cfu per gram of tissue. Statistically significant differences in colonization (with a P value <0.05) were determined by using an unpaired nonparametric Mann–Whitney test (InStat; GraphPad, San Diego, CA). All animal protocols were approved by the University Committee on Use and Care of Animals at the University of Michigan Medical School.
In Vivo Bioluminescent Imaging of Ascending UTI.
For in vivo imaging of experimentally infected mice, hair was completely removed from both ventral and dorsal sides of the animals below the scapula including the abdominal and pelvic areas using a razor and cosmetic hair removal treatment (Veet, Reckitt Benckiser, Parsippany, NJ). Infected mice were imaged at various times after infection by using an IVIS 200 imaging system (Xenogen), as described (29). Mice were anesthetized in chambers containing 2.0% isofluorane inhalant (Baxter, Deerfield, IL). The anesthetic was sustained during imaging by using 2.0% isofluorane delivered via an IVIS manifold placed within the imaging chamber. Both the dorsal and ventral sides of the animals (as well as bladder and kidney tissue that was harvested ex vivo) were imaged separately for a maximum of 5 min. Mice infected with CFT073 containing the Pneg-lux reporter were also imaged at each time point to determine the level of background luminescent signal. Photon emissions were obtained from defined fixed regions of interest by using the Living Image software (Xenogen). Photon flux quantified for the bladders and kidneys was determined from ventral and dorsal sides of mice, respectively. The significance of PfliC-lux and Pem7-lux reporter luminescence over background at each time point was assessed by using a Mann–Whitney statistical test (InStat; GraphPad). Immediately after the final imaging time point, mice were killed, and bladders and individual kidneys were surgically removed and homogenized to enumerate bacterial viable counts.
Additional Methods.
Detailed methods, protocols, and primers for the construction of the bioluminescent reporters for the isolation of RNA and cDNA from in vitro and in vivo samples and for the comparative qPCR studies can be found in SI Text and SI Table 2.
Supplementary Material
Acknowledgments
We thank G. Luker and K. Luker for invaluable help with the mouse imaging experiments, C. Contag (Stanford University, Palo Alto, CA) for pUT-Tn5-EM7-lux-Km1, J. Galen (University of Maryland, Baltimore, MD) for pGEN222, the University of Michigan Center for Molecular Imaging, and M. O'Riordan for critical review of the manuscript. This work was supported by Public Health Service Grants AI43363 and AI059722 from the National Institutes of Health.
Abbreviations
- UPEC
uropathogenic E. coli
- UTI
urinary tract infection
- qPCR
quantitative PCR
- hpi
hours postinoculation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607898104/DC1.
References
- 1.Josenhans C, Suerbaum S. Int J Med Microbiol. 2002;291:605–614. doi: 10.1078/1438-4221-00173. [DOI] [PubMed] [Google Scholar]
- 2.Ottemann KM, Miller JF. Mol Microbiol. 1997;24:1109–1117. doi: 10.1046/j.1365-2958.1997.4281787.x. [DOI] [PubMed] [Google Scholar]
- 3.Bacheller CD, Bernstein JM. Med Clin North Am. 1997;81:719–730. doi: 10.1016/s0025-7125(05)70542-3. [DOI] [PubMed] [Google Scholar]
- 4.Warren JW. Urinary Tract Infections: Molecular Pathogenesis and Clinical Management. Washington, DC: Am Soc Microbiol Press; 1996. [Google Scholar]
- 5.Faro S, Fenner DE. Clin Obstet Gynecol. 1998;41:744–754. doi: 10.1097/00003081-199809000-00030. [DOI] [PubMed] [Google Scholar]
- 6.Lane MC, Lockatell V, Monterosso G, Lamphier D, Weinert J, Hebel JR, Johnson DE, Mobley HL. Infect Immun. 2005;73:7644–7656. doi: 10.1128/IAI.73.11.7644-7656.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright KJ, Seed PC, Hultgren SJ. Infect Immun. 2005;73:7657–7668. doi: 10.1128/IAI.73.11.7657-7668.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snyder JA, Haugen BJ, Buckles EL, Lockatell CV, Johnson DE, Donnenberg MS, Welch RA, Mobley HL. Infect Immun. 2004;72:6373–6381. doi: 10.1128/IAI.72.11.6373-6381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SH, Butler SM, Camilli A. Proc Natl Acad Sci USA. 2001;98:6889–6894. doi: 10.1073/pnas.111581598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cummings LA, Wilkerson WD, Bergsbaken T, Cookson BT. Mol Microbiol. 2006;61:795–809. doi: 10.1111/j.1365-2958.2006.05271.x. [DOI] [PubMed] [Google Scholar]
- 11.Andersen-Nissen E, Hawn TR, Smith KD, Nachman A, Lampano AE, Uematsu S, Akira S, Aderem A. J Immunol. 2007;178:4717–4720. doi: 10.4049/jimmunol.178.8.4717. [DOI] [PubMed] [Google Scholar]
- 12.Andersen-Nissen E, Smith KD, Strobe KL, Barrett SL, Cookson BT, Logan SM, Aderem A. Proc Natl Acad Sci USA. 2005;102:9247–9252. doi: 10.1073/pnas.0502040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feuillet V, Medjane S, Mondor I, Demaria O, Pagni PP, Galan JE, Flavell RA, Alexopoulou L. Proc Natl Acad Sci USA. 2006;103:12487–12492. doi: 10.1073/pnas.0605200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doyle TC, Burns SM, Contag CH. Cell Microbiol. 2004;6:303–317. doi: 10.1111/j.1462-5822.2004.00378.x. [DOI] [PubMed] [Google Scholar]
- 15.Hardy J, Francis KP, DeBoer M, Chu P, Gibbs K, Contag CH. Science. 2004;303:851–853. doi: 10.1126/science.1092712. [DOI] [PubMed] [Google Scholar]
- 16.Kadurugamuwa JL, Modi K, Yu J, Francis KP, Purchio T, Contag PR. Infect Immun. 2005;73:3878–3887. doi: 10.1128/IAI.73.7.3878-3887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright JS, III, Jin R, Novick RP. Proc Natl Acad Sci USA. 2005;102:1691–1696. doi: 10.1073/pnas.0407661102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hopkins WJ, Hall JA, Conway BP, Uehling DT. J Infect Dis. 1995;171:462–465. doi: 10.1093/infdis/171.2.462. [DOI] [PubMed] [Google Scholar]
- 19.Mobley HL, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren JW. Infect Immun. 1990;58:1281–1289. doi: 10.1128/iai.58.5.1281-1289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan K, Kim CC, Falkow S. Infect Immun. 2005;73:5438–5449. doi: 10.1128/IAI.73.9.5438-5449.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galen JE, Nair J, Wang JY, Wasserman SS, Tanner MK, Sztein MB, Levine MM. Infect Immun. 1999;67:6424–6433. doi: 10.1128/iai.67.12.6424-6433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. J Bacteriol. 1999;181:7500–7508. doi: 10.1128/jb.181.24.7500-7508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harshey RM, Matsuyama T. Proc Natl Acad Sci USA. 1994;91:8631–8635. doi: 10.1073/pnas.91.18.8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiles S, Pickard KM, Peng K, MacDonald TT, Frankel G. Infect Immun. 2006;74:5391–5396. doi: 10.1128/IAI.00848-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson JR. Infect Immun. 1998;66:6063–6064. doi: 10.1128/iai.66.12.6063-6064.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunther NW, IV, Snyder JA, Lockatell V, Blomfield I, Johnson DE, Mobley HL. Infect Immun. 2002;70:3344–3354. doi: 10.1128/IAI.70.7.3344-3354.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sperandio V, Torres AG, Kaper JB. Mol Microbiol. 2002;43:809–821. doi: 10.1046/j.1365-2958.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- 28.Johnson DE, Lockatell CV, Hall-Craigs M, Mobley HL, Warren JW. J Urol. 1987;138:632–635. doi: 10.1016/s0022-5347(17)43287-3. [DOI] [PubMed] [Google Scholar]
- 29.Kadurugamuwa JL, Sin LV, Yu J, Francis KP, Kimura R, Purchio T, Contag PR. Antimicrob Agents Chemother. 2003;47:3130–3137. doi: 10.1128/AAC.47.10.3130-3137.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.