Abstract
Sperm show patterns of rapid and divergent evolution that are characteristic of sexual selection. Sperm competition has been proposed as an important selective agent in the evolution of sperm morphology. However, several comparative analyses have revealed evolutionary associations between sperm length and female reproductive tract morphology that suggest patterns of male–female coevolution. In the dung beetle Onthophagus taurus, males with short sperm have a fertilization advantage that depends on the size of the female's sperm storage organ, the spermatheca; large spermathecae select for short sperm. Sperm length is heritable and is genetically correlated with male condition. Here we report significant additive genetic variation and heritability for spermatheca size and genetic covariance between spermatheca size and sperm length predicted by both the “good-sperm” and “sexy-sperm” models of postcopulatory female preference. Our data thus provide quantitative genetic support for the role of a sexually selected sperm process in the evolutionary divergence of sperm morphology, in much the same manner as precopulatory female preferences drive the evolutionary divergence of male secondary sexual traits.
Keywords: male–female coevolution, postcopulatory female choice, sperm length
Female mating preferences are widely recognized as being responsible for the rapid and divergent evolution of male secondary sexual traits (1). Indicator or good gene models envisage genetic coupling between male sexual trait expression and offspring fitness, so that females with a preference for male traits produce offspring of greater viability (2). Once a preference becomes established, females choosing males with elaborate secondary sexual traits will produce sons that carry alleles for the trait and daughters that carry alleles for the preference, generating genetic coupling that will drive self-reinforcing coevolution of both trait and preference because of the mating advantage to males with the trait. Thus, the original viability benefits associated with the preference can be undermined by a runaway Fisherian sexy sons process (2). Analogous models have been proposed for postcopulatory female preferences (3). “Good-sperm” models predict positive genetic associations between a male's sperm competitiveness and the general viability of his offspring (4), whereas “sexy-sperm” models predict that multiply mating females produce sons successful in sperm competition and daughters that incite sperm competition through multiple mating (5, 6). As with precopulatory processes, postcopulatory models predict that the trait in males that determines fertilization success will become genetically coupled with the mechanism by which females bias sperm use toward preferred males (7).
Although spermatozoa are well known for their rapid and divergent morphological variation (8, 9), little is known of the selective processes that drive sperm evolution. Patterns of divergent evolution are characteristic of strong sexual selection, and researchers have suggested that postcopulatory sexual selection via sperm competition may be responsible for evolutionary divergence in sperm morphology (10, 11). Thus, among frogs (12), birds (13), and butterflies (14) there are evolutionary associations between the strength of selection acting via sperm competition and sperm length. These patterns, however, are by no means consistent. In fish both positive (15) and negative (16) evolutionary associations have been found, whereas in mammals no evolutionary associations have been found (17). In birds the association between sperm competition and sperm length appears to be indirect, via an effect of sperm competition on the length of sperm storage tubules in the female reproductive tract and evolutionary covariation between sperm storage tubule and sperm lengths (18).
Associations between sperm lengths and the lengths of female reproductive ducts and/or sperm storage organs are well documented in the insects (19–23). These patterns of correlated evolution implicate selection processes imposed by females during the evolution of sperm morphology. Using populations of Drosophila melanogaster artificially selected for long or short sperm, Miller and Pitnick (24) showed that males with long sperm had a fertilization advantage over males with short sperm when both were mated to females artificially selected to have long seminal receptacles. The heads of long sperm lie closer to the exit of the seminal receptacle, giving them precedence over short sperm at the time of fertilization (25). Miller and Pitnick (24) also showed that artificial selection for increased seminal receptacle length in females resulted in a correlated response in sperm length, suggesting that seminal receptacle length imposed directional selection on sperm length. Although the selective pressures driving female seminal receptacle evolution are unknown, the data for D. melanogaster implicate a sexually selected sperm process in the evolution of sperm gigantism in this group of flies (24).
Here we provide a critical test of the key prediction underlying both good-sperm (4) and sexy-sperm (5) models of postcopulatory sexual selection, that there is a genetic correlation between a sperm trait that contributes to male fertilizations success and the mechanism used by females to select sperm. Previously, we documented patterns of quantitative genetic variation in sperm competition traits in the dung beetle Onthophagus taurus. We found significant levels of additive genetic variation in sperm length and the high heritability for this trait that is necessary for a sexually selected sperm process. Moreover, we found significant additive genetic variation in male condition (26) and strong genetic covariance between male condition and sperm length; males in high condition produced shorter sperm (27). These patterns of genetic (co)variance are necessary for viability indicator and/or sexy son processes and could provide an avenue for postcopulatory sexual selection in this beetle; females that selectively fertilize their eggs using short sperm could produce offspring of greater condition because of the genetic covariances between these traits (the good-sperm process) and sons with short sperm who would be more successful in sperm competition (the sexy-sperm process) (27). García-González and Simmons (28) have recently shown that short sperm do have a competitive fertilization advantage and that this advantage depends on the size of the female's sperm storage organ, the spermatheca. Larger spermathecae select shorter sperm for fertilization (28). A sexually selected sperm process predicts the correlated evolution of sperm trait and preference and thus a genetic correlation between sperm length in sons and spermatheca size in daughters. Specifically, because small sperm are preferred by large spermathecae, we would expect a process of postcopulatory sexual selection to generate a negative genetic correlation between these traits. We found strong empirical support for this prediction.
Results
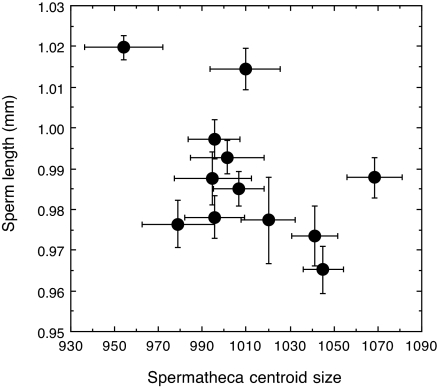
We found significant additive genetic variation in spermatheca size (centroid size) that was due to sires and significant heritability (Tables 1 and 2). The effect of sire on spermatheca shape (RW1; see Figs. 1 and 2) was smaller and not statistically significant (Tables 1 and 2). The heritability of spermatheca shape was considerably less than spermatheca size (Table 2). Sperm length also exhibited significant variance due to sires (27). Restricted maximum likelihood methods returned estimates of genetic variance and heritability consistent with our previous ANOVA approach (27) (Table 2). Coefficients of additive genetic variance in sperm length and spermatheca size were of similar magnitude. Both were relatively low, indicative of a history of directional selection. The heritability of sperm length was close to the maximum possible value of 1.0. Heritabilities were calculated by assuming autosomal inheritance. However, heritabilities ≈1.0 implicate Y-linked inheritance and should be halved to obtain minimum estimates of the true heritability (29, 30). Consistent with predictions from sexy-sperm and good-sperm processes, there was a significant genetic correlation between spermatheca size and sperm length (−0.851 ± 0.114, t11 = 7.46, P < 0.0001). Fathers that sired sons with short sperm also sired daughters with large spermathecae (Fig. 3). There was no genetic correlation between sperm length and spermatheca shape (RW1) (0.016 ± 0.292, t11 = 0.95, P = 0.362).
Table 1.
Nested analysis of variance for sermathecal size and shape in O. taurus
| Trait | Source | SS | df | MS | F value | P value |
|---|---|---|---|---|---|---|
| Centroid size | Sire | 0.0197 | 11 | 0.0018 | 3.91 | 0.002 |
| Dam (Sire) | 0.0108 | 24 | 0.0005 | 0.71 | 0.838 | |
| Residual | 0.0804 | 126 | 0.0006 | |||
| RW1 | Sire | 0.0927 | 11 | 0.0084 | 1.74 | 0.120 |
| Dam (Sire) | 0.1173 | 24 | 0.0049 | 1.17 | 0.280 | |
| Residual | 0.0042 | 126 | 0.0042 |
SS, sums of squares; MS, mean squares.
Table 2.
Descriptive phenotypic and genetic statistics for spermatheca size and shape and sperm length for O. taurus
| Trait | Mean | VP | VA | h2 (SE) | CVP | CVA |
|---|---|---|---|---|---|---|
| Centroid size | 1,005.57 | 3,552.31 | 599.30 | 0.68 (0.15) | 5.92 | 2.43 |
| RW1* | 0.00 | 4.7 × 10−3 | 3.3 × 10−4 | 0.28 (0.03) | — | — |
| Sperm length,† mm | 0.99 | 6.7 × 10−4 | 1.9 × 10−4 | 1.14 (0.47) | 2.61 | 1.39 |
VP, phenotypic variance; VA, additive genetic variance; h2, narrow sense heritability; CVP, coefficient of phenotypic variation; CVA, coefficient of additive genetic variation.
*RW scores have zero mean, precluding the calculation of coefficients of variation.
†Data reanalyzed from Simmons and Kotiaho (27).
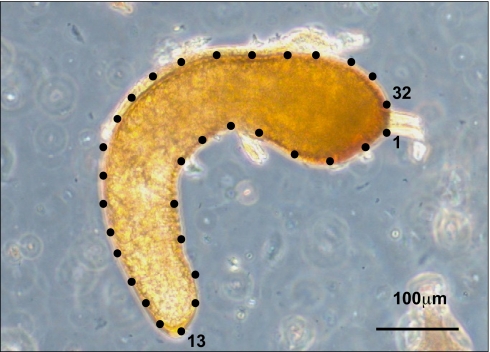
Fig. 1.
Image of the spermatheca of O. taurus with 32 landmarks placed around its periphery. For geometric morphometric analysis, landmarks 1, 13, and 32 were assigned as fixed, and the remaining were assigned as sliding semilandmarks.
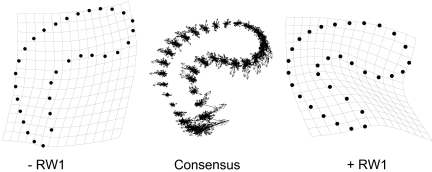
Fig. 2.
The consensus shape (Center) from geometric morphometric analysis is shown with partial warps shown as vectors with their origin at the consensus position of each landmark. Thin-plate splines show variation in shape along the first relative warp, with extreme negative scores shown in Left and extreme positive scores shown in Right.
Fig. 3.
Plot of the sire family mean (±SE) sperm lengths against sire family mean (±SE) spermatheca centroid size. The data show a genetic correlation across the sexes for these sperm and sperm storage traits.
Discussion
Good-sperm (4) and sexy-sperm (5) models of postcopulatory sexual selection were originally developed to explain the evolution of costly multiple mating by females. Genetic linkage between the fitness of offspring sired by superior sperm competitors and the tendency for females to incite sperm competition can theoretically promote the evolution of polyandry. However, the evolutionary consequences of these models need not be restricted to female mating behavior, because any female trait that facilitates the biasing of fertilization toward preferred males is expected to undergo coevolution with the male trait that influences paternity (6, 31). Thus, good-sperm and sexy-sperm processes can generate coevolutionary cycles between sperm and the structure and function of female reproductive tracts. Although plausible, empirical support for these sexually selected sperm models has been slow in accumulating.
Our finding of genetic covariance between sperm length and spermatheca size in O. taurus provides empirical support for a sexually selected sperm process. Competitive fertilization trials have shown that increasing spermatheca size is associated with an increasing strength of directional selection for shorter sperm (28), and the genetic coupling of these traits is expected to generate reinforcing selection for short sperm in males and large spermathecae in females until checked by opposing selection. Spermatheca shape has also been shown to influence paternity in O. taurus, but independent of sperm length (28). As such we would not expect to find genetic covariance between spermatheca shape and sperm length, an expectation borne out by our quantitative genetic analysis. Previously, sperm competition has been argued to favor increasingly numerous and small sperm and to drive the evolution of anisogamy (32, 33). Our data suggest an additional role for females in the evolutionary reduction in male gamete size, at least in Onthophagus.
Before our study, the only other quantitative genetic evidence in favor of a sexually selected sperm process came from studies of Drosophila. In contrast to our findings, female D. melanogaster with longer seminal receptacles favor males producing longer sperm so that selection drives the evolutionary exaggeration of sperm length (24, 25). Genetic covariances between female morphologies that bias paternity toward males with particular sperm characteristics are likely to underlie the increasing number of comparative analyses that are revealing evolutionary associations between sperm morphology and female reproductive tract morphology (19–23, 34). The contrasting findings for Onthophagus and Drosophila illustrate how postcopulatory female preferences can generate divergent patterns of evolution across taxa and contribution to the rapid and divergent variation that is characteristic of sperm morphology.
Previously, we found male condition in O. taurus to be heritable and genetically correlated with sperm length; males of high condition produce shorter sperm, suggesting that short sperm may be costly to produce (27). Thus, the costs of producing shorter sperm may counter the continued reduction in sperm length imposed by postcopulatory female preferences. Moreover, the condition-dependent nature of sperm length in this species means that females could gain indirect benefits from their selection of short sperm via an indicator process if short-spermed males sire offspring of higher viability. Such a relationship is implicit in the genetic covariances between male condition and both courtship rate and sperm length, traits that contribute to male attractiveness in precopulatory female choice (26) and paternity in postcopulatory choice (28), respectively. In other species, phenotypic studies have revealed correlations between a male's success in sperm competition and life history characteristics of offspring, such as development speed in dung flies (35), fecundity in bulb mites (36), and viability in Antechinus (37), that are consistent with an indicator process. However, differential maternal allocation by females after copulations with males found attractive during precopulatory mate choice (38) offers a viable alternative explanation for some of these findings (39). Quantitative genetic approaches such as ours offer greater power for testing models of preference evolution (40), and, if females of these species are exercising postcopulatory preferences, then we expect to see genetic covariance between competitive fertilization success, measures of offspring performance, and the behavior and/or morphology of females that biases paternity (7).
In conclusion, spermatozoa are the most morphologically diverse cells in nature, offering a reliable toolkit for the construction of animal phylogenies (8, 9). It has long been argued that sperm competition plays an important role in the evolution of sperm form and function (10, 11). Our quantitative genetic data provide support for a role of postcopulatory female preferences in driving evolutionary divergence in sperm morphology, in much the same way as precopulatory preferences drive the evolutionary divergence of male secondary sexual traits.
Methods
Breeding Design.
We used a standard half-sibling breeding design (41). Beetles were collected from fresh cattle dung from a field in Margaret River, Western Australia. Beetles were maintained in mixed-sex cultures for 1 week before females were established in individual breeding chambers (PVC pipe, 25 cm in length, 6 cm in diameter, three-quarters filled with damp sand topped with 200 ml of cow dung). Chambers were left at 28°C for 1 week and sieved, and brood masses were buried en masse in 6-liter boxes containing moist sand. When adult beetles emerged, females were maintained in single-sex cultures for 3 weeks with constant access to fresh dung. These unmated females were used as dams for our breeding design.
We collected 12 adult male beetles from the field and housed each with four laboratory-reared dams for 5 days in 50-ml plastic boxes containing moist sand as substrate and fresh dung. Females were then established in individual breeding chambers that were sieved every 7 days. Brood masses were removed and incubated in full-sibling family groups. On emergence, females were frozen and preserved in ethanol. Males were housed individually for 2–3 weeks before assessment of sperm length as described by Simmons and Kotiaho (27). To ensure that our design remained balanced, for each sire we chose the three dams that produced the greatest number of offspring. For these dams the numbers of sons ranged from four to six (upper and lower 95%: four and five) and the number of daughters from one to nine (4.5 and five). Although the quantitative genetic analysis of sperm length has been published previously (27), we present these data again here both for completeness and because we use different methods for extracting variance components and the calculation of standard errors.
Spermatheca Size and Shape.
Spermathecae were dissected and placed on individual cavity slides, and an image was captured by using an Axio Imager microscope and AxioCam MRc5 (Zeiss). We used geometric morphometrics to quantify variation in size and shape (42). We placed 32 landmarks around the periphery of the spermatheca. The spermatheca provides few distinct locations on which to place fixed landmarks, but rather presents a curved perimeter that makes defining comparable points along the structure difficult (Fig. 1). We therefore used the method of sliding semilandmarks (42). Thus, we placed a total of 32 points around the periphery of the spermatheca using tpsDig v2.04 (http://life.bio.sunysb.edu/morph; F. James Rohlf, Department of Ecology and Evolution, Stony Brook University, Stony Brook, NY). We assigned points 1 and 32, which clearly lie on either side of the spermathecal duct entrance to the spermatheca, and point 13, which lies at the distal apex of the spermatheca, as fixed landmarks (Fig. 1). The remaining landmarks were assigned as sliding semilandmarks.
We generated relative warp scores and centroid sizes using the software package tpsDig v2.04. Briefly, partial warp scores were generated that describe variation in spermatheca morphometry among spermathecae from different females as deviations in shape from a consensus shape. Visualizations of variation in shape are obtained as deformations of the thin-plate spline (Fig. 2). Scores were subject to relative warp analysis, which corresponds to a principal component analysis and serves to reduce the multivariate shape data to one or a few variables that allow differences in shape among individuals to be examined (42). Centroid size provides a measure of the size of the structure independent of shape and is calculated as the square root of the summed square distances between each landmark and the centroid of the structure being measured (42).
Relative warp analysis returned 29 relative warps. The first relative warp (RW1) explained 61% of the variance in spermatheca shape and described a narrowing of the spermatheca and a bending at its midpoint (Fig. 2). RW2 explained a further 15% of the variance, and RW3 explained a further 7%. We used scores on RW1 to examine genetic variation in spermatheca shape and centroid size to examine genetic variation in spermatheca size.
We assessed the repeatability of geometric morphometric analysis as applied to spermatheca size and shape by placing landmarks on two replicate images of spermathecae from each of 10 females. RW1 was highly repeatable (RW1: F9, 10 = 185.72, P < 0.0001, r = 0.995), as was centroid size (formal statistical analysis precluded because variance between repeated measures of the same spermatheca was zero). All spermatheca measurements were made blind with respect to the measurements of sperm lengths.
Quantitative Genetic Analysis.
For hypothesis testing, we used mixed-model nested analyses of variance, with dams nested within sires, and Satterthwaite's approximation of the error term to account for unequal sample sizes of offspring, as recommended by Lynch and Walsh (41). Analyses of genetic variation were conducted by using sire and dam variance components estimated from restricted maximum likelihood procedures in S-Plus (43). Narrow sense heritabilities were calculated from the sire and dam variance components (41). Coefficients of phenotypic and additive genetic variation were calculated following Houle (44). Analyses of genetic covariance between sperm length and spermatheca size and shape were conducted by using method 3 in Via (45); within dams, each brother was arbitrarily paired with a sister to calculate genetic covariances by using the procedures in S-Plus outlined by Roff (43). Centroid size was log-transformed before analyses of covariance, so that variables were on the same scale. Standard errors for heritabilities and genetic correlations were estimated by jackknifing across paternal half-sibling families (43). All means are presented ±1 SE.
Acknowledgments
We thank John Hunt and Joseph Tomkins for help with breeding the beetles; Julie Wernham, who measured the length of sperm; and Kate Harvey, who dissected the spermathecae and placed landmarks on images for the geometric morphometric analysis. L.W.S. was funded by the Australian Research Council and the West Australian Centres of Excellence in Science and Innovation program, and J.S.K. was funded by the Academy of Finland and the Finnish Centre of Excellence in Evolutionary Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Andersson M. Sexual Selection. Princeton: Princeton Univ Press; 1994. [Google Scholar]
- 2.Mead LS, Arnold SJ. Trends Ecol Evol. 2004;19:264–271. doi: 10.1016/j.tree.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Andersson M, Simmons LW. Trends Ecol Evol. 2006;21:296–302. doi: 10.1016/j.tree.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Yasui Y. Am Nat. 1997;149:573–584. [Google Scholar]
- 5.Curtsinger JW. Am Nat. 1991;138:93–102. [Google Scholar]
- 6.Keller L, Reeve HK. Adv Stud Behav. 1995;24:291–315. [Google Scholar]
- 7.Evans JP, Simmons LW. Genetica. 2007 Jul 7; doi: 10.1007/s10709-007-9162-5. [DOI] [Google Scholar]
- 8.Jamieson BGM. Fish Evolution and Systematics: Evidence from Spermatozoa. Cambridge, UK: Cambridge Univ Press; 1991. [Google Scholar]
- 9.Jamieson BGM, Dallai R, Afzelius BA. Insects: Their Spermatozoa and Phylogeny. Enfield, NH: Science Publishers; 1999. [Google Scholar]
- 10.Sivinski J. In: Sperm Competition and the Evolution of Animal Mating Systems. Smith RL, editor. London: Academic; 1984. pp. 86–115. [Google Scholar]
- 11.Simmons LW. Sperm Competition and Its Evolutionary Consequences in the Insects. Princeton: Princeton Univ Press; 2001. [Google Scholar]
- 12.Byrne PG, Simmons LW, Roberts JD. Proc R Soc London Ser B. 2003;270:2079–2086. doi: 10.1098/rspb.2003.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Briskie JV, Montgomerie R. Proc R Soc London Ser B. 1992;247:89–95. doi: 10.1098/rspb.1992.0013. [DOI] [PubMed] [Google Scholar]
- 14.Gage MJG. Proc R Soc London Ser B. 1994;258:247–254. [Google Scholar]
- 15.Balshine S, Leach BJ, Neat F, Werner NY, Montgomerie R. Behav Ecol. 2001;12:726–731. [Google Scholar]
- 16.Stockley P, Gage MJG, Parker GA, Møller AP. Am Nat. 1997;149:933–954. doi: 10.1086/286031. [DOI] [PubMed] [Google Scholar]
- 17.Gage MJG, Freckleton RP. Proc R Soc London Ser B. 2003;270:625–632. doi: 10.1098/rspb.2002.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Briskie JV, Montgomerie R, Birkhead TR. Evolution (Lawrence, Kans) 1997;51:937–945. doi: 10.1111/j.1558-5646.1997.tb03674.x. [DOI] [PubMed] [Google Scholar]
- 19.Dybas LK, Dybas HS. Evolution (Lawrence, Kans) 1981;35:168–174. doi: 10.1111/j.1558-5646.1981.tb04869.x. [DOI] [PubMed] [Google Scholar]
- 20.Presgraves DC, Baker RH, Wilkinson GS. Proc R Soc London Ser B. 1999;266:1041–1047. [Google Scholar]
- 21.Minder AM, Hosken DJ, Ward PI. J Evol Biol. 2005;18:60–69. doi: 10.1111/j.1420-9101.2004.00799.x. [DOI] [PubMed] [Google Scholar]
- 22.Morrow EH, Gage MJG. Proc R Soc London Ser B. 2000;267:307–313. doi: 10.1098/rspb.2000.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasakawa K. Naturwissenschaften. 2007;94:384–391. doi: 10.1007/s00114-006-0200-4. [DOI] [PubMed] [Google Scholar]
- 24.Miller GT, Pitnick S. Science. 2002;298:1230–1233. doi: 10.1126/science.1076968. [DOI] [PubMed] [Google Scholar]
- 25.Pattarini JM, Starmer WT, Bjork A, Pitnick S. Evolution (Lawrence, Kans) 2006;60:2064–2080. [PubMed] [Google Scholar]
- 26.Kotiaho JS, Simmons LW, Tomkins JL. Nature. 2001;410:684–686. doi: 10.1038/35070557. [DOI] [PubMed] [Google Scholar]
- 27.Simmons LW, Kotiaho JS. Evolution (Lawrence, Kans) 2002;56:1622–1631. doi: 10.1111/j.0014-3820.2002.tb01474.x. [DOI] [PubMed] [Google Scholar]
- 28.García-González F, Simmons LW. Evolution (Lawrence, Kans) 2007;61:816–824. doi: 10.1111/j.1558-5646.2007.00084.x. [DOI] [PubMed] [Google Scholar]
- 29.Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. Harlow, Essex, UK: Longman; 1996. [Google Scholar]
- 30.Roff DA. Evolutionary Quantitative Genetics. New York: Chapman & Hall; 1997. [Google Scholar]
- 31.Eberhard WG. Female Control: Sexual Selection by Cryptic Female Choice. Princeton: Princeton Univ Press; 1996. [Google Scholar]
- 32.Parker GA, Baker RR, Smith VGF. J Theor Biol. 1972;36:529–513. doi: 10.1016/0022-5193(72)90007-0. [DOI] [PubMed] [Google Scholar]
- 33.Parker GA. Proc R Soc London Ser B. 1993;253:245–254. [Google Scholar]
- 34.Pitnick S, Miller GT, Schneider K, Markow TA. Proc R Soc London Ser B. 2003;270:1507–1512. doi: 10.1098/rspb.2003.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hosken DJ, Garner TWJ, Tregenza T, Wedell N, Ward PI. Proc R Soc London Ser B. 2003;270:1933–1938. doi: 10.1098/rspb.2003.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konior M, Radwan J, Kolodziejczyk M. Evolution (Lawrence, Kans) 2001;55:1893–1896. doi: 10.1111/j.0014-3820.2001.tb00838.x. [DOI] [PubMed] [Google Scholar]
- 37.Fisher DO, Double MC, Blomberg SP, Jennions MD, Cockburn A. Nature. 2006;444:89–92. doi: 10.1038/nature05206. [DOI] [PubMed] [Google Scholar]
- 38.Head ML, Hunt J, Brooks R. Biol Lett. 2006;2:341–344. doi: 10.1098/rsbl.2006.0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simmons LW. Annu Rev Ecol Evol Syst. 2005;36:125–146. [Google Scholar]
- 40.Partridge L. In: Quantitative Genetic Studies of Behavioral Evolution. Boake CRB, editor. Chicago: Univ of Chicago Press; 1994. pp. 126–141. [Google Scholar]
- 41.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 42.Zelditch ML, Swiderski DL, Sheets HD, Fink WL. Geometric Morphometrics for Biologists: A Primer. London: Elsevier Academic; 2004. [Google Scholar]
- 43.Roff DA. Introduction to Computer-Intensive Methods of Data Analysis in Biology. Cambridge, UK: Cambridge Univ Press; 2006. [Google Scholar]
- 44.Houle D. Genetics. 1992;130:195–204. doi: 10.1093/genetics/130.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Via S. Evolution (Lawrence, Kans) 1984;38:896–905. doi: 10.1111/j.1558-5646.1984.tb00360.x. [DOI] [PubMed] [Google Scholar]